Significance
We describe an unprecedented type of intramolecular cross-link in a protein molecule, which we have found in the repetitive domains of a cell surface adhesin from the Gram-positive organism Clostridium perfringens. From high-resolution crystal structures of the protein, coupled with MS, we show that these domains contain intramolecular ester bonds joining Thr and Gln side chains. These bonds are generated autocatalytically by a serine protease-like mechanism and provide the long, thin protein with greatly enhanced mechanical strength and protection from proteolytic attack. The bonds provide an intriguing parallel with the internal isopeptide bonds that stabilize Gram-positive pili. Bioinformatics analysis suggests that these intramolecular ester bonds are widespread and common in cell surface adhesion proteins from Gram-positive bacteria.
Keywords: intramolecular ester bond, protein stability
Abstract
Gram-positive bacteria are decorated by a variety of proteins that are anchored to the cell wall and project from it to mediate colonization, attachment to host cells, and pathogenesis. These proteins, and protein assemblies, such as pili, are typically long and thin yet must withstand high levels of mechanical stress and proteolytic attack. The recent discovery of intramolecular isopeptide bond cross-links, formed autocatalytically, in the pili from Streptococcus pyogenes has highlighted the role that such cross-links can play in stabilizing such structures. We have investigated a putative cell-surface adhesin from Clostridium perfringens comprising an N-terminal adhesin domain followed by 11 repeat domains. The crystal structure of a two-domain fragment shows that each domain has an IgG-like fold and contains an unprecedented ester bond joining Thr and Gln side chains. MS confirms the presence of these bonds. We show that the bonds form through an autocatalytic intramolecular reaction catalyzed by an adjacent His residue in a serine protease-like mechanism. Two buried acidic residues assist in the reaction. By mutagenesis, we show that loss of the ester bond reduces the thermal stability drastically and increases susceptibility to proteolysis. As in pilin domains, the bonds are placed at a strategic position joining the first and last strands, even though the Ig fold type differs. Bioinformatic analysis suggests that similar domains and ester bond cross-links are widespread in Gram-positive bacterial adhesins.
A striking feature of globular proteins is that despite the chemical diversity inherent in the side chains of their constituent amino acids, chemical reactions between these side chains are very rare. This may be explained by evolutionary selection, which minimizes reactions that could prejudice proper protein folding. Thus, the only common example of a covalent cross-link between protein side chains is the disulfide bond, which forms only in an appropriate redox environment when two Cys residues are brought together by protein folding. Nevertheless, some surprising examples of unexpected cross-links have been brought to light by protein structure analysis or by the observation of unusual spectroscopic or biophysical properties. Examples include the Cys-Tyr bond in galactose oxidase (1), which provides a radical center; similar bonds in some catalases (2); the His-Tyr bond in cytochrome C oxidase (3); and the remarkable chromophore of GFP (4). These, and other examples, arise through intramolecular reactions facilitated by particular local environments.
The recent discovery of isopeptide bonds joining Lys and Asn side chains in the proteins that make up pili on the Gram-positive bacterium Streptococcus pyogenes (5), as well as on other Gram-positive pathogens (6), has highlighted a class of proteins in which intramolecular cross-links seem to be remarkably prevalent. It includes not only Gram-positive pili but a number of other cell surface adhesins, known as microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) (7). Examples of the latter include the collagen-binding A domain and repetitive B domains from the Staphylococcus aureus collagen-binding surface protein Cna (8, 9), the fibronectin-binding protein FbaB from S. pyogenes (10), and the adhesin SspB from Streptococcus gordonii (11). In contrast to the Gram-positive pili, which are assembled from discrete protein subunits (pilins) by sortase enzymes (12), the MSCRAMMs are typically single polypeptides folded into many domains. What both pili and MSCRAMMs have in common is that they are very long and thin but also subject to large mechanical shear stresses and protease-rich environments.
The pilus components and MSCRAMMs share a common domain organization: an N-terminal signal sequence followed by the protein segment that is to be displayed; a sorting motif (LPXTG or similar) that is processed by a sortase that attaches the protein to the cell wall or incorporates it into pili; and a C-terminal hydrophobic transmembrane segment and short, positively charged tail (13). MSCRAMMs commonly possess an N-terminal functional region followed by a repetitive series of domains that provide a supporting “stalk” that holds the functional domain(s) away from the cell surface (9).
Isopeptide bonds, both Lys-Asn and Lys-Asp, now appear to be common in the Ig-like domains that make up the shafts, or stalks, of these structures, providing tensile strength and stability along the length of the assembly (14). These bonds form spontaneously on protein folding; the hydrophobic environment lowers the pKa of the lysine residue, enabling its nucleophilic attack on the Cγ of the Asn/Asp, aided by proton transfer via an adjacent Glu or Asp. The latter also polarizes the C = O bond of the Asn or Asp side chain, resulting in a partial positive charge on Cγ (10, 14). This is essentially a one-turnover autocatalytic reaction dependent on the polarity of the environment and the proximity of the reacting groups. So far, the bonds are found in just two types of Ig-like domain, labeled CnaB and CnaA, and appear in characteristic positions in each (14).
In an effort to find how prevalent intramolecular isopeptide bonds are in bacterial cell surface proteins, we carried out a bioinformatic analysis of ∼100 sequences for putative cell wall-anchored proteins (identified by their LPXTG motifs) from a variety of Gram-positive organisms, seeking potential MSCRAMMs. Among these was a putative surface-anchored protein from Clostridium perfringens (GenBank accession no. EDT23863.1), which we refer to as Cpe0147 in the following discussion. This protein has an N-terminal domain that resembles, at the sequence level, the thioester-containing adhesin domain from S. pyogenes pili (15). This domain is followed by a series of repetitive domains of ∼150 residues each that share remarkably high sequence similarity, more than 85% identity between any pair of domains.
Mass spectral analysis of a two-domain fragment showed a loss of 34 Da from the expected molecular mass, suggesting the formation of two isopeptide bonds (a loss of 2 × 17 Da), yet no sequence pattern characteristic of isopeptide bonds could be found. Further mass spectrometric and crystallographic studies, reported here, show that these domains contain unprecedented ester bonds joining Thr and Gln side chains, formed by an esterification reaction in a solvent-exposed environment. A focused search of sequence databases further suggests that these intramolecular ester bonds are a widespread and common feature of cell surface adhesion proteins in Gram-positive bacteria.
Results
Structure Determination.
Cpe0147 is an ∼220 kDa cell wall-anchored adhesion protein from C. perfringens. Bioinformatic analysis predicts that it comprises an N-terminal adhesin domain tethered to the cell wall by a shaft composed of 11 repeating domains terminating with a C-terminal cell wall-anchoring motif (LPKTG) (Fig. 1A). The 11 repeat domains are predicted to each have an all β-strand IgG-like fold such as is commonly found in both Gram-positive pili and other MSCRAMMs (6, 16). The stalk domain sequences are highly conserved with a minimum pairwise sequence identity of 85%. In an effort to define the stalk domain boundaries, a construct spanning the first two predicted repeats was expressed in Escherichia coli and purified. This construct (C2) encompasses residues 292–625 of Cpe0147; however, for convenience, we number the start of C2 from residue 1. An additional construct comprising a single putative domain (residues 8–152 of the C2 construct) was also expressed and purified in E. coli. For simplicity, the single- and two-domain constructs are referred to herein as C1 and C2, respectively.
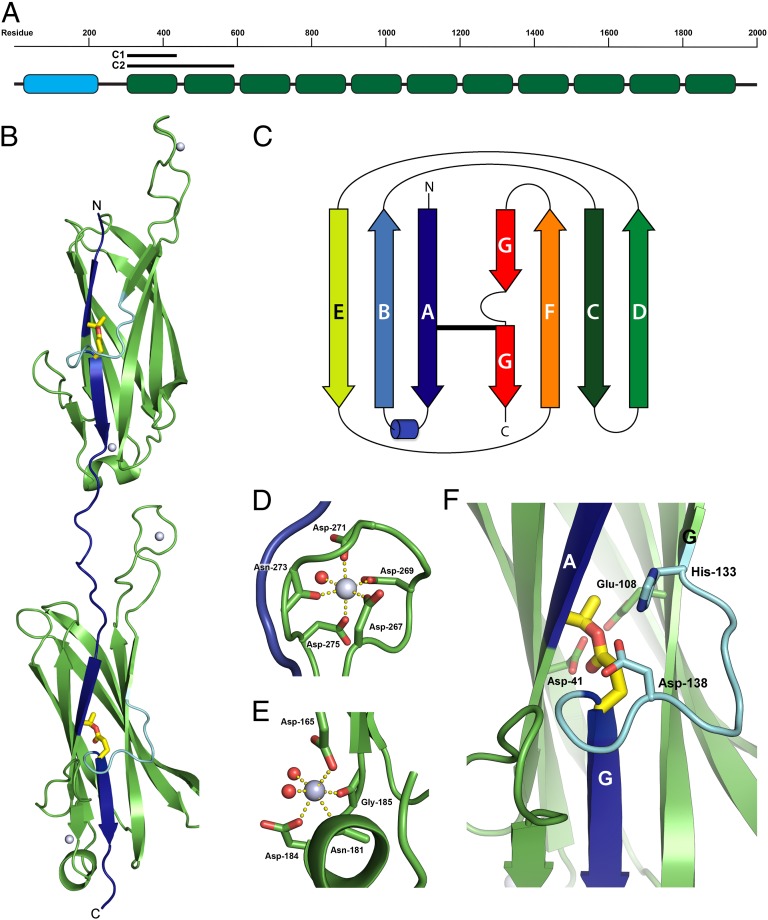
Fig. 1.
Structure of Cpe0147. (A) Domain structure of Cpe0147 with 11 repeat domains (green) and an adhesin domain (blue). Location and size of C1 and C2 constructs are indicated. (B) Two-domain C2 structure. Strands A and G are linked by an internal ester bond (yellow sticks) such that an extended covalent connectivity (indicated in blue) is propagated along the 11 repeat domains from the cell wall to the adhesin domain. Strand G is interrupted by a loop (cyan), which is shown in detail in F. Calcium ions are shown as light blue spheres. (C) Topology of the repeat domain highlights the IgG-like fold, the location of the ester bond (thick black line), and the loop interrupting strand G. (D) Calcium site in the linker region. The Ca2+ ion is shown as a light blue sphere, the coordinated water is shown as a small red sphere, and metal–ligand bonds are shown as dashed yellow lines. (E) Second calcium site (inside repeat domain) shows two coordinated waters. (F) Close-up view of the internal ester bond between Thr-11 and Gln-141 side chains. Side chains involved in ester bond formation or stabilization are labeled. The loop-interrupting strand G is highlighted in cyan and contains two of the amino acids critical to ester bond formation.
Domain boundaries of C2 were determined by limited proteolysis, revealing a highly stable trypsin-resistant fragment of ∼32 kDa (a loss of ∼6 kDa). Crystals of proteolyzed selenomethionine (SeMet)-substituted C2 protein were subsequently grown, and the structure was solved by single-wavelength anomalous dispersion (SAD). The crystal structure has one C2 molecule per asymmetric unit and was refined at a resolution of 1.90 Å (R = 19.1%, Rfree = 21.9%). Data collection and refinement statistics are shown in Table S1.
Stalk Domain Structure.
C2 is an elongated molecule that is ∼104 Å long and ∼17 Å wide (Fig. 1B). It contains two IgG-like domains connected by a long linker. The two repeat IgG-like domains are virtually identical with an rmsd of 0.59 Å over 143 aligned Cα atoms. The protein forms a β-sandwich consisting of opposing three- and four-stranded β-sheets, similar to an IgG constant domain (IgGC). The C2 domain fold differs from the IgGC fold, however, in that one edge strand (D) is switched from the first sheet of the β-sandwich to the second; the two β-sheets have the strand order A-B-E and G-F-C-D (Fig. 1C). This corresponds to the switched-type Ig fold, as defined by Bork et al. (17).
The interdomain linker is ∼35 Å long comprising residues 140–152. The length of the linker implies interdomain flexibility and may allow large domain motions. The linker is predominantly stabilized by an extended loop, residues 256–274, that projects ∼20 Å up from the C-terminal domain between β-strands F and G. This F-G loop stacks against the linker peptide and makes primarily hydrophobic interactions. A metal ion is bound at the end of the extended loop, octahedrally coordinated by Asp-267 (Oδ1), Asp-269 (Oδ1), Asp-271 (Oδ1), Asn-273 (O), Asp-275 (Oδ2), and a water molecule (Fig. 1D). The coordination environment and average metal–ligand bond length (2.3 Å) are indicative of a Ca2+ ion, and although the C2 crystals were grown in the presence of both 30 mM Mg2+ and 30 mM Ca2+, the metal ions are modeled as calcium (18). A second Ca2+ ion is coordinated by Asp-165 (Oδ1) from β-strand A, Asn-181 (O), Asp-184 (Oδ1), and Gly-185 (O) on a short helix between β-strands A and B, and two water molecules (Fig. 1E).
Both domains of C2 contain equivalent Ca2+ binding sites (Fig. 1B). In the single-domain C1 structure, however, which was crystallized without either Ca2+ or Mg2+, neither site contains a bound metal ion (Fig. S1). We conclude that neither site is of high affinity. Thus, although bound calcium ions appear to be a common stabilizing feature of both pilin proteins, for example, both SpaA from Corynebacterium diphtheriae (19) and GBS80 from Streptococcus agalactiae (20), and multidomain adhesins, such as S. gordonii SspB (11), the Ca2+ binding sites in Cpe0147 seem unlikely to play a major role in overall stability. Ca2+ binding may enhance local stability, however, as shown by ordering of the F-G loop in C2.
The most striking feature of the C2 structure is the presence of two clearly defined Thr-Gln covalent bonds, one in each domain, joining the side chains of Thr-11 and Gln-141 in the first domain and Thr-160 and Gln-290 in the second domain. In each case, the Thr residue is located on the first β-strand and the Gln residue is located on the last β-strand of the domain, reminiscent of the isopeptide bonds found in Gram-positive pili (5, 14, 21, 22) (Fig. 1F). However, this apparently spontaneous self-catalytic bond forms in an entirely different environmental context from that of autocatalytic isopeptide bonds.
Intramolecular Ester Bonds.
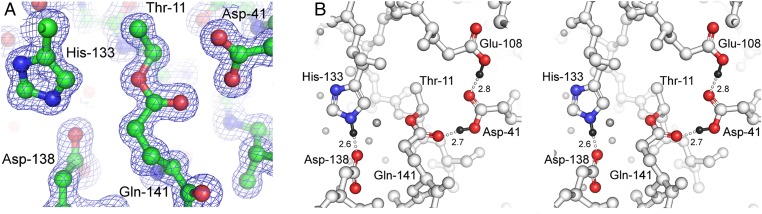
To aid characterization of the covalent linkage we observed, and to probe its formation, a single-domain construct (C1) was produced. The crystal structure of C1 was solved by molecular replacement with a partial C2 model and refined at a resolution of 1.1 Å (R = 18.2%, Rfree = 21.0%). Data collection and refinement statistics are shown in Table S1. The structure of this single-domain protein (Fig. S1) is very similar to that of the same domain in the C2 protein, with an rmsd over 136 Cα positions of 0.52 Å. The only significant difference is the loss of the two Ca2+ ions, although this has little effect on the protein conformation. As in the C2 structure, there is clearly defined electron density linking the side chains of Thr-11 and Gln-141 (Fig. 2A). A careful analysis of bond geometry and interatomic contacts in this atomic resolution structure suggests that the covalent linkage is an ester bond formed between Thr-11 Oγ1 and Gln-141 Cδ; the side chain amino group of Gln-141 has been eliminated (Fig. 2B). The ester bond is stabilized by hydrogen bonding between Gln-141 Oε1 and a protonated Asp-41 Oδ2. Asp-41 is itself stabilized by hydrogen bonding with Glu-108, which is buried in the hydrophobic core of the domain. Both Asp-41 and Glu-108 appear protonated as judged by the hydrogen bonding interactions.
Fig. 2.
Internal ester bond. (A) Electron density map (2Fo-Fc omit map contoured at 1σ) shows continuous density unambiguously assigned as an internal ester bond. (B) Stereo view of side chains involved in ester bond formation and stabilization. The hydrogen atoms on His-133, Asp-41, and Glu-108 are shown as small black spheres, and hydrogen bonds are shown as dashed white lines (H-bond distances in angstroms). Water molecules are shown as small light gray spheres.
The presence of the ester bonds in both the C1 and C2 protein constructs was independently confirmed by electrospray ionization (ESI) TOF MS. The molecular masses of C1 and C2 were found to be 16,646 Da and 33,298 Da, respectively, which are 17 Da and 33 Da less than the theoretical masses and consistent with the elimination of one NH3 molecule from each domain (Table S2). The specific location of the ester bond was confirmed by proteolytic digestion of the C1 protein and analysis by liquid chromatography tandem MS (MS/MS). A peptide was identified that gave a parent ion with an m/z of 676.93+ containing the ester bond joining Thr-11 and Gln-141, in conformity with the crystal structures (Fig. 3 and Table S3).
Fig. 3.
MS/MS spectrum of the peptide generated after trypsin digest of the C1 construct. Fragmentation spectra of the peptides containing the ester bond are shown. A full list of the assigned structures is provided in Table S3. The observed peaks indicate a stable Thr-11/Gln-141 cross-linked fragment. amu, atomic mass units.
Like isopeptide bonds in CnaB folds (14), the ester bonds provide a covalent cross-link between the first and last β-strands of each domain, and as with isopeptide bonds, the ester bonds contribute to the proteolytic stability of the protein. C1 protein digested with trypsin at 37 °C for 24 h is found to be completely intact when analyzed by SDS/PAGE. In contrast, mutant proteins in which the bond is eliminated (T11A or Q141A) were completely digested after 6 h (Fig. S2A).
To investigate the requirements for ester bond formation, the following protein variants were made: T11A, T11S, D41A, E108A, H133A, D138A, and Q141A. All mutants were successfully expressed and purified, although they eluted as broad peaks on size-exclusion chromatography, suggestive of multiple species, possibly aggregated. With the exception of D138A, none of the mutant proteins contained an ester bond, as determined by MS analysis. The D138A mutation produces a mixed population of cross-linked and non–cross-linked protein; immediately after purification from E. coli, ∼50% of the protein has an intact ester bond (Fig. S3A). Incubation of the D138A protein at 37 °C, however, reduces the proportion with an intact ester bond to ∼40% after 48 h (Fig. S3B), and further to <20% of total protein after 150 h (Fig. S3C).
The contribution the ester bond makes to the stability of the C1 construct was examined by CD spectroscopy and differential scanning fluorimetry (DSF). WT type C1 gave a CD spectrum typical of a well-folded all-β protein, whereas all mutants gave CD spectra characteristic of unfolded proteins (Fig. S2B). Melting curves measured by DSF showed that the WT C1 has a single unfolding curve with a melting temperature (Tm) of 68 °C, indicating that the protein is well folded and stable (Fig. S2C). In contrast, the D138A mutant has a much broader unfolding curve, reflecting the heterogeneous ester bond formation, with the species lacking an ester bond starting to unfold at 25 °C. All other mutants display characteristics of unfolded or aggregated protein (23).
Discussion
Proposed Mechanism of Ester Bond Formation.
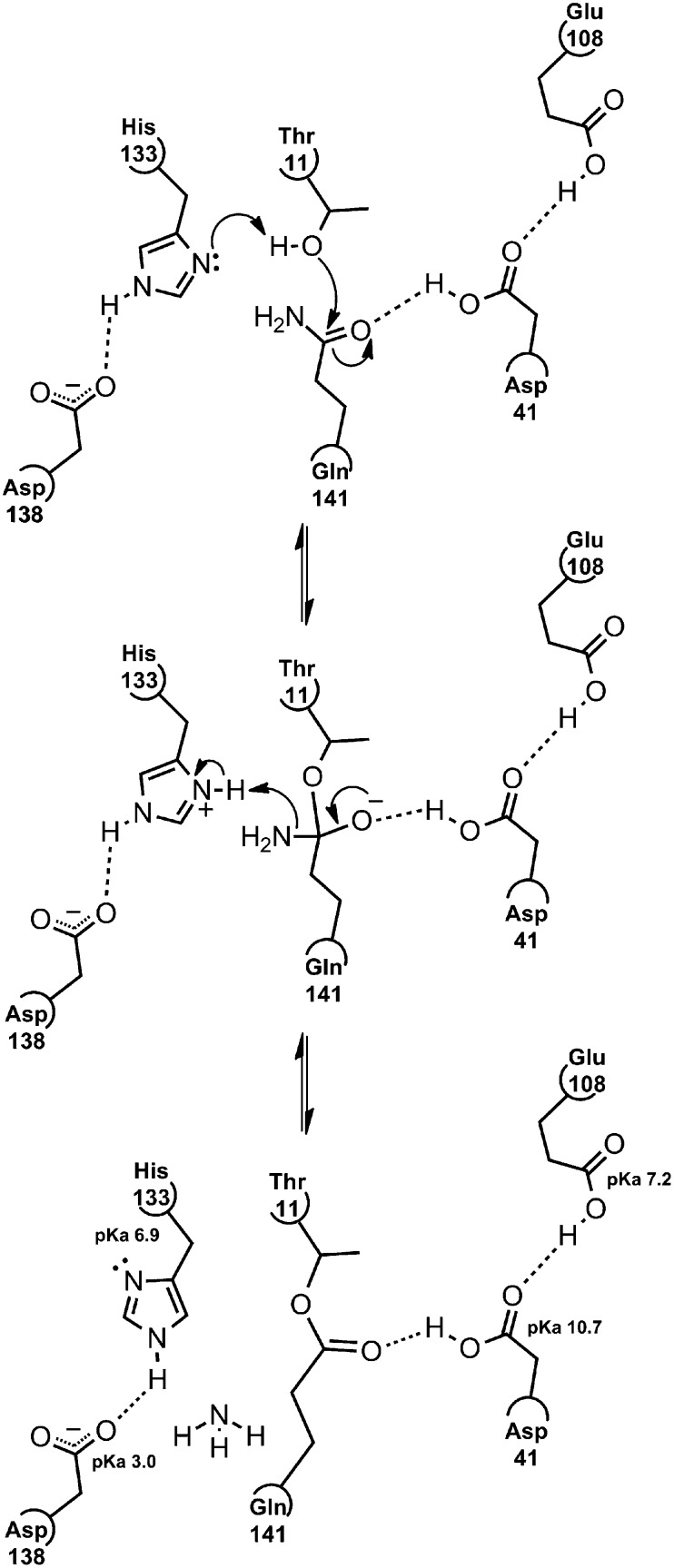
A distinguishing feature of the C2 fold is the presence of a seven-residue insertion in the middle of the last β-strand (G) of each domain (Fig. 1F). Taking the first domain as the example, the insertion of these seven residues between His-133 and Gln-141 forms a loop that positions His-133 and Asp-138 adjacent to Thr-11 and Gln-141. In this arrangement, Thr-11, His-133, and Asp-138 form a triad similar to that seen in serine proteases. We therefore propose a mechanism for ester bond formation that is similar to that of the serine protease family (24), in which the hydroxyl oxygen of Thr-11 acts as a nucleophile in attacking the side chain carbonyl carbon of Gln-141; in this case, the Gln side chain is analogous to the peptide substrate of serine proteases (Fig. 4). The reaction appears to be specific to Thr, because no bond is formed when Thr-11 is substituted by Ser in the T11S mutant protein. We assume that steric factors in the state that exists before bond formation are responsible for this discrimination.
Fig. 4.
Proposed mechanism of ester bond formation. The first step is nucleophilic attack of Thr-11 on Gln-141, proton abstraction by His-133, and bond polarization by the Asp-41/Glu-108 pair. The next step highlights an oxyanion intermediate that rearranges, abstracts a proton from His-133, and results in the elimination of NH3. The final state (crystal structure) shows an internal ester bond stabilized by the Asp-41/Glu-108 pair and highlights the unusual pKa values of these residues. The ester bond is prevented from hydrolysis by the His-133/Asp-138 interaction.
Additional residues promote the reaction, as shown by the fact that the mutant proteins H133A, D41A, and E108A do not form the ester bond and D138A forms the bond, but to a reduced extent. His-133 is positioned adjacent to Thr-11 Oγ1, where it is presumed to act as a catalytic base, accepting the hydrogen from the Thr-OH group, and thus enhancing the nucleophilic potential of the Oγ1 oxygen. Although we only see the catalytic site structure after bond formation, we propose that His-133 would also hold the Thr-11 side chain in a suitable geometry for catalysis. Asp-138 hydrogen-bonds to His-133, and we propose that it plays a dual role in holding His-133 in a catalytically ideal orientation and in making the nonprotonated His-133 nitrogen more electronegative for proton abstraction. Asp-138 is clearly not essential for the reaction, however, because the D138A mutant is still capable of bond formation, albeit less effectively. On the other side of the active site, a pair of acidic residues also promotes catalysis; both Glu-108 and Asp-41 are buried in the protein interior, and hydrogen bonding considerations suggest that both are protonated. Glu-108 hydrogen-bonds to Asp-41, which, in turn, hydrogen-bonds to the carbonyl oxygen of the Gln-141 side chain, increasing the electrophilic potential of the carbonyl carbon.
Nucleophilic attack should produce a tetrahedral intermediate, an adduct of Thr-11 and Gln-141 side chains with a high-energy oxyanion that is stabilized by the “proton shuttle” arrangement of the Glu-108/Asp-41 pair. The pKa of Asp-41 as calculated by PROPKA (25) from the crystal structure coordinates has an unusually high value of 10.7. Although this number may not be completely reliable, it indicates that at biological pH, the side chain is exclusively protonated as is required for stabilizing the oxyanion species; no other chemistry in the active site appears to stabilize the oxyanion. The tetrahedral intermediate breaks down with the reformation of the carbonyl oxygen double bond and a concerted attack by the Gln-141 amino group on the now protonated His-133 residue to abstract a proton; one molecule of ammonia (NH3) is eliminated, resulting in the 17-Da or 33-Da loss of mass observed by MS of the single-domain and double-domain constructs.
The formation of the new bond and the elimination of ammonia produce the equivalent of the acyl intermediate in serine proteases. However, unlike the acyl intermediate of a protease, which is then attacked by water to break the ester bond and regenerate the catalytic site, the ester bond of the C1 protein, as previously noted, is stable; it does not react further, and the ester bond species is essentially trapped. In serine proteases, the histidine equivalent to His-133 mediates the hydrolysis of the ester bond (24). In our C1 structure, however, the hydrogen bond between Asp-138 (pKa 3.0 as calculated by PROPKA) and His-133 sequesters the histidine in a conformation in which it is unable to interact with the ester bond. The Asp-138/His-133 hydrogen-bonded pair is also in a position where it could block the entry of a water molecule into an appropriate location for ester bond hydrolysis.
Site-directed mutagenesis of the catalytic residues (as outlined above) supports our proposed catalytic mechanism, showing that in all but one case, ester bond formation is eliminated in mutant proteins. Mutation of Asp-138 to alanine produces a mixed population of ester-bonded and nonbonded protein as outlined above. Does this fit the proposed mechanism? A time-course analysis of ester bond presence in the D138A protein shows that over time, the proportion of ester bond-formed species is reduced (Fig. S3). Consistent with our hypothesis that the Asp-138/His-133 pair, in essence, traps the ester bond species, we believe that elimination of this critical interaction allows water-mediated hydrolysis to occur, again mirroring the serine proteases. In our proposed hydrolysis mechanism for the D138A mutant (Fig. S4), water binds between an unrestricted His-133 and the ester bond and His-133 functions as a base in abstracting a water proton and enhancing the nucleophilic attack of the water oxygen on the Gln-141 carbonyl carbon. A tetrahedral intermediate results, with a high-energy oxyanion species that is again stabilized by hydrogen bonding to the Glu-108/Asp-41 pair. The intermediate collapses as the threonine oxygen accepts a proton from His-133 and the oxyanion coalesces back into a carbonyl species; the ester bond has broken, the threonine is regenerated, and we presume that Gln-141 is converted to Glu-141.
Conservation of Side Chain Ester Bonds in Bacterial Cell Surface Proteins.
A sequence search suggests that ester bonds of this type are a conserved feature in many cell surface proteins of Gram-positive bacteria. Despite low overall sequence identities between C2 and other proteins, all residues that are presumed to be involved in bond formation appear conserved (Fig. S5).
Both the CD and DSF data show that the ester bonds confer significant stability to the proteins. The WT C1 protein has high thermal stability, unfolding in a single transition at a Tm of 68 °C, whereas the mutants that lack the ester bond are destabilized to the point of being unfolded when measured by these techniques. This contrasts with the case of isopeptide bonds in pilin as described by Kang and Baker (22), where the loss of the isopeptide bond thermally destabilized the protein by ∼25 °C but the protein remained demonstrably folded as measured by DSF. The proteolytic stabilities of the C1 mutants are similarly compromised. The WT protein remains intact over 24 h, whereas the mutants that lack ester bonds are almost fully degraded after a few hours of incubation with trypsin.
Several factors may contribute toward the stability of Cpe0147. The addition of an intramolecular covalent bond provides inherent stability, but the positioning of these cross-links is highly significant (14). Like the intramolecular isopeptide bonds of bacterial surface proteins, the ester bonds join the first and last β-strands of each domain at a point where molecular dynamics simulations and force distribution analysis suggest the critical point of stress concentration exists (26). Mutagenesis studies of Ig-like domains, coupled with atomic force microscopy analysis, come to similar conclusions (27). In these multidomain proteins, the repeated ester (or isopeptide) bonds essentially produce a covalent connectivity that extends from the cell wall anchor to adhesin along the axis of the protein and provides the main force-bearing feature of these cell surface proteins: greatly increased tensile strength.
Bond-Forming Chemistry.
Why do some cell surface proteins use ester bonds rather than isopeptide bonds to provide strength? We suggest that this relates to the specific environment at the cross-linking site. For isopeptide bond formation, a hydrophobic environment is critical in manipulating side chain pKa, whereas in domains such as those in Cpe0147, the chemistry that forms the cross-linking ester bond occurs in a relatively solvent-exposed environment and uses a well-established mechanism similar to that of serine/threonine proteases, an example of convergent evolution. We hypothesize that Asp-138 is a key residue that separates the chemistry of Cpe0147 from that of serine proteases. The Asp-138/His-133 interaction prevents the hydrolysis of the ester bond after its formation, in essence trapping an acyl intermediate.
Materials and Methods
Bioinformatics.
To investigate how widespread intramolecular isopeptide bonds are in Gram-positive bacteria, the Jpred server (www.compbio.dundee.ac.uk/jpred) (28) was searched using major pilin sequences to produce multiple sequence alignments. Based on sequence alignment and on secondary and tertiary structure prediction (Jpred/I-TASSER) (28, 29), two sequences were chosen as potentially containing intramolecular isopeptide bonds, one of which was Cpe0147.
Gene Synthesis, Protein Expression, and Purification.
The C2 construct was synthesized by GENEART (Life Technologies) from the GenBank sequence EDT23863.1 (residues 292–625) with codon optimization. The single-domain C1 construct (residues 8–152 of C2) was PCR-amplified from the C2 gene. Both constructs were subcloned into pET22b, overexpressed in E. coli as C-terminal His-tagged proteins, and purified by nickel-affinity chromatography followed by size-exclusion chromatography according to Kang et al. (30). SeMet-substituted C2 was produced according to a protocol from Sreenath et al. (31) and purified similar to the native protein. Full details of protein expression and purification are given in SI Materials and Methods.
Crystallization and Structure Determination.
Crystallization conditions for native and SeMet C2 were screened after incubation with trypsin at a 1:20,000 trypsin/protein (wt/wt) ratio at 310 K for 2 h. Cleavage was monitored by SDS/PAGE. Crystals were grown in sitting drops. The best native C2 crystals were obtained using a precipitant solution comprising 12.5% (vol/vol) PEG 1000, 12.5% (vol/vol) PEG 3350, 12.5% (vol/vol) 2-Methyl-2,4-pentanediol, 0.03 M MgCl2, 0.03 M CaCl2, and 0.1 M bicine/Trizma base (pH 8.5). The SeMet C2 crystals were macroseeded from native C2 crystals. The best C1 crystals were obtained with 10% (vol/vol) PEG 20,000, 20% (vol/vol) PEG monomethyl ether 550, 0.02 M amino acids (0.02 M sodium l-glutamate, 0.02 M dl-alanine, 0.02 M glycine, 0.02 M dl-lysine, 0.02 M dl-serine), and 0.1 M 3-morpholinopropane-1-sulfonic acid (MOPS)/HEPES (pH 7.5). All three proteins were crystallized at 100 mg/mL. Full details of crystallization protocols are given in SI Materials and Methods. Crystals were flash-cooled without further cryoprotection. X-ray diffraction data were collected at the Australian Synchrotron (MX1 beamline) to a resolution of 1.9 Å for SeMet C2 and to a resolution of 1.1 Å for C1. Data were processed and scaled with X-ray Detector (XDS) (32) and SCALA (Scala, Inc.) (33) software. The structure of C2 was solved by SAD phasing. Phase determination, density modification, and model building used PHENIX software (34). Model building was completed with Crystallographic Object-Oriented Toolkit (Coot) software (35). The SeMet C2 structure was refined with BUSTER software (36). The C1 structure was solved by molecular replacement using the C2 structure and refined using REFMAC software (37). Final validation used MOLPROBITY (38). Data collection and refinement statistics are shown in Table S1.
MS.
SDS/PAGE gel bands containing recombinant protein were excised and trypsinized. Peptides were analyzed using a Q-STAR XL Hybrid MS/MS system (Applied Biosystems) and identified using the MASCOT search engine v.2.0.05 (Matrix Science). Unmatched peptides were manually inspected to identify cross-linked sequences. Accurate protein molecular mass was determined by ESI TOF MS. Samples were analyzed in the positive ionization mode on a Q-STAR XL Hybrid MS/MS system. Data were acquired in the m/z range of 500–1,600. Raw data were deconvoluted to give accurate molecular mass using the Bayesian Protein Reconstruct tool from the Bioanalyst extensions within Analyst QS 1.1 (Applied Biosystems).
Mutagenesis.
All mutations were made on the C1 construct using 5′-phosphorylated primers (Table S4). PCR-amplified products were gel-purified and then ligated at 18 °C overnight. Mutagenesis was sequence-verified. All mutants were expressed and purified similar to WT. Bond formation for each mutant was confirmed by MS. Mutants were subjected to proteolysis as for the native protein.
DSF.
Protein thermal unfolding was monitored by the increase in fluorescence of SyproOrange (Sigma), using a real-time PCR device (7900HT Fast RT PCR System; Applied Biosystems). A reaction volume of 50 μL contained 30 μM protein, 5 μL of 25× SyproOrange, and 15 μL of protein storage buffer. Experiments were performed in triplicate in a 96-well plate with a temperature gradient from 25–95 °C in steps of 1 °C/min. Fluorescence emission at 600 nm was plotted as a function of temperature. Tm values were fitted to the Boltzmann equation using Microsoft Excel (39).
CD.
WT and mutant C1 proteins were buffer-exchanged into 5 mM sodium phosphate buffer (pH 8.0) and 50 mM sodium fluoride to a final concentration of 2.5 μM. CD spectra were recorded on a PiStar-180 (Applied Photophysics) spectrometer. To obtain overall CD spectra, wavelength scans between 180 and 320 nM were collected at 20 °C using a 2-nm bandwidth, 1-nm step size, and time per step of 2 s. The data were collected over five accumulations and averaged.
Supplementary Material
Acknowledgments
We thank Martin Middleditch for help with MS. This research was undertaken on the MX1 beamline at the Australian Synchrotron. This work was supported by the Marsden Fund of New Zealand, the Health Research Council of New Zealand, and the Maurice Wilkins Centre for Molecular Biodiscovery.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.J.R. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4NI6 and 4MKM).
See Commentary on page 1229.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316855111/-/DCSupplemental.
References
- 1.Firbank SJ, et al. Crystal structure of the precursor of galactose oxidase: An unusual self-processing enzyme. Proc Natl Acad Sci USA. 2001;98(23):12932–12937. doi: 10.1073/pnas.231463798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Díaz A, Horjales E, Rudiño-Piñera E, Arreola R, Hansberg W. Unusual Cys-Tyr covalent bond in a large catalase. J Mol Biol. 2004;342(3):971–985. doi: 10.1016/j.jmb.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 3.Kaila VRI, Johansson MP, Sundholm D, Laakkonen L, Wikstrom MR. The chemistry of the CuB site in cytochrome c oxidase and the importance of its unique His-Tyr bond. Biochim Biophys Acta, Bioenerget. 2009;1787(4):221–233. doi: 10.1016/j.bbabio.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Barondeau DP, Putnam CD, Kassmann CJ, Tainer JA, Getzoff ED. Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures. Proc Natl Acad Sci USA. 2003;100(21):12111–12116. doi: 10.1073/pnas.2133463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318(5856):1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- 6.Kang HJ, Baker EN. Structure and assembly of Gram-positive bacterial pili: Unique covalent polymers. Curr Opin Struct Biol. 2012;22(2):200–207. doi: 10.1016/j.sbi.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Patti JM, Allen BL, McGavin MJ, Höök M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 8.Symersky J, et al. Structure of the collagen-binding domain from a Staphylococcus aureus adhesin. Nat Struct Biol. 1997;4(10):833–838. doi: 10.1038/nsb1097-833. [DOI] [PubMed] [Google Scholar]
- 9.Deivanayagam CCS, et al. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure. 2000;8(1):67–78. doi: 10.1016/s0969-2126(00)00081-2. [DOI] [PubMed] [Google Scholar]
- 10.Hagan RM, et al. NMR spectroscopic and theoretical analysis of a spontaneously formed Lys-Asp isopeptide bond. Angew Chem Int Ed Engl. 2010;49(45):8421–8425. doi: 10.1002/anie.201004340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forsgren N, Lamont RJ, Persson K. Two intramolecular isopeptide bonds are identified in the crystal structure of the Streptococcus gordonii SspB C-terminal domain. J Mol Biol. 2010;397(3):740–751. doi: 10.1016/j.jmb.2010.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in gram-positive pathogens. Nat Rev Microbiol. 2006;4(7):509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 13.Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70(1):192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang HJ, Baker EN. Intramolecular isopeptide bonds: Protein crosslinks built for stress? Trends Biochem Sci. 2011;36(4):229–237. doi: 10.1016/j.tibs.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 15.Pointon JA, et al. A highly unusual thioester bond in a pilus adhesin is required for efficient host cell interaction. J Biol Chem. 2010;285(44):33858–33866. doi: 10.1074/jbc.M110.149385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vengadesan K, Narayana SVL. Structural biology of Gram-positive bacterial adhesins. Protein Sci. 2011;20(5):759–772. doi: 10.1002/pro.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bork P, Holm L, Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J Mol Biol. 1994;242(4):309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 18.Harding MM. Small revisions to predicted distances around metal sites in proteins. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 6):678–682. doi: 10.1107/S0907444906014594. [DOI] [PubMed] [Google Scholar]
- 19.Kang HJ, Paterson NG, Gaspar AH, Ton-That H, Baker EN. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc Natl Acad Sci USA. 2009;106(40):16967–16971. doi: 10.1073/pnas.0906826106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vengadesan K, Ma X, Dwivedi P, Ton-That H, Narayana SVL. A model for group B Streptococcus pilus type 1: The structure of a 35-kDa C-terminal fragment of the major pilin GBS80. J Mol Biol. 2011;407(5):731–743. doi: 10.1016/j.jmb.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang HJ, Middleditch M, Proft T, Baker EN. Isopeptide bonds in bacterial pili and their characterization by X-ray crystallography and mass spectrometry. Biopolymers. 2009;91(12):1126–1134. doi: 10.1002/bip.21170. [DOI] [PubMed] [Google Scholar]
- 22.Kang HJ, Baker EN. Intramolecular isopeptide bonds give thermodynamic and proteolytic stability to the major pilin protein of Streptococcus pyogenes. J Biol Chem. 2009;284(31):20729–20737. doi: 10.1074/jbc.M109.014514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavinder JJ, Hari SB, Sullivan BJ, Magliery TJ. High-throughput thermal scanning: A general, rapid dye-binding thermal shift screen for protein engineering. J Am Chem Soc. 2009;131(11):3794–3795. doi: 10.1021/ja8049063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102(12):4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61(4):704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 26.Wang B, Xiao S, Edwards SA, Gräter F. Isopeptide bonds mechanically stabilize spy0128 in bacterial pili. Biophys J. 2013;104(9):2051–2057. doi: 10.1016/j.bpj.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Carrion-Vazquez M, Oberhauser AF, Marszalek PE, Fernandez JM. Point mutations alter the mechanical stability of immunoglobulin modules. Nat Struct Biol. 2000;7(12):1117–1120. doi: 10.1038/81964. [DOI] [PubMed] [Google Scholar]
- 28.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36(Web Server issue) Suppl 2:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9(1):40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang HJ, Paterson NG, Baker EN. Expression, purification, crystallization and preliminary crystallographic analysis of SpaA, a major pilin from Corynebacterium diphtheriae. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65(Pt 8):802–804. doi: 10.1107/S1744309109027596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sreenath HK, et al. Protocols for production of selenomethionine-labeled proteins in 2-L polyethylene terephthalate bottles using auto-induction medium. Protein Expr Purif. 2005;40(2):256–267. doi: 10.1016/j.pep.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W. Automatic indexing of rotation diffraction patterns. J Appl Cryst. 1988;21(1):67–72. [Google Scholar]
- 33.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 34.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Smart OS, et al. Exploiting structure similarity in refinement: Automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: Structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32(Web Server issue) Suppl 2:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown AM. A step-by-step guide to non-linear regression analysis of experimental data using a Microsoft Excel spreadsheet. Comput Methods Programs Biomed. 2001;65(3):191–200. doi: 10.1016/s0169-2607(00)00124-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.