Significance
The Kai system is a widely studied model in theoretical biology and systems biology. It is to date the only known circadian clock that can be reconstituted in vitro. Essential to the rhythmicity of the system is the formation of the KaiC–KaiB complex. Many aspects of this interaction, such as the mode of binding of KaiB, the stoichiometry of the interaction, and the exact binding interfaces have long remained ambiguous. We present a mass spectrometry-based structural model of the KaiC–KaiB interaction that answers many of these outstanding questions on the basis of direct experimental evidence. This structural model sheds light on the intricate workings of the in vitro oscillator.
Keywords: protein–protein docking, ion mobility spectrometry, native MS
Abstract
Circadian timing in cyanobacteria is determined by the Kai system consisting of KaiA, KaiB, and KaiC. Interactions between Kai proteins change the phosphorylation status of KaiC, defining the phase of circadian timing. The KaiC–KaiB interaction is crucial for the circadian rhythm to enter the dephosphorylation phase but it is not well understood. Using mass spectrometry to characterize Kai complexes, we found that KaiB forms monomers, dimers, and tetramers. The monomer is the unit that interacts with KaiC, with six KaiB monomers binding to one KaiC hexamer. Hydrogen–deuterium exchange MS reveals structural changes in KaiC upon binding of KaiB in both the CI and CII domains, showing allosteric coupling upon KaiB binding. Based on this information we propose a model of the KaiB–KaiC complex and hypothesize that the allosteric changes observed upon complex formation relate to coupling KaiC ATPase activity with KaiB binding and to sequestration of KaiA dimers into KaiCBA complexes.
Cyanobacteria represent some of the simplest organisms known to have an autonomous circadian timing mechanism. The core circadian clock of cyanobacteria does not rely on transcription–translation feedback loops (TTFLs) as it does in higher organisms (1–4). This clock is known as the Kai system and it produces oscillations with an approximate 24-h period that are based on phosphorylation and assembly dynamics of the three Kai proteins: KaiA, KaiB, and KaiC (5). Phosphorylation and assembly of Kai proteins determine their interactions with circadian output kinases, which translate circadian timing information into 24-h rhythms of global gene expression (6–8). The Kai proteins can go through the same 24-h rhythm of phosphorylation and assembly when the system is reconstituted in vitro, simply by incubating KaiA, KaiB, and KaiC in the presence of ATP (9). The in vitro oscillator is robust and can maintain a stable rhythm for weeks on end. It is temperature-compensated and subject to temperature-step entrainment (9–12). It bears all of the hallmark features of a true circadian clock and hence provides a unique opportunity to study biological timing to a high level of detail.
There are a large number of studies that describe the phosphorylation dynamics and interactions between Kai proteins. KaiC consists of two domains, CI and CII, which possess ATPase and phosphorylation activity, respectively. KaiC forms hexamers and is the central hub of the system (13–15). The default activity of the CII domain is autodephosphorylation. Unphosphorylated KaiC binds KaiA dimers, which triggers sequential autophosphorylation of KaiC at residues T432 and S431 (12, 16–19). This “hyperphosphorylated” KaiC binds KaiB, which results in a switch back to autodephosphorylation, first of T432, then S431. Dephosphorylation proceeds through ATP/ADP phosphotranferase activity of KaiC (20, 21). The phosphorylation status of KaiC and the assembly with KaiA and KaiB define the phase of circadian timing.
We recently proposed a theoretical model for Kai that accurately described experimental phosphorylation dynamics, temperature-step entrainment, and assembly dynamics of Kai protein complexes (22, 23). In this model, the formation of a phosphorylated KaiC–KaiB complex counteracts the stimulation of KaiC autophosphorylation by KaiA. Phosphorylated KaiC–KaiB complexes sequester KaiA in KaiCBA complexes, thus preventing formation of “productive” KaiC–KaiA complexes. This allows KaiC to switch back to its default autodephosphorylation activity. KaiA sequestration was demonstrated by semiquantitative monitoring of Kai protein assembly with native MS. Using this approach, we could confirm the formation of ternary Kai complexes bearing a high number of KaiA dimers, concomitant with the peak in KaiC phosphorylation. The formation of phosphorylated KaiCB complexes is thus a crucial step in producing stable oscillations of phosphorylation, but the mechanism by which these complexes trigger sequestration of KaiA is still unknown (17, 22–24).
The structures of KaiC and KaiB have both been described individually to atomic-level resolution, but structural details of their interaction remain ambiguous. KaiC forms a hexamer with a double ring structure, the CI and CII domain each forming one ring, stacked together (4, 25, 26). Free KaiB has been crystallized as a tetramer in four studies and has once been described as a dimer (27–32). It was proposed to bind to KaiC as a tetramer (33), dimer (27), or, more recently, as a monomer (32). From single-particle EM reconstructions of KaiCB complexes, KaiB was suggested to bind to the CII domain of KaiC, which was supported by biochemical experiments where KaiCB interactions were assessed on the isolated CI and CII domains and by negative-stain EM analysis of nanogold-labeled KaiCB (27, 32, 34, 35). However, it was recently reported that KaiB does not interact with the isolated CII domain of KaiC, but interacts with an engineered monomeric form of the CI domain instead (36). It was suggested in this study that CI–CII stacking interactions wedge the CI domains apart, exposing the putative KaiB binding site. The EM reconstruction of KaiCB complexes is ambiguous with respect to the stoichiometry of the interaction and the exact binding interface. Several small-angle X-ray scattering (SAXS) reconstructions of KaiCB suffer the same ambiguity in describing structural details of the interaction (33, 34). KaiB has been proposed to bind to KaiC with a final stoichiometry of either four or six copies of KaiB to six of KaiC (27, 32, 33). Moreover, there is currently no direct experimental evidence that pinpoints the KaiC-interacting region of KaiB.
Here, we describe the MS-based structural characterization of the KaiC–KaiB interaction, together with protein–protein docking predictions using HADDOCK (37, 38). We show that KaiB exists as monomers, in addition to tetramers. We demonstrate that KaiB binds KaiC with a final stoichiometry of 6:6 and that KaiB monomers are the basic interacting unit. Using hydrogen–deuterium exchange (HDX) MS, we have determined the structural changes in both KaiC and KaiB upon complex formation that revealed allosteric coupling between the CI and CII domains of KaiC upon KaiB binding. Using this information to drive modeling with HADDOCK, we tested binding of KaiB to CI or CII against experimental ion mobility mass spectrometry constraints (39). Allosteric transitions are observed in both KaiC and KaiB that provide insight into the mechanism of KaiA sequestration and coupling between KaiB-binding and ATPase activity in the CI domain.
Results
Defining the Stoichiometry of the KaiC–KaiB Complex.
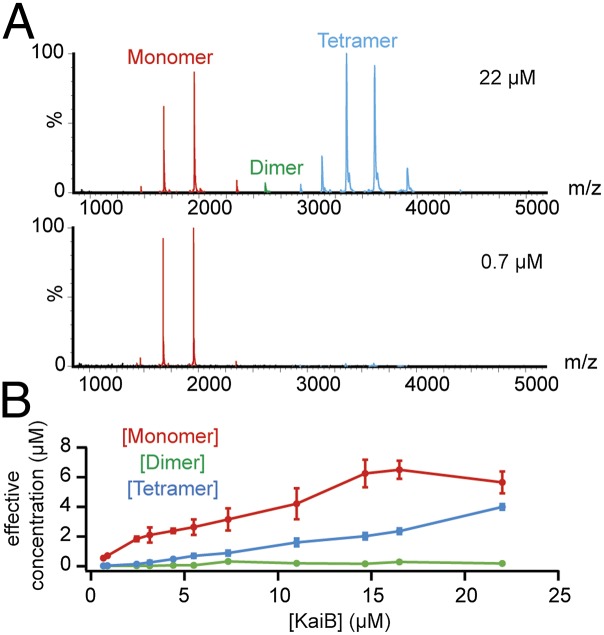
The stoichiometries of KaiB and KaiC as well as KaiCB complexes were studied using native MS, which allows for the semiquantitative monitoring of complex stoichiometries with unmatched precision (40). The masses of all detected Kai proteins and protein complexes are listed in Table S1. Three KaiB stoichiometries could be detected: tetramers, dimers, and monomers (Fig. 1A). Tetramer and monomer account for most of the observed signal and the dimer is a minor component. At lower concentration, most of the tetramer signal disappears. Under the standard concentration of the in vitro oscillator (3 µM) (9), KaiB forms mostly monomers (Fig. 1B). Owing to concentration-dependent tetramerization of KaiB, the effective KaiB monomer concentration is rather robust to changes in total KaiB concentration.
Fig. 1.
KaiB forms monomers, dimers, and tetramers. (A) Native MS of KaiB at high (22 µM) and low (0.7 µM) concentration. A mix of monomers, dimers, and tetramers is formed. The tetramer almost completely disappears at lower concentrations. (B) Titration experiment of KaiB stoichiometry with native MS. The fraction of the total MS signal is used to calculate the effective concentration of monomers, dimers, and tetramers. The averages of two experiments are plotted; error bars represent SDs. The effective KaiB monomer concentration is much more stable as a function of total KaiB concentration, compared with the hypothetical situation where no tetramers can form, which would correspond to a linear curve that is 1:1 for effective KaiB concentration as a function of total KaiB concentration.
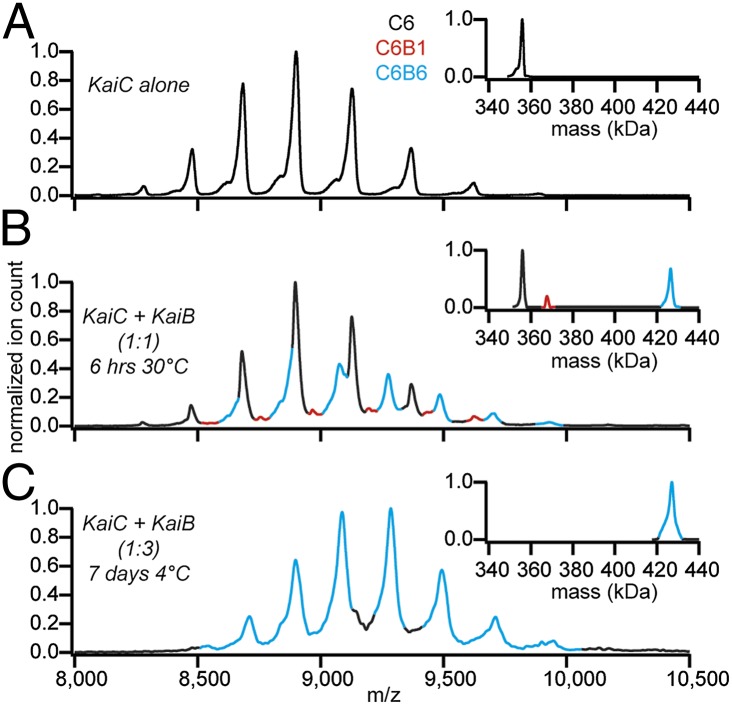
The formation of KaiC hexamers could be unambiguously confirmed (Fig. 2A). Next, the stoichiometry of KaiCB complexes was determined upon mixing KaiC and KaiB under standard conditions of the in vitro oscillator (3 µM KaiC and 3 µM KaiB incubated for 6 h at 30 °C in the presence of ATP). In this preparation, three distinct components could be detected (Fig. 2B). There was still a substantial amount of free KaiC, and two different KaiCB complexes were detected: KaiC6B6 and KaiC6B1. Both stoichiometries could be unambiguously confirmed using tandem MS, whereby selected precursor ions are dissociated in the instrument and their mass is deduced from the observed dissociation products (41) (Fig. S1). In addition, the experiment shows that no intermediate stoichiometries, such as KaiC6B4, are formed. The absence of any intermediate stoichiometries of KaiCB complexes suggests that assembly of the KaiC6B6 complex is cooperative, whereby one binding or unbinding event triggers complete assembly or disassembly of the full KaiC6B6 complex. The presence of the KaiC6B1 complex clearly shows that KaiB interacts with KaiC as a monomer, not the dimer or tetramer, as has previously been suggested (27, 33, 34).
Fig. 2.
Six KaiB monomers bind one phosphorylated KaiC hexamer. Native MS of KaiC and KaiCB complexes. (A) KaiC forms hexamers. (B) Upon mixing KaiB and KaiC under standard conditions of the in vitro oscillator, KaiC6B6 and KaiC6B1 complexes are formed. (C) Nearly complete formation of KaiC6B6 is achieved when an excess of KaiB is added and incubated at reduced temperature. (Insets) Transformed spectra, where m/z is converted to mass.
KaiB has been shown to bind preferentially to phosphorylated KaiC (18, 19, 42, 43). It was recently also reported that KaiC switches from its default autodephosphorylation activity at 30 °C to autophosphorylation activity when incubated at 4 °C (21, 34). To confirm that KaiB binds phosphorylated KaiC, a mixture of 3 µM KaiC with 9 µM KaiB was incubated in the presence of ATP for 1 wk at 4 °C. Indeed, near-complete formation of KaiC6B6 complexes was observed and the KaiC6B1 complex could no longer be detected (Fig. 2C). The formation of KaiCB complexes and the enhanced formation of the complex after long incubation at 4 °C were confirmed using native PAGE (Fig. S2). Native MS and SDS/PAGE analyses confirmed higher phospholevels in KaiC after incubation at 4 °C (mostly pSpT) and in KaiCB complexes compared with KaiC input material and unbound KaiC (Fig. S2). SDS/PAGE analysis of KaiC after 1 wk at 4 °C, followed by a 6-h incubation at 30 °C, revealed that KaiC had retained its dephosphorylation activity after 1 wk at reduced temperature (Fig. S2).
Determining the Binding Interface of the KaiC6B6 Complex.
The structural changes in KaiC and KaiB upon KaiCB assembly were determined with HDX-MS (44). The levels of deuterium uptake of free KaiC and KaiB were compared with KaiCB mixtures with either KaiC or KaiB in excess after 0, 1, 10, and 60 min. The coverage of KaiB (96%) and KaiC (92%) sequence in the HDX-MS experiment are shown in Figs. S3 and S4, respectively. Overviews of the levels of deuterium uptake are presented in Tables S2 and S3 (KaiB and KaiC, respectively).
Upon binding to KaiC, protection from deuterium uptake was observed on the three alpha-helices of the KaiB monomer (Fig. 3, blue). This indicates that the helical face of the KaiB monomer is the KaiC binding region. There are several residues of KaiB that are known to abolish or alter the circadian rhythm of Kai when mutated (5, 24, 29, 31). Three of these residues are part of the KaiC-binding region of KaiB we identified (residues R22, K66, and R74, numbered according to wild-type Synechococcus elongatus sequence). The HDX-MS data thus provide a rationale for their effect on circadian timing, because mutation of these residues will affect the KaiC–KaiB binding by removing electrostatic interactions. An allosteric change on the opposite face of the KaiB monomer is also observed that results in increased deuterium uptake of the N terminus of KaiB (Fig. 3, red), attributable to a small-scale conformational change.
Fig. 3.
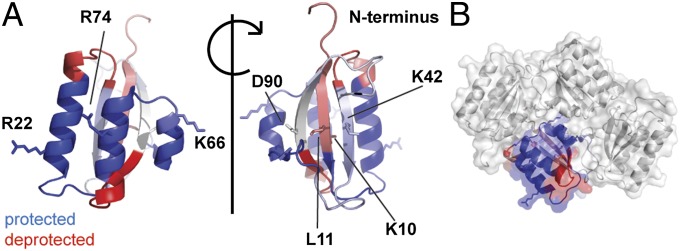
The alpha-helices of KaiB bind to KaiC. (A) Changes in deuterium uptake observed between free and bound KaiB. The changes in deuterium uptake are filtered to P < 0.01 and plotted onto chain D from the crystal structure of S. elongatus KaiB (PDB ID code 4kso). Regions in blue become protected upon binding KaiC, regions in red show increased deuterium uptake, and regions in white show no change or are not detected. Darker colors represent a stronger change upon binding. Labeled residues are known to affect the rhythm of the circadian clock when mutated. (B) One KaiB monomer colored as described in A is shown in the context of the tetramer.
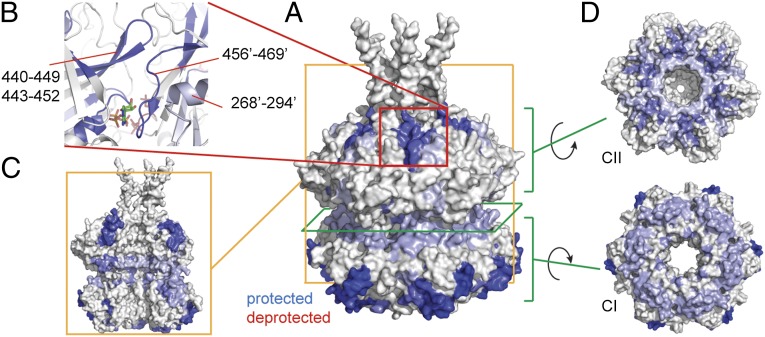
Upon binding of KaiB to KaiC, many changes are observed in the deuterium uptake of KaiC (Fig. 4A). All observed changes reflect a decrease in deuterium uptake. The most strongly protected regions in KaiC are residues 440–452 and 456–469 in the CII domain and residues 92–99 and 126–132 in the CI domain (numbering according to wild-type S. elongatus sequence). The protected regions within the CII domain from two adjacent subunits together form one continuous patch spanning the subunit interface (Fig. 4B). This region sits directly on top of the ATP-binding pocket of the CII domain. Weaker protection is observed for additional regions on the subunit interface of the hexamer (Fig. 4C) and the interface where the CI and CII domains stack upon each other (Fig. 4D). From our HDX-MS data we can conclude that KaiB binds on either the subunit interface of the CII domain or on residues 92–99 and 126–132 in the CI domain. In either case, there seems to be an allosteric coupling between CI and CII upon binding of KaiB, which further induces a conformational change that results in protection of the rest of the subunit interface and the contact region between the CI and CII domains.
Fig. 4.
KaiB binding induces allosteric changes in KaiC. (A) Changes in deuterium uptake observed between free and bound KaiC. Only protected regions are detected. The changes in deuterium uptake are filtered to P < 0.01 and colored blue when protected in the KaiCB complex. Darker colored regions show stronger protection, and regions in white show no change or are not detected. The changes are plotted on a model of the KaiC hexamer of the crystal structure of KaiC (PDB ID code 3dvl); a hexamer of chain A was reconstructed as its entire C terminus is modeled in the PDB. (B) Zoom-in of the protected region on the CII domain. The indicated residues (a prime indicates residues from the adjacent chain) form one continuous binding surface that spans the interface between two subunits. The binding surface sits very close to the ATP-binding pocket. (C) Side view of three chains from the KaiC hexamer showing protection along the interface between KaiC subunits. (D) Views of the contact region between the CI and CII domains that becomes protected upon binding of KaiB.
A Structural Model of the KaiC–KaiB Interaction Using HDX-MS Restraints in HADDOCK.
We used the HDX-MS data to drive protein–protein docking of KaiB on KaiC using HADDOCK (37, 38). For S. elongatus KaiB, we generated an ensemble of four models, based on the four monomers described in the crystal structure (PDB code: 4kso, see Fig. S5). As input for HADDOCK, we used this ensemble of models, together with a dimer of KaiC (two adjacent CI and CII domains) extracted from the crystal structure (25) of S. elongatus KaiC (PDB ID code 3dvl) and the HDX-MS defined interface. The full hexamer was then reconstructed from the docking model using sixfold symmetry.
Two HADDOCK runs were performed, where the protected regions in either the CI or the CII domain were specified as active residues. The resulting models were then compared with experimental ion mobility mass spectrometry (IM-MS) data on the KaiCB complex (Fig. 5 A and B). MS-based ion mobility spectrometry provides information on the shape of protein complexes in the form of their collisional cross-section (CCS) (39). We tested the validity of this approach by comparing the experimental CCS of KaiC to the calculated CCS from the known structure of KaiC, which were in good agreement (Fig. S6). We analyzed the four best-scoring HADDOCK clusters of CI and CII binding, an overview of which is presented in Fig. S7. When the CI domain is specified as the binding region, all four best-scoring clusters yield theoretical CCS that lie outside the 95% confidence interval of the experimentally determined cross-section. When the CII domain is specified as the KaiB-interacting domain, all but the second ranking cluster lie within the experimentally determined range of the CCS. There is also a substantial difference in the HADDOCK scores between the top-ranked clusters: −76 a.u. for CII binding vs. −48 a.u. for CI binding (Table S4). Our modeling approach, using HDX-MS–driven HADDOCK and IM-MS, therefore favors a model in which KaiB monomers bind on the CII domain of KaiC. It should be noted, however, that the HDX experiments indicated allosteric changes in both KaiC and KaiB upon binding. Therefore, based on our data we cannot exclude CI as a possible binding site for KaiB because conformation changes upon KaiB binding might affect the CCS of the complex.
Fig. 5.
HADDOCK-based model of the KaiC6B6 complex. (A) Surface rendering of the four clusters with the highest scores (Table S4), with KaiB docked on either the CI or CII domain of KaiC. The theoretically predicted CCS of each cluster is indicated. (B) Bar representation of the CCSs of HADDOCK models. The 95% confidence interval of the experimental CCS is indicated with dotted lines. (C) Detailed view of the four CII-docked clusters. KaiB is colored blue, KaiC white-gray, and the important residues of KaiB and KaiC that are involved in electrostatic contacts are colored red and green, respectively. Only residue pairs that make contact (<4 Å proximity) are labeled; a prime indicates residues from the adjacent chain. (D) Reconstructed KaiC6B6 complex based on the best-scoring HADDOCK cluster. (E) Detailed view of the proposed interaction between KaiC and KaiB, showing alignment of the two protected regions. The changes in deuterium uptake as detected with HDX-MS are plotted onto the structure as described in Figs. 3 and 4.
A closer inspection of the four clusters where KaiB binds to the CII domain (Fig. 5C) reveals that the experimentally determined binding region of KaiB is barely buried in the binding interface of clusters 2 and 3. These clusters therefore do not accurately reflect the KaiCB interaction. In contrast, the binding region of KaiB is mostly buried in the first- and fourth-ranking models. Beside the worse HADDOCK score of the fourth-ranking cluster compared with the first-ranking cluster, the main difference between them is an approximate 180° rotation of the KaiB monomer with respect to KaiC. The three positively charged residues in KaiB that have been shown to be crucial for standard rhythmicity of the oscillator were also used to assess the quality of the model. In the fourth-scoring model, two of the three residues are involved in electrostatic interactions with KaiC, whereas in the top-ranked cluster all three residues are involved in intermolecular electrostatic interactions. This top-ranking cluster according to the HADDOCK score indeed best matches the binding region determined experimentally by HDX-MS for both KaiB and KaiC (Fig. 5 D and E). Table S5 lists intermolecular KaiC–KaiB contacts predicted from the model.
An allosteric transition in KaiB that results in increased deuterium exposure of the N terminus is also observed in the HDX experiments (Fig. 3). The N terminus of KaiB is part of a beta-sheet on the opposite face of the KaiC-binding region (Fig. 5E). This beta-sheet, including the N terminus of KaiB, contains several additional residues that are known to affect circadian timing. This would mean that they are exposed in the KaiC–KaiB complex and could thus be involved in other steps within the circadian cycle, such as KaiA sequestration or interactions with circadian output kinases.
Discussion
Our experiments demonstrate the formation of a KaiC6B6 complex, which MS indicates is the relevant stoichiometry in the in vitro oscillator as seen from the stoichiometry of KaiCBA complexes (20). In contrast to our report, a stoichiometry of KaiC6B4 has been proposed from various studies of the KaiC–KaiB interaction (27, 28, 33, 34). Given the ambiguity of reported EM- and SAXS-based shape reconstructions of KaiCB, this stoichiometry was mostly inspired by the tetrameric (dimer of dimers) crystal structures reported for KaiB. Most recently, a cryo-EM reconstruction of KaiCB indicated that a KaiC6B6 complex is formed, and our native MS data provide an independent line of evidence to unambiguously demonstrate that this is the stoichiometry of the KaiCB complex (32).
The tetrameric structure of KaiB was determined on several accounts with protein crystallography (27, 29, 31, 32). In these reports, the tetrameric state of KaiB could also be confirmed with a number of solution-phase techniques. A SAXS-based shape reconstruction was also shown to be consistent with tetramers (34). The native MS experiments described here show that the tetramer is a concentration-dependent assembly of KaiB and that it is less abundant than the monomer at concentrations more relevant to the in vitro oscillator (3 µM). This is still consistent with the previous findings mentioned above: Those experiments were performed in the concentration range of 40 µM to 10 mM.
The observation that the KaiB monomer is the interacting unit with KaiC does raise the question of why KaiB forms tetramers. It was reported that the in vitro oscillator produces stable oscillations over a wide range of KaiB concentrations (45). At the same time, KaiC and KaiCB concentrations fluctuate in a circadian manner and KaiB is sequestered at the membrane, depending on the time of day (46). In the case that KaiB binding is in fact a rate-limiting step in circadian oscillation of the Kai system, the KaiB tetramer possibly contributes to the robustness of the system by acting as a sink for excess KaiB monomers, thus effectively dampening large fluctuations in total KaiB concentration. Binding of tetramers is also not easily reconciled with the observed stoichiometry, KaiC6B6. The hypothetical cases of either KaiB dimers or tetramers binding imply that the stoichiometry of the interaction is determined by steric restraints, rather than by the number of available binding sites. The proposed modes of binding of dimers and tetramers are further complicated by the rotational symmetry of KaiC hexamers: If multiple subunits from one particular dimer/tetramer would be in contact with KaiC simultaneously, the rotational symmetry of the hexamer would require the binding interface of each interacting subunit within the dimer/tetramer to be unique. In the KaiC6B6 model, no such issues arise.
Whereas the first structural studies on the KaiCB interaction showed that KaiB binds to the CII domain of KaiC, recent studies have indicated otherwise (27, 34, 36). No interaction between isolated CII domains and KaiB could be confirmed, and instead an interaction between KaiB and isolated CI domains was demonstrated. Notably, the interaction could only be demonstrated on an engineered KaiB variant that can no longer form tetramers. Our native MS results suggest that the requirement for this KaiB variant is the direct result of the high concentrations that were used in this study: Wild-type KaiB is forced into a tetrameric state that does not interact with KaiC. In addition, no direct evidence for an interaction between KaiB and KaiC–CI was demonstrated in the context of the full hexamer; only an interaction with an engineered monomeric variant of KaiC-CI was demonstrated. Our modeling data favor KaiB binding to the CII domain of KaiC, but we nevertheless still observe protection of CI in our HDX experiment as well, near a region (residues 116–123) that was recently suggested to be involved in KaiB binding (47).
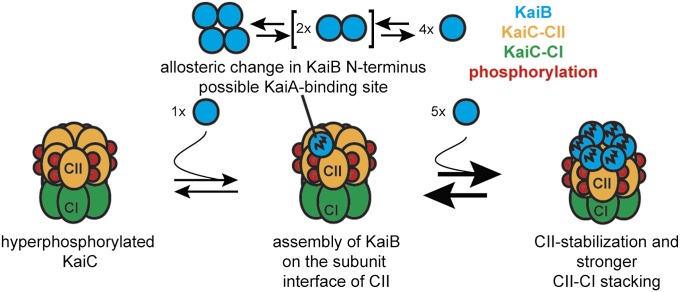
The protected region on the KaiC CII ring observed here with HDX-MS was recently predicted to be the KaiC–KaiB binding interface, based on observations of the electrostatic surface properties of the two proteins (28). The three positively charged alpha-helices of the KaiB monomer bind onto the negatively charged cleft that spans the interface between two adjacent KaiC subunits. A recent docking study, driven by a Cryo-EM reconstruction at moderate resolution, placed KaiB at approximately the same site as described here (32). As one KaiB monomer effectively bridges between two KaiC subunits, binding of KaiB to KaiC could conceivably stabilize the intrahexamer interaction of KaiC. This might offer a possible explanation for the protection of the KaiC subunit–subunit interface observed in the HDX-MS experiments. The ATPase activity of the CI domain of KaiC determines circadian timing of cell division, and KaiB was also reported to inhibit the ATPase activity of the CI domain in KaiC (48, 49). Moreover, catalytic activity in the CI domain was recently shown to be crucial for the KaiC–KaiB interaction (50). Phosphorylation is a known determinant of the ATPase activity of the CI domain (51). It has recently been shown that enhanced interactions between subunits in the CII ring in doubly phosphorylated and S431 phosphorylated KaiC stimulate stacking of the CI and CII rings (43). The increased contacts between the CI and CII rings were proposed to inhibit ATPase activity. Our HDX-MS data confirm that there is an allosteric coupling between KaiB binding and the CI domain, and the data suggest a possible mechanism by which ATPase activity is inhibited: KaiB binding enhances subunit interactions within the CII ring, which promotes stacking contacts between the CI and CII rings (Fig. 6). Direct binding of KaiB to CI could also explain its effect on ATPase activity, in which case an allosteric transition is observed that affect residues near the ATP-binding site of the CII domain, which could conceivably influence the phosphorylation activity of the domain.
Fig. 6.
KaiB binding enhances KaiC stability by stabilizing the CII ring and stimulating CII–CI stacking. Schematic of the structural changes associated with the formation of KaiC6B6.
The allosteric change in KaiB upon KaiC binding may be involved in sequestration of KaiA into ternary Kai complexes. The beta-sheet of the KaiB monomer is exposed in the KaiCB complex, providing sufficient space for KaiA dimers to bind. It has been shown that the region on KaiC where KaiA binds in the KaiC6A2 complex is not required for the formation of ternary KaiCBA complexes (24, 43). We propose that the N terminus of KaiB, as well as adjacent regions concentrated around the beta-sheet of KaiB monomers, together form the alternative binding site for KaiA in ternary Kai complexes.
In summary, we show that KaiB monomers are the interacting units with KaiC. The previously ambiguous stoichiometry of the KaiC:KaiB complex and the unknown KaiC-binding site of KaiB could be determined. Our results indicate that KaiB binds on the CII domain of KaiC, but, on the basis of our HDX data, binding to the CI domain cannot be excluded. Additional changes were also observed in both KaiC and KaiB that have important implications for how KaiC ATPase activity is coupled to KaiB binding and how KaiB induces sequestration of KaiA dimers in ternary Kai complexes.
Experimental Procedures
A full description of the experimental procedures can be found in Supporting Information.
Purification and Gel Analyses of KaiC and KaiB.
GST-fusion proteins of KaiC and KaiB were expressed in Escherichia coli BL21 strains, kindly provided by T. Kondo, Nagoya University, Nagoya, Japan. GST-fusion proteins were purified on gluthathion agarose resin, cleaved using PreScission protease, and further purification was carried out with ion-exchange chromatography. Kai proteins were incubated in Reaction Buffer (RB) [20 mM Tris (pH 8.0), 150 mM NaCl, 0.5 mM EDTA, 5 mM MgCl2, and 1 mM ATP] before SDS/PAGE and native PAGE analysis (concentration, temperature, and time as indicated in Results).
Native MS.
KaiB, KaiC, and KaiCB mixtures were prepared and incubated in RB as indicated in Results. Before MS, samples were transferred to an MS-compatible buffer of 75 mM ammonium acetate, pH 6.8. Samples were loaded into gold-coated boro-silicate capillaries prepared in-house. KaiB tetramerization was studied on an LCT1 (Micromass; Waters, U.K.) (52). KaiC/KaiCB complexes were analyzed on a modified QToF II (MS Vision; Waters, U.K.). Ion mobility spectrometry was performed on a Synapt G1 (Waters, U.K.). The CCS was calibrated using denatured ubiquitin, cytochrome C, and myoglobin, as well as native GroEL (53, 54). Theoretical cross-sections were calculated with the Driftscope Projection Approximation algorithm (Waters, U.K.).
HDX-MS.
The HDX reaction was started by diluting the Kai protein solutions in RB 20-fold in D2O. The reaction was carried out on ice before quenching by 2:1 dilution into ice-cold 6 M guanidine⋅HCl, 300 mM Tris (2-carboxyethyl) phosphine, with pH adjusted to give a final pH of 2.5. Immediately after quenching, the sample was injected into a Waters HDX/ nanoAcquity system for digestion on an online pepsin column, followed by separation on a RP-UPLC gradient at 0 °C and MS on a Waters Xevo QToF G2. For peptide identification, samples were prepared under identical conditions in H2O and analyzed using MSe data acquisition. Data for peptide identification was processed with ProteinLynx Global Server 2.5 software. Deuterium uptake was calculated using Waters DynamX 1.0.0 software.
Structural Modeling and Analysis.
Crystal structures of S. elongatus KaiB (PDB ID code 4kso) and KaiC (PDB ID code 3dvl) were used for docking as described in Results and Supporting Information. The docking simulations were performed using the HADDOCK web server (37) (http://haddock.science.uu.nl/services/HADDOCK). The HDX-MS information was mapped onto KaiB and KaiC solvent-accessible surfaces, resulting in two lists of active residues used to derive ambiguous interaction restraints (55).
Supplementary Material
Acknowledgments
We thank Nancy Sauer for assistance with protein purification. This work was supported by the Netherlands Proteomics Centre, embedded in the Netherlands Genomics Initiative (J.S., R.J.B., and A.J.R.H.), by the Deutsche Forschungsgemeinschaft (A.W. and I.M.A.), and by the German Ministry for Education and Research through FORSYS partner program Grant 0315294 (to I.M.A.). A.M.J.J.B. acknowledges support from the Netherlands Organization for Scientific Research VICI Grant 700.56.442.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314326111/-/DCSupplemental.
References
- 1.Dong G, Kim YI, Golden SS. Simplicity and complexity in the cyanobacterial circadian clock mechanism. Curr Opin Genet Dev. 2010;20(6):619–625. doi: 10.1016/j.gde.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307(5707):251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 3.Xu Y, Mori T, Johnson CH. Cyanobacterial circadian clockwork: Roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. EMBO J. 2003;22(9):2117–2126. doi: 10.1093/emboj/cdg168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Y, et al. Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses. Proc Natl Acad Sci USA. 2004;101(38):13933–13938. doi: 10.1073/pnas.0404768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishiura M, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281(5382):1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 6.Taniguchi Y, Takai N, Katayama M, Kondo T, Oyama T. Three major output pathways from the KaiABC-based oscillator cooperate to generate robust circadian kaiBC expression in cyanobacteria. Proc Natl Acad Sci USA. 2010;107(7):3263–3268. doi: 10.1073/pnas.0909924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito H, et al. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc Natl Acad Sci USA. 2009;106(33):14168–14173. doi: 10.1073/pnas.0902587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hertel S, Brettschneider C, Axmann IM. Revealing a two-loop transcriptional feedback mechanism in the cyanobacterial circadian clock. PLOS Comput Biol. 2013;9(3):e1002966. doi: 10.1371/journal.pcbi.1002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308(5720):414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida T, Murayama Y, Ito H, Kageyama H, Kondo T. Nonparametric entrainment of the in vitro circadian phosphorylation rhythm of cyanobacterial KaiC by temperature cycle. Proc Natl Acad Sci USA. 2009;106(5):1648–1653. doi: 10.1073/pnas.0806741106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rust MJ, Golden SS, O’Shea EK. Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science. 2011;331(6014):220–223. doi: 10.1126/science.1197243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori T, et al. Elucidating the ticking of an in vitro circadian clockwork. PLoS Biol. 2007;5(4):e93. doi: 10.1371/journal.pbio.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi F, Iwase R, Uzumaki T, Ishiura M. Hexamerization by the N-terminal domain and intersubunit phosphorylation by the C-terminal domain of cyanobacterial circadian clock protein KaiC. Biochem Biophys Res Commun. 2006;348(3):864–872. doi: 10.1016/j.bbrc.2006.07.143. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi F, et al. ATP-induced hexameric ring structure of the cyanobacterial circadian clock protein KaiC. Genes Cells. 2003;8(3):287–296. doi: 10.1046/j.1365-2443.2003.00633.x. [DOI] [PubMed] [Google Scholar]
- 15.Mori T, et al. Circadian clock protein KaiC forms ATP-dependent hexameric rings and binds DNA. Proc Natl Acad Sci USA. 2002;99(26):17203–17208. doi: 10.1073/pnas.262578499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishiwaki T, et al. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 2007;26(17):4029–4037. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318(5851):809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kageyama H, et al. Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol Cell. 2006;23(2):161–171. doi: 10.1016/j.molcel.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 19.Kim YI, Dong G, Carruthers CW, Jr, Golden SS, LiWang A. The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2008;105(35):12825–12830. doi: 10.1073/pnas.0800526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egli M, et al. Dephosphorylation of the core clock protein KaiC in the cyanobacterial KaiABC circadian oscillator proceeds via an ATP synthase mechanism. Biochemistry. 2012;51(8):1547–1558. doi: 10.1021/bi201525n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishiwaki T, Kondo T. Circadian autodephosphorylation of cyanobacterial clock protein KaiC occurs via formation of ATP as intermediate. J Biol Chem. 2012;287(22):18030–18035. doi: 10.1074/jbc.M112.350660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brettschneider C, et al. A sequestration feedback determines dynamics and temperature entrainment of the KaiABC circadian clock. Mol Syst Biol. 2010;6:389. doi: 10.1038/msb.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clodong S, et al. Functioning and robustness of a bacterial circadian clock. Mol Syst Biol. 2007;3:90. doi: 10.1038/msb4100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin X, et al. Intermolecular associations determine the dynamics of the circadian KaiABC oscillator. Proc Natl Acad Sci USA. 2010;107(33):14805–14810. doi: 10.1073/pnas.1002119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pattanayek R, et al. Analysis of KaiA-KaiC protein interactions in the cyano-bacterial circadian clock using hybrid structural methods. EMBO J. 2006;25(9):2017–2028. doi: 10.1038/sj.emboj.7601086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pattanayek R, et al. Visualizing a circadian clock protein: Crystal structure of KaiC and functional insights. Mol Cell. 2004;15(3):375–388. doi: 10.1016/j.molcel.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 27.Pattanayek R, et al. Structural model of the circadian clock KaiB-KaiC complex and mechanism for modulation of KaiC phosphorylation. EMBO J. 2008;27(12):1767–1778. doi: 10.1038/emboj.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murakami R, et al. The roles of the dimeric and tetrameric structures of the clock protein KaiB in the generation of circadian oscillations in cyanobacteria. J Biol Chem. 2012;287(35):29506–29515. doi: 10.1074/jbc.M112.349092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwase R, et al. Functionally important substructures of circadian clock protein KaiB in a unique tetramer complex. J Biol Chem. 2005;280(52):43141–43149. doi: 10.1074/jbc.M503360200. [DOI] [PubMed] [Google Scholar]
- 30.Garces RG, Wu N, Gillon W, Pai EF. Anabaena circadian clock proteins KaiA and KaiB reveal a potential common binding site to their partner KaiC. EMBO J. 2004;23(8):1688–1698. doi: 10.1038/sj.emboj.7600190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hitomi K, Oyama T, Han S, Arvai AS, Getzoff ED. Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm. J Biol Chem. 2005;280(19):19127–19135. doi: 10.1074/jbc.M411284200. [DOI] [PubMed] [Google Scholar]
- 32.Villarreal SA, et al. CryoEM and molecular dynamics of the circadian KaiB-KaiC complex indicates that KaiB monomers interact with KaiC and block ATP binding clefts. J Mol Biol. 2013;425(18):3311–3324. doi: 10.1016/j.jmb.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akiyama S, Nohara A, Ito K, Maéda Y. Assembly and disassembly dynamics of the cyanobacterial periodosome. Mol Cell. 2008;29(6):703–716. doi: 10.1016/j.molcel.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 34.Pattanayek R, et al. Combined SAXS/EM based models of the S. elongatus post-translational circadian oscillator and its interactions with the output His-kinase SasA. PLoS ONE. 2011;6(8):e23697. doi: 10.1371/journal.pone.0023697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pattanayek R, Yadagiri KK, Ohi MD, Egli M. Nature of KaiB-KaiC binding in the cyanobacterial circadian oscillator. Cell Cycle. 2013;12(5):810–817. doi: 10.4161/cc.23757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang YG, Tseng R, Kuo NW, LiWang A. Rhythmic ring-ring stacking drives the circadian oscillator clockwise. Proc Natl Acad Sci USA. 2012;109(42):16847–16851. doi: 10.1073/pnas.1211508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5(5):883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 38.Dominguez C, Boelens R, Bonvin AMJJ. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 39.Uetrecht C, Rose RJ, van Duijn E, Lorenzen K, Heck AJR. Ion mobility mass spectrometry of proteins and protein assemblies. Chem Soc Rev. 2010;39(5):1633–1655. doi: 10.1039/b914002f. [DOI] [PubMed] [Google Scholar]
- 40.Heck AJR. Native mass spectrometry: A bridge between interactomics and structural biology. Nat Methods. 2008;5(11):927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 41.Benesch JLP. Collisional activation of protein complexes: Picking up the pieces. J Am Soc Mass Spectrom. 2009;20(3):341–348. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 42.Xu Y, et al. Intramolecular regulation of phosphorylation status of the circadian clock protein KaiC. PLoS ONE. 2009;4(11):e7509. doi: 10.1371/journal.pone.0007509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang YG, Kuo NW, Tseng R, LiWang A. Flexibility of the C-terminal, or CII, ring of KaiC governs the rhythm of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2011;108(35):14431–14436. doi: 10.1073/pnas.1104221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konermann L, Pan J, Liu YH. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem Soc Rev. 2011;40(3):1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 45.Nakajima M, Ito H, Kondo T. In vitro regulation of circadian phosphorylation rhythm of cyanobacterial clock protein KaiC by KaiA and KaiB. FEBS Lett. 2010;584(5):898–902. doi: 10.1016/j.febslet.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 46.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22(9):2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tseng R, et al. KaiA assists the KaiB-KaiC interaction and KaiB/SasA competition in the circadian clock of cyanobacteria. J Mol Biol. 2013 doi: 10.1016/j.jmb.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terauchi K, et al. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2007;104(41):16377–16381. doi: 10.1073/pnas.0706292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong G, et al. Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell. 2010;140(4):529–539. doi: 10.1016/j.cell.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phong C, Markson JS, Wilhoite CM, Rust MJ. Robust and tunable circadian rhythms from differentially sensitive catalytic domains. Proc Natl Acad Sci USA. 2013;110(3):1124–1129. doi: 10.1073/pnas.1212113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murayama Y, et al. Tracking and visualizing the circadian ticking of the cyanobacterial clock protein KaiC in solution. EMBO J. 2011;30(1):68–78. doi: 10.1038/emboj.2010.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tahallah N, Pinkse M, Maier CS, Heck AJR. The effect of the source pressure on the abundance of ions of noncovalent protein assemblies in an electrospray ionization orthogonal time-of-flight instrument. Rapid Commun Mass Spectrom. 2001;15(8):596–601. doi: 10.1002/rcm.275. [DOI] [PubMed] [Google Scholar]
- 53.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung SJ, Robinson CV. Ion mobility-mass spectrometry analysis of large protein complexes. Nat Protoc. 2008;3(7):1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 54.Bush MF, et al. Collision cross sections of proteins and their complexes: A calibration framework and database for gas-phase structural biology. Anal Chem. 2010;82(22):9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- 55.de Vries SJ, et al. HADDOCK versus HADDOCK: New features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69(4):726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.