Significance
Understanding the neuronal functions of diverse RNA pathways will lead to treatments of human neurological diseases that are caused by perturbations in RNA metabolism. Two proteins, Drosha and Pasha/DGCR8, play important roles in neurons, where they are responsible for the biogenesis of many microRNAs. Here, we show that Pasha/DGCR8 also promotes the morphogenesis of neurons in developing fruit flies independently of Drosha and therefore of most microRNA production. These studies therefore illuminate a novel function of Pasha that is medically relevant, because loss of the human ortholog of Pasha may contribute to cognitive and behavioral disorders associated with DiGeorge syndrome.
Keywords: miRNA, RNA metabolism, neurodevelopment
Abstract
Cleavage of microRNAs and mRNAs by Drosha and its cofactor Pasha/DGCR8 is required for animal development, but whether these proteins also have independent roles in development has been unclear. Known phenotypes associated with loss of either one of these two proteins are very similar and consistent with their joint function, even though both cofactors are involved with additional distinct RNA biogenesis pathways. Here, we report clear phenotypic differences between drosha and pasha/dgcr8 null alleles in two postembryonic lineages in the Drosophila brain: elimination of pasha/dgcr8 leads to defects that are not shared by drosha null mutations in the morphology of gamma neurons in the mushroom body lineage, as well as many neurons in the anterodorsal projection neuron lineage. These morphological defects are not detected in neurons that are genetically depleted of two additional microRNA pathway components, dicer-1 and argonaute1, indicating that they are not due to loss of microRNA activity. They are, however, phenocopied by a newly identified recessive gain-of-function allele in drosha that probably interferes with the microRNA independent functions of Pasha/DGCR8. These data therefore identify a general Drosha-independent DGCR8/Pasha pathway that promotes proper morphology in multiple neuronal lineages. Given that reduction of human DGCR8/Pasha may contribute to the cognitive and behavioral characteristics of DiGeorge syndrome patients, disruption of this newly described pathway could underlie human neurological disease.
MicroRNA (miRNA) biogenesis factors are required for nervous system development, likely in part because of their various miRNA-independent functions. In addition to their well-defined roles in miRNA processing (1), the three core miRNA biogenesis factors, Drosha, DiGeorge critical region 8 (DGCR8), and Dicer, process other RNAs as well. For example, the Drosha RNase III enzyme and its binding partner DGCR8 together form the microprocessor complex (2–4) that recognizes and cleaves a variety of RNAs, including mRNAs (5, 6). Drosha and DGCR8 also have microprocessor-independent roles: Drosha is involved in pre-rRNA processing (7), whereas DGCR8 reportedly regulates small nucleolar RNA (snoRNA) biogenesis (8). The Dicer RNase III enzymes likewise have multiple substrates: they cleave canonical miRNA precursors (9, 10) and endogenous siRNAs, Alu RNA elements, and noncanonical miRNAs like mirtrons (11–14). However, the relative contribution of these various RNA classes to nervous system formation remains unclear.
Phenotypic comparisons among miRNA pathway mutants delineate these distinct pathways. Phenotypic similarities of drosha, dgcr8, and dicer mutants indicate the requirements for canonical miRNAs, whereas phenotypic differences imply roles for alternate RNA pathways (15). However, few studies have reported such phenotypic differences. For example, although the brains of dicer mutant mice are smaller and more malformed than dgcr8 mutants (16), no studies have reported phenotypic differences between dgcr8 and drosha mutants that would indicate the biological relevance of their microprocessor-independent functions.
Drosophila neurodevelopment provides an unrivaled system for detailed phenotypic analysis, in part because single genetically manipulated neurons or neuronal lineages can be visualized using the mosaic analysis with a repressible cell marker (MARCM) technique (17). Comparative analysis in the anterodorsal projection neuron (adPN) lineage has stressed the similarities between the axon targeting phenotypes of pasha and dicer-1 (dcr-1) mutations (18). Surprisingly, mutations in the miRNA-specific argonaute-1 (ago1) are reported not to affect adPN neurons (18). Analysis of drosha function in this and other lineages would therefore clarify the involvement of canonical miRNAs in neuronal dcr-1 and pasha phenotypes.
Using unique drosha alleles identified from a genetic screen, we present a detailed phenotypic comparison of drosha, pasha, dicer, and argonaute alleles in both the adPN lineage and another postembryonic lineage, the mushroom body (MB) lineage. We show that canonical miRNAs control self-renewal of MB progenitor cells, whereas a Drosha-independent Pasha pathway is required for morphogenesis in both lineages. This unique function of Pasha is medically relevant, because loss of the human ortholog of Pasha is reported to contribute to cognitive and behavioral disorders associated with DiGeorge syndrome (3, 19, 20).
Results
Eye Color Variegation Screen Yields Two Unique drosha Alleles.
To identify mutations affecting neural let-7-Complex (let-7-C) miRNAs, we performed an F1 clonal screen for ethylmethylsulfonate (EMS)-induced mutations that caused eye color variegation of flies harboring the broad 3′UTR reporter (Fig. S1). We screened approximately one million flies for mutations on the right arm of the second chromosome and identified ∼1,000 variegators. Progeny from 75 of these variegators also displayed variegation, consistent with results that ∼10% of EMS-induced mutations are transmitted through the germ line (21). Balanced stocks were established from those 75 flies and were used for subsequent analysis. The mutagenized chromosomes contained at least one lethal mutation in 54 of these stocks. Thirty of the mutants had no effect on a negative control sensor containing the white 3′UTR, suggesting that our screen had successfully recovered mutations that disrupted let-7-C miRNAs.
Two of our lethal variegator mutations failed to complement each other, as well as droshaR662X, a null allele of drosha (22). The lethality of both mutations was rescued by a drosha genomic fragment, confirming them as drosha alleles. To assess the strength of these alleles relative to each other and to the null allele, we determined their terminal phases. To eliminate possible second site mutation effects, we analyzed their lethal phases in trans to Df(2R) Exel6055. We refer to these trans-heterozygous animals by their allele name. drosha∆,E859K animals died at the larval-to-pupal transition: ∼10% died as late-stage larvae, whereas the remainder attempted unsuccessfully to pupariate. Like the reported null droshaR662X mutants, which display an identical lethal phase (22), these animals had small CNSs and lacked imaginal discs. The droshaR1113X allele was slightly weaker: all animals died as tanned larvae with rudimentary imaginal discs. Thus, the terminal phase and phenotype of drosha∆,E859K was virtually indistinguishable from the null droshaR662X allele, whereas droshaR1113X acted as a hypomorphic allele.
To molecularly characterize the alleles, we sequenced the drosha locus of both and found mutations in the drosha ORF. The droshaR1113X allele carried a C to T base pair change, creating a premature stop codon after the second RNase III domain (Fig. 1A). The drosha∆,E859K allele was more complex. It contained both a G to A change, creating a glutamic acid to lysine missense mutation in a conserved residue of the first RNase III domain, as well as a 441-bp deletion, removing amino acids 561–707. Although this highly conserved region may be partially responsible for Drosha’s interaction with Pasha/DGCR8 and itself (19, 23), coimmunoprecipitations indicated that removal of these amino acids did not affect either interaction (Fig. S2). Thus, our analysis added two more drosha alleles to an existing allelic series that includes two reported presumptive nulls: droshaR662X and droshaQ884X (22, 24).
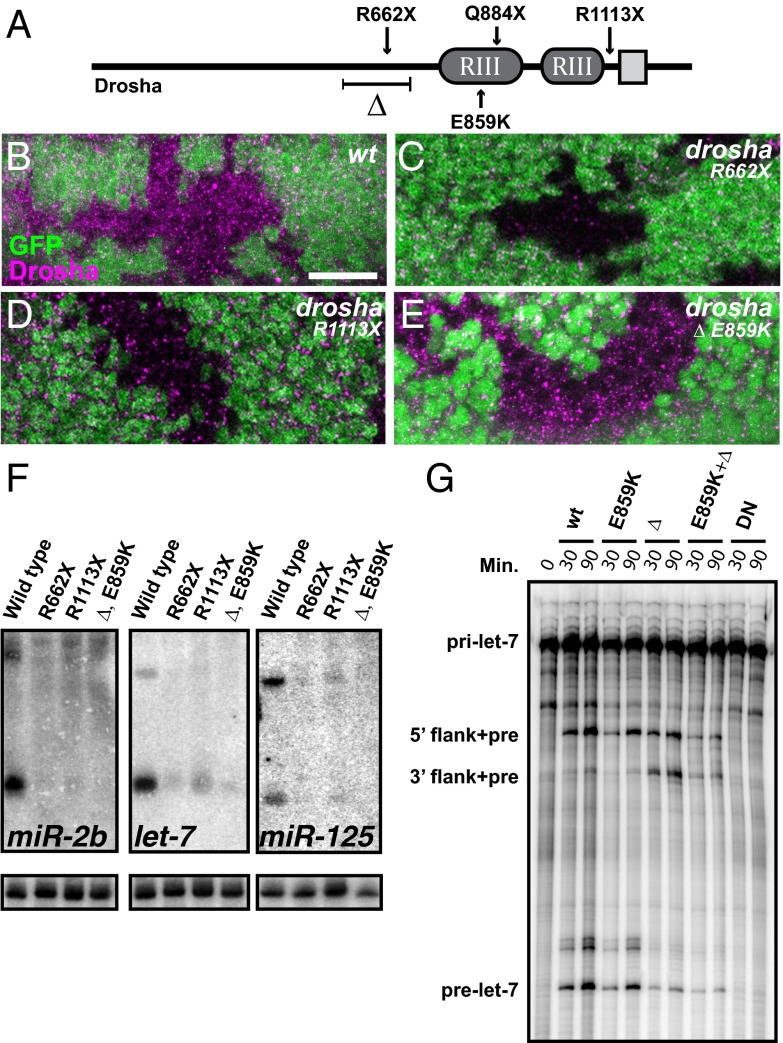
Fig. 1.
Genetic screen yields two unique drosha alleles that are defective in miRNA biogenesis. (A) Schematic of Drosha protein indicates two RNase III domains (RIII) and double-stranded RNA binding domain (gray square), as well as locations of mutations. The drosha∆,E859K allele deletes 147 amino acids (∆) and contains a missense mutation at residue 859. (B–E) Eye discs from third-instar larvae containing WT (B), droshaR662X (C), droshaR1113X (D), and drosha∆,E859K (E) clones stained with anti-Drosha antibodies (magenta). Clones are indicated by the absence of GFP. (Scale bar, 10 μm.) (F) Northern blots of total RNA from third-instar larvae of indicated genotypes probed for miR-2b, let-7, miR-125, and U6 snRNA (loading control). (G) In vitro processing of pri-let-7 by WT, E859K, ∆, E859K+∆, or dominant negative (DN) Drosha proteins. Length of incubation is indicated in minutes.
To further assess the effects of these new drosha alleles on Drosha protein, we analyzed Drosha expression in third-instar larval eye imaginal discs containing clones of homozygous WT or mutant cells. Using a C-terminal antibody (2), Drosha was detected in a punctate pattern throughout WT clones but not in clones of nonsense droshaR662X or droshaR1113X alleles (Fig. 1 B–D). However, Drosha staining was only mildly affected in drosha∆,E859K mutant clones (Fig. 1E). Thus, a truncated and mutated form of the Drosha protein is likely expressed at physiologically relevant levels in drosha∆,E859K mutants, raising the possibility that it may not be a null allele despite its lethal phase.
Unique drosha Alleles Are Defective in RNA Processing.
To evaluate the effect of the droshaR1113X and drosha∆,E859K mutations on RNA processing, we analyzed total RNA from late third-instar larvae harboring drosha alleles in trans to the Df(2R) Exel6055 deficiency for expression of a subset of processed miRNAs, including miR-2b, let-7, and miR-125. We found that mature miRNA levels were substantially reduced in all mutant larvae (Fig. 1F). Slightly more let-7 was detected in droshaR1113X than droshaR662X larvae, consistent with findings that the C-terminally located RNA-binding domain is not absolutely required for pri-miRNA binding (25). Because Drosha also cleaves mRNAs (5, 6), including pasha transcript (22), we also analyzed Pasha levels and found that they were elevated in drosha∆,E859K clones (Fig. S3), indicating that mRNA cleavage was also defective. Consistent with the terminal phase analysis, these data indicated that both drosha∆,E859K and droshaR1113X mutations disrupted RNA processing, although the effect of droshaR1113X on miRNA processing was weaker than either droshaR662X or drosha∆,E859K.
The strong reduction in miRNAs in drosha∆,E859K mutants suggested that Drosha function was crippled in these animals. This defect could be the consequence of the in-frame deletion, the E859K mutation in the RNase III domain, or a combination of both mutations. To distinguish among these possibilities, we analyzed the let-7 miRNA processing ability of WT or mutant FLAG-tagged Drosha proteins purified from transfected BG3-c2 cells. For a negative control, we prepared a dominant negative mutant: a FLAG-tagged version of Drosha that contained point mutations in both RNase III domains. The expression levels of all of the mutants were comparable to that of WT (Fig. S4). The E859K mutant processed pri-let-7 at a kinetic efficiency comparable to WT, the ∆ mutant processed pri-let-7 at a reduced efficiency that was similar to the ∆ E859K double mutant, and the dominant negative (DN) mutant was not capable of producing pre-let-7 at all (Fig. 1G). We noted an accumulation of an intermediary product that contained both pre-let-7 and flanking 3′ sequence specifically in the Drosha∆-containing assays, indicating that absence of prelet-7 was due to defective cleavage at the 3′ side of the stem-loop. These data indicated that amino acids 561–707 of Drosha were required for efficient let-7 processing and suggested that deletion of these amino acids was responsible for the sharp reduction of processed miRNAs in drosha∆,E859K mutant larvae.
Canonical miRNA Pathway Is Required for NB Maintenance.
To characterize the role of drosha in neurodevelopment, we performed MARCM analysis of multiple drosha alleles, including the null nonsense alleles, in the well-characterized MB lineages. Four MB progenitors, or neuroblasts (NBs), in each hemisphere of the fly CNS generate these lineages by dividing continuously from embryogenesis through pharate adulthood, producing three subtypes of MB neurons in an invariant order (ɣ → α′/β′ → α/β) (26). These neurons populate the ɣ, α′/β′, and α/β lobes of the adult MB, which are distinguishable based on their morphology and Fasciclin II (FasII) expression (Fig. 2A). Two miRNAs, let-7 and miR-125, participate in MB cell fate determination (27), and analysis of drosha phenotypes would indicate whether additional miRNAs might also be involved in MB development. The droshaR662X, droshaR1113X, and drosha∆,E859K alleles all displayed similar MB phenotypes. These mutant MBs contained significantly fewer neurons in adult flies, and those present were predominantly early-born ɣ neurons (Fig. 2 B–D), which stain weakly for FasII. They also contained a handful of α′/β′ neurons but no α/β neurons, which were distinguishable based on the absence and presence of FasII, respectively. Axonal extensions of the rare α′/β′ neurons were frequently mistargeted, located inappropriately near α/β lobe termini (arrowheads in Fig. 2D). Collectively, these data indicated that drosha is required for the production of later born α′/β′ and α/β neurons and suggested that Drosha processing of RNAs in addition to let-7 and miR-125 promoted NB maintenance.
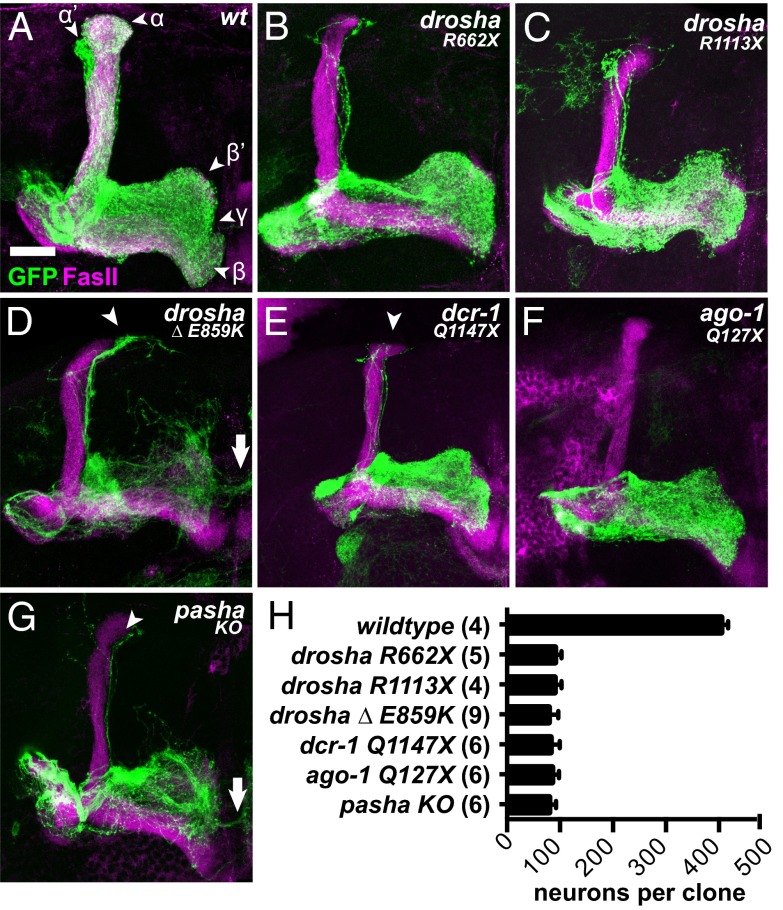
Fig. 2.
Core microRNA pathway components are required for NB maintenance in the mushroom body lineages. OK107-Gal4, UAS-mCD8::GFP-labeled WT (A), droshaR662X (B), droshaR1113X (C), drosha∆,E859K (D), dcr-1Q1147X (E), ago1Q127X (F), and pashaKO (G) adult MB neuroblast clones generated in newly hatched larvae and stained with anti-FasII antibodies. Arrowheads indicate α′/β′ neuron mistargeting and arrows indicate ɣ neuron overextension. (H) Average number of cells in adult MB clones of indicated genotypes. Differences between WT and all mutants are statistically significant (P < 0.0001). (Scale bar for A–G, 25 μm.)
To determine whether canonical miRNAs were involved in MB NB maintenance, we analyzed loss-of-function alleles in three additional members of the miRNA pathway, including pashaKO, dcr-1Q1147X, and ago1Q127X. Like droshaR662X mutants, pashaKO null mutants die as malformed pupae with severely reduced CNSs (22, 28), although this neural phenotype has not been investigated at cellular resolution. dcr-1Q1147X and ago1Q127X nonsense alleles are presumptive nulls that are embryonic lethal (24, 29). We found that homozygous mutant dcr-1Q1147X, ago1Q127X, and pashaKO adult MB clones displayed generally similar phenotypes to the drosha clones, including a reduction in cell number and absence of α′/β′ and α/β neurons (Fig. 2 E–G). To quantify this reduction, we counted the total number of cell bodies in adult WT and mutant MB clones. WT MB clones contained ∼404 neurons, whereas mutant MB clones contained ∼79–90 neurons (Fig. 2H). To test whether the reduction in neuron number in mutant clones was due to premature absence of MB NBs, we analyzed NB cell number in late larval MB lineages (Fig. S5 E–G). In all WT MB clones (n = 10), we detected one NB. In sharp contrast, no NBs were detected in dcr-1Q1147X (n = 6), ago1Q127X (n = 5), pashaKO (n = 4), droshaR662X (n = 5), and drosha∆,E859K (n = 5) mutant larval MB lineages. Thus, four core miRNA pathway components are required for maintenance of third-instar larval MB NBs, indicating that loss of canonical miRNAs leads to progenitor cell cycle exit in early third-instar larvae when the transition from ɣ to α′/β′ neuron production occurred.
pasha Null Phenotypes Are More Severe Than drosha Null Phenotypes in Both MBs and adPNs.
In addition to the phenotypic similarities among mutations in these four genes, we also noted some important differences. In particular, pashaKO mutant MBs (n = 6) displayed ɣ neuron disorganization and mistargeting, as evidenced by inappropriate extension across the midline (arrow in Fig. 2G), whereas droshaR662X (n = 7), dcr-1Q1147X (n = 17), and ago1Q127X (n = 5) MBs did not display either of these morphological defects (Fig. 2 B, E, and F). These data indicated a role for Pasha in ɣ neuron morphogenesis that was not shared with other miRNA pathway members. The reported similarity between drosha and pasha null phenotypes in the female germ line (30) further suggested that this miRNA-independent function of Pasha might be specific to neurons.
To determine whether these phenotypic differences between pasha and other miRNA pathway mutants were displayed in additional neuronal subtypes, we used our expanded allelic battery to generate Acj6-Gal4–labeled NB clones in the adPN lineage. The adPN NB exits the cell cycle at the larval-to-pupal transition, having generated ∼40 morphologically diverse neuronal subtypes whose dendrites target specific adult glomeruli (31). The Acj6-Gal4 driver labels all these adPN types, whereas a second driver, GH146-Gal4, labels just a subset of ∼25 embryonically and early larval born adPNs. Phenotypic differences have been previously described among pasha, dcr-1, and ago1 mutant GH146-Gal4–labeled adPN clones: defects in adPN targeting and dendritic density were highly penetrant in clones of the lethal, protein-null pashaLL03660 allele, moderately penetrant in clones of the lethal, transcript-null dcr-1LL06357 allele, and absent in clones of the hypomorphic ago1K08121 allele (18). These phenotypes were interpreted as evidence for miRNA function in neurons, because the differences could have been due to the relative strengths of the alleles used: none were molecular nulls, and neither pashaLL03660 nor ago1K08121 acted like genetic nulls in subsequent analyses (24, 30). This interpretation of neuronal miRNA function would be supported if drosha null alleles displayed adPN morphogenesis defects similar to pasha, dcr-1, and ago1 null alleles.
To stringently evaluate the role of canonical miRNAs in this lineage, we compared the adPN phenotypes of the molecular-null pashaKO allele with the nonsense droshaR662X, dcr-1Q1147X, and ago1Q127X alleles. As a prelude to morphological analysis, we first assessed whether these mutations affected adPN neurogenesis by analyzing adPN cell number in mutant clones. We found varying degrees of cell number reduction (Fig. 3A). The dcr-1 and ago1 alleles had the strongest effect: WT clones contained 59.3 ± 2.3 (n = 4) cells, whereas dcr-1Q1147X and ago1Q127X clones contained 21.0 ± 1.2 (n = 3) and 34.3 ± 0.9 (n = 6) cells, respectively. pashaKO and droshaR662X clones displayed more modest effects, containing 45.8 ± 1.8 (n = 6) and 45.0 ± 0.8 (n = 5) cells, respectively. Analogously to the MB lineage, these reductions likely reflected premature adPN NB cell cycle exit during mid- to late larval stages, because cell number defects were not found in GH146-labeled clones of dcr-1LL06357, pashaLL03660, and ago1K08121 (18) or dcr-1Q1147X, pashaKO, and ago1Q127X (Fig. S6A). These data indicated a role for canonical miRNAs in adPN neurogenesis, but the difference between dcr-1 and drosha/pasha phenotypes also suggested possible involvement of miRNAs like mirtrons that are dcr-1 dependent but drosha/pasha independent (13, 14).
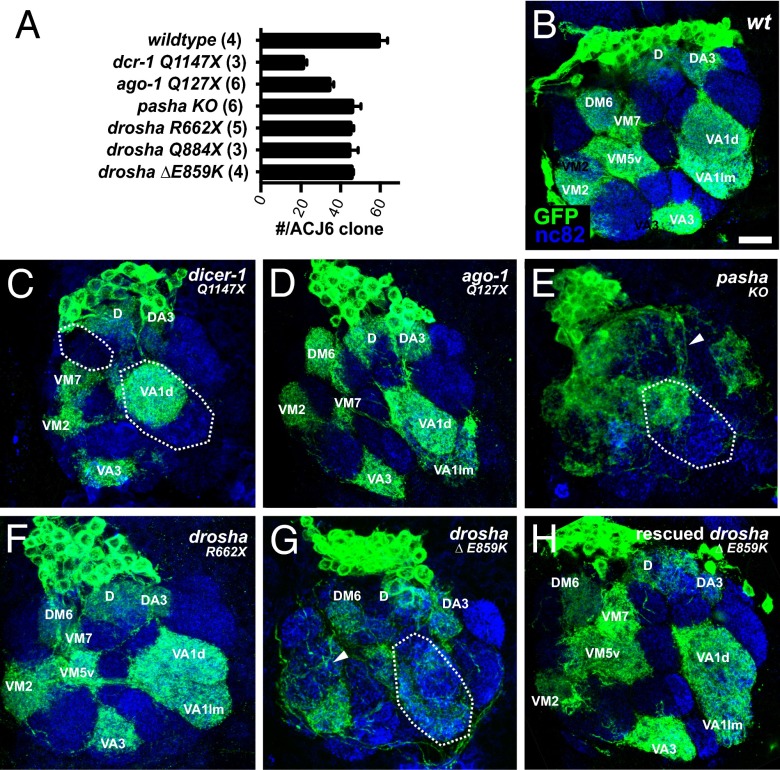
Fig. 3.
Distinct roles for miRNA processing components in the adPN lineage. (A) Average number of cells in adult Acj6-labeled adPN clones of indicated genotypes. Numbers of clones analyzed are indicated in parentheses. Acj6-Gal4, UAS-mCD8::GFP-labeled adult WT (B), dcr-1Q1147X (C), ago1Q127X (D), pashaKO (E), droshaR662X (F), drosha∆,E859K (G), and rescued drosha∆,E859K (H) adPN NB clones generated in newly hatched larvae and stained with anti-GFP (green) and anti-nc82 (magenta) antibodies. Glomeruli names are labeled in white. Areas exhibiting targeting defects are circled and arrowheads point to dendritic mistargeting. (Scale bar for B–H, 10 μm.)
Because of this absence of later-born neurons in mutant clones, we focused our analysis on the morphology of early born adPNs that predominantly innervate anteriorly located glomeruli. Like the previous analysis with GH146-Gal4 (18), Acj6-Gal4–labeled pashaKO clones (Fig. 3E) displayed more severe defects in adPN neuron morphology than either dcr-1Q1147X (Fig. 3C) or ago1Q127X clones (Fig. 3D), which appeared very similar to WT clones (Fig. 3B). These defects included dendritic mistargeting, as indicated by spillover of dendritic branches into incorrect glomerular classes (arrowhead in Fig. 3E). In stark contrast, droshaR662X null allele adPN clones appeared grossly normal (Fig. 3F). Confirming this result, adPN clones of a second reported drosha null allele, droshaQ884X, which contains a nonsense mutation in the first RNase III (24), as well as droshaR1113X, appeared grossly normal (Fig. S6 B and C). Thus, neither the ribonuclease activity of Drosha nor the miRISC-silencing activity of Ago1 was required for proper morphogenesis of most of the adPN neurons, strongly suggesting a miRNA-independent function for Pasha in neuronal morphogenesis.
droshaΔ,E859K Is a Recessive Gain-of-Function Allele That Phenocopies pashaKO in Multiple Neuronal Lineages.
The absence of morphological defects in drosha null adPN clones indicated that Drosha was not required for proper dendritic targeting. However, adPN clones of drosha∆,E859K, the allele in which we had detected Drosha protein, displayed severe dendritic density and mistargeting defects (Fig. 3G). Unlike droshaR662X, this allele also affected axonal morphology, because the ɣ lobes of drosha∆,E859K mutant MB clones (n = 14) were disorganized and extended across the midline (Fig. 2D). These difference between drosha null and drosha∆,E859K indicated that drosha∆,E859K displayed gain-of-function phenotypes.
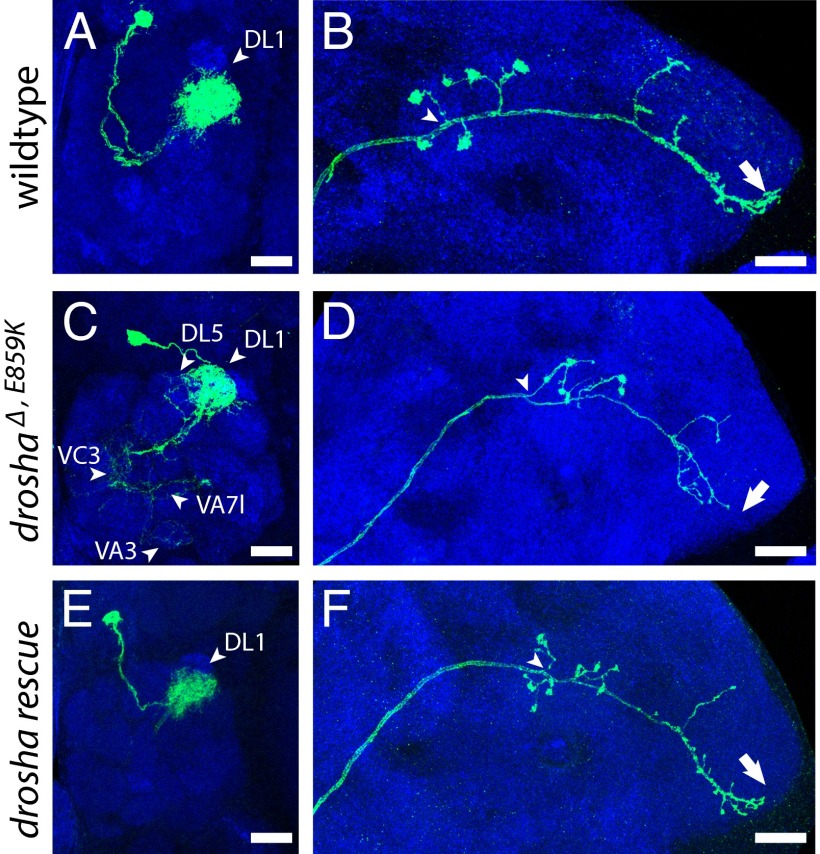
These gain-of-function drosha∆,E859K phenotypes, however, appeared very similar to those displayed by pashaKO. To carefully compare and quantify this similarity, we examined single-cell clones in a specific adPN neuron: the DL1 neuron. Although WT DL1 single cell clones target a posterior, dorsolateral glomerulus with dendritic branches (Fig. 4A), all drosha∆,E859K DL1 single neurons innervated this DL1 glomerulus more sparsely and inappropriately targeted additional glomeruli (n = 7; arrowheads in Fig. 4C). In addition, axons of WT DL1 single cell clones terminated their main branches at the lateral edge of the lateral horn (LH) (Fig. 4B), whereas all drosha∆,E859K mutant DL1 axons failed to reach the LH lateral edge (n = 7; arrow in Fig. 4D). These drosha∆,E859K DL1 phenotypes precisely matched the severity and penetrance of previously reported pasha mutant phenotypes (18). In contrast, no droshaR662X DL1 single neuron clones and only one of eight dcr-1Q1147X DL1 mutant cells displayed mistargeting (Fig. S7). The difference between drosha∆,E859K and droshaR662X phenotypes suggested that they were not due to loss of miRNAs, because both alleles disrupted miRNA production, whereas the similarity to pashaKO phenotypes suggested that drosha∆,E859K interfered with the microprocessor-independent function of Pasha.
Fig. 4.
DL1 drosha∆,E859K mutant neurons display phenotypes that are rescued by WT drosha. Acj6-Gal4, UAS-mCD8::GFP-labeled WT (A and B), drosha∆,E859K (C and D), and rescued drosha∆,E859K (E and F) adult DL1 clones generated in newly hatched larvae and stained with anti-GFP (green) and anti-nc82 (blue) antibodies. (Scale bar: A, C, and E, 10 μm; B, D, and F, 20 μm.)
To test whether these gain-of-function drosha∆,E859K MB and adPN phenotypes were dominant, we analyzed OK107- and GH146-Gal4–labeled cells in adult drosha∆,E859K heterozygotes. We did not observe morphological defects in the MBs or adPNs of these animals (Fig. S8), suggesting that the drosha∆,E859K gain-of-function phenotypes were suppressed by WT Drosha. To test this, we labeled homozygous drosha∆,E859K adPN and MB NBs as well as DL1 neurons in flies harboring an extra, transgenic copy of drosha and found that they did not display the morphological defects associated with drosha∆,E859K (Figs. 3H and 4 E and F). Thus, MB and adPN drosha∆,E859K phenotypes were detectable only in the absence of WT Drosha, indicating that these gain-of-function phenotypes were recessive.
Overexpression of DroshaΔ,E859K Leads to Neuronal Overgrowth.
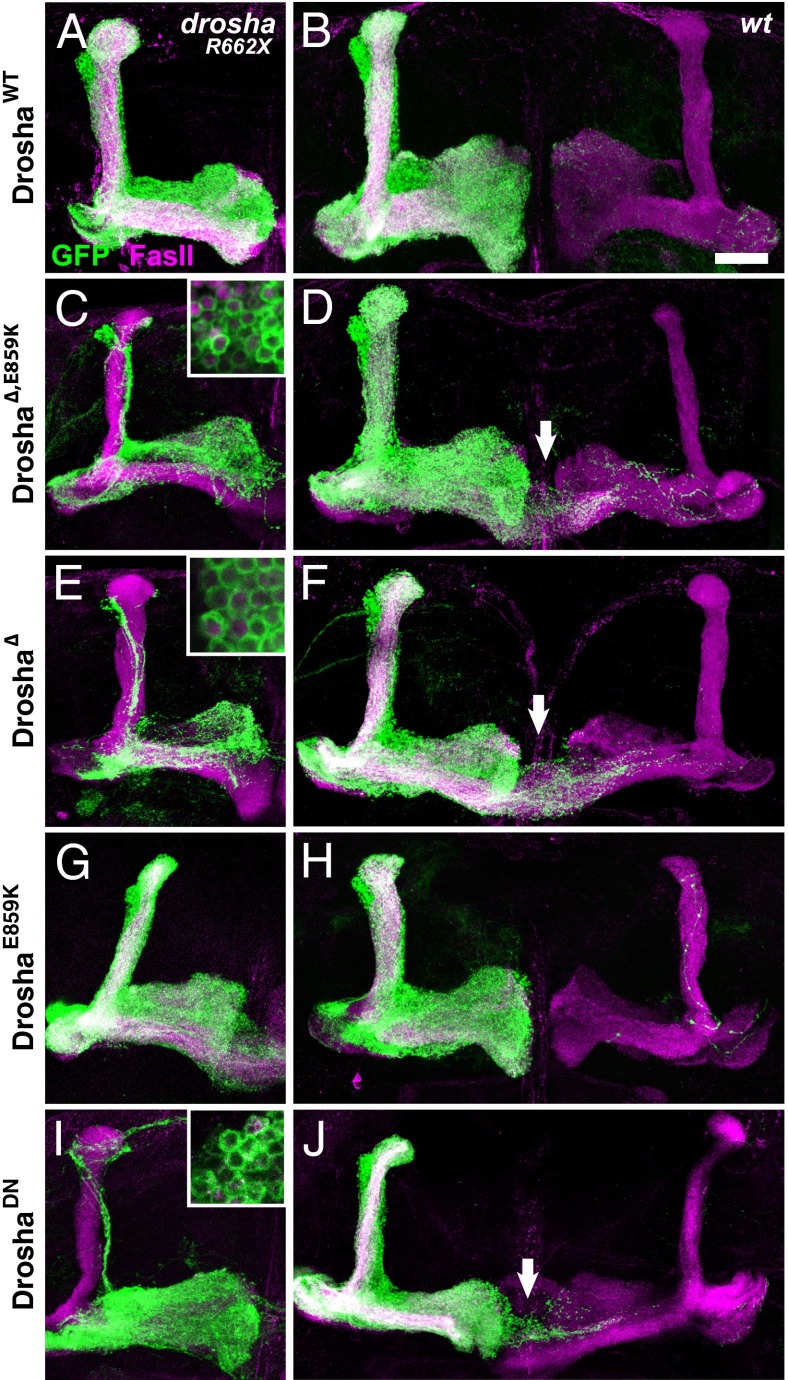
The finding that drosha∆,E859K phenotypes were suppressed by a wild type copy of drosha suggested that elevated levels of Drosha∆,E859K might lead to neuronal defects even in WT cells. To test this possibility, we used the strong OK107-Gal4 MB driver to direct expression of UAS-transgenes encoding WT Drosha or mutant forms of Drosha that we had analyzed in our in vitro processing assay (Fig. 1G). To verify these transgenes, we tested whether they could rescue MB phenotypes associated with the droshaR662X null allele. As expected, transgenes encoding DroshaWT and DroshaE859K substantially rescued the drosha R662X MB defects (Fig. 5 A and G), whereas transgenes encoding Drosha∆, Drosha∆+E859K, and DroshaDN did not, despite comparable transgene expression as detected by Flag staining (Fig. 5 C, E, and I). The rescue by DroshaE859K was not complete, however, because α/β neuron overextension was detected in both cases (Fig. 5G). We then tested whether expression of any of these transgenes elicited neuronal defects in a WT background (Fig. 5 B, D, F, H, and J). Forced expression of Drosha∆+E859K always resulted in overgrowth of α/β neurons (10/10 clones) and forced Drosha∆ or DroshaDN frequently did as well (17/24 or 10/17 clones, respectively). In contrast, most MB clones expressing either DroshaWT or DroshaE859K were normal (19/21 or 20/23, respectively). These data indicated that overexpression of Drosha proteins missing amino acids 561–707 led to penetrant defects in axonal targeting, supporting the view that the gain-of-function drosha∆,E859K phenotypes are dependent on the relative concentration of mutant Drosha protein.
Fig. 5.
Forced expression of Drosha∆,E859K leads to neuronal overgrowth. DroshaR662X (A, C, E, G, and I) or WT (B, D, F, H, and J) OK107-Gal4, UAS-mCD8::GFP-labeled adult MB clones expressing UAS-DroshaWT (A and B), UAS-Drosha∆+E859K (C and D) UAS-Drosha∆ (E and F), UAS-DroshaE859K (G and H), or UAS-DroshaDN (I and J) and stained with anti-FasII antibody (magenta) and anti-Flag antibody (magenta; Insets in C, E, and I). Arrows point to mistargeted neurons. (Scale bar, 25 μm.)
Discussion
We delineated the function of two RNA pathways in neuronal lineages. The first, the canonical miRNA pathway, maintains neural progenitors. The relevant miRNAs may include miR-124, a neural-specific miRNA that contributes to neural progenitor proliferation (32). The second, a unique Pasha-dependent and Drosha- and Ago1-independent pathway, promotes dendritic and axonal targeting independent of canonical miRNAs. The RNA effectors of this pathway are not known but may include snoRNAs, because human Pasha is reported to regulate snoRNA biogenesis by an unknown mechanism (8, 16). We cannot exclude the possibility that this pathway involves Ago1-independent noncanonical miRNAs, because some dcr-1 adPN NB and DL1 clones display mistargeting. However, the low penetrance of mistargeting defects in dcr-1 adPNs and the absence of phenotypes in dcr-1 MB ɣ neurons indicates that any such contribution to the morphogenesis function of Pasha is minor. Because loss of the human ortholog of Pasha/DGCR8 is reported to contribute to DiGeorge syndrome, this unique Pasha pathway may be directly relevant to the cognitive and behavioral disorders associated with this syndrome. Our results strongly suggest that the dendritic defects associated with dgcr8 heterozygosity in mice involve this unique, canonical miRNA-independent function of Pasha/DGCR8 (33).
The gain-of-function adPN phenotypes of drosha∆,E859K illuminate this Pasha pathway, likely because the mutant form of Drosha incapacitates its binding partner and blocks not only miRNA processing but Pasha’s other functions as well. Drosha∆ does not efficiently cleave both strands of pri-miRNA, leading to the accumulation of partially processed intermediates. These intermediates likely sequester Pasha, because current biochemical and in vivo imaging data suggest that pri-miRNA cleavage causes a conformational change in the microprocessor that leads to rapid release of Drosha and a slower release of Pasha (25). Trapped Pasha would therefore not be available to bind either to other pri-miRNAs or additional RNA targets. The morphological defects in ɣ MB and adPN neurons are likely due to aberrant metabolism of these other RNAs, because drosha null ɣ MB and adPN neurons that lack canonical miRNAs are mostly normal. Identifying additional phenotypic differences between drosha∆,E859K and drosha null alleles will reveal other contexts where Pasha’s miRNA-independent roles are relevant.
The gain-of-function drosha∆,E859K phenotypes are also recessive, a relatively rare occurrence that provides genetic support that the microprocessor complex contains two independent Drosha subunits. Biochemical purification and reconstitution experiments indicate that the large microprocessor complex, which is ∼650 kDa in humans and ∼500 kDa in flies, contains at least one ∼70-kDa Pasha/DGCR8 subunit and one ∼150-kDa Drosha subunit, although it has been unclear whether the complex contains additional Pasha/Drosha subunits and/or other auxiliary proteins (2–4, 19). Recessive gain-of-function mutations have previously been identified in genes whose products contribute several subunits to a protein complex, such as an ion channel (34). In these cases, the effects of the mutant form are suppressed by redundant WT versions in heterozygotes but lead to phenotypes in homozygotes that are stronger than complete elimination of the protein. Isolation of recessive gain-of-function alleles in drosha suggests that the fly microprocessor complex functions analogously and contains two functionally independent Drosha subunits, either one of which is sufficient for its activity.
Materials and Methods
All flies were cultured on standard cornmeal medium at 25 °C. Tissues were stained with primary antibodies, including rabbit anti-Drosha (1:1,000; from G. Hannon, Cold Spring Harbor, NY), rabbit anti-Pasha (1:200; from G. Hannon), chicken anti-GFP (1:500; Rockland Immunochemicals), mouse anti-nc82 [1:5; Developmental Studies Hybridoma Bank (DSHB)], mouse anti-Fas II (1:5; DSHB), mouse anti-Dac (1:100; DSHB), and guinea pig anti-Dpn (1:1,000; from J. Skeath, Washington University, St. Louis), and secondary AlexaFluor 488–, 568–, or 633–conjugated secondary antibodies (1:1,000; Life Technologies). Images were collected on a Leica SP5 confocal microscope (Light Microscopy Imaging Center, Indiana University). Statistical analysis was performed using GraphPad Prism software, and P values were calculated using a two-tailed unpaired t test. Values are presented as mean ± SEM. Details of fly strains, clone induction, animal staging, mutagenesis screen, transgenes, plasmids, and molecular techniques are included in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank R. Carthew, P. Geyer, G. Hannon, E. Lai, T. Lee, J. Skeath, the Drosophila Genome Resource Center, and the Developmental Studies Hybridoma Bank for reagents and K. Cook and A. Zelhof for helpful comments on the manuscript. This work was supported by National Institute of Mental Health Award R01MH087511.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318445111/-/DCSupplemental.
References
- 1.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 2.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 3.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 5.Chong MM, et al. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24(17):1951–1960. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han J, et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136(1):75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu H, Xu H, Miraglia LJ, Crooke ST. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J Biol Chem. 2000;275(47):36957–36965. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- 8.Macias S, et al. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat Struct Mol Biol. 2012;19(8):760–766. doi: 10.1038/nsmb.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 10.Hutvágner G, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 11.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22(20):2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaneko H, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471(7338):325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130(1):89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448(7149):83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43(6):892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babiarz JE, et al. A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus Dicer1 knockouts and small RNA sequencing. RNA. 2011;17(8):1489–1501. doi: 10.1261/rna.2442211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22(3):451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 18.Berdnik D, Fan AP, Potter CJ, Luo L. MicroRNA processing pathway regulates olfactory neuron morphogenesis. Curr Biol. 2008;18(22):1754–1759. doi: 10.1016/j.cub.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14(23):2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Ashburner M, Golic KG, Hawley RS. Drosophila: A Laboratory Handbook. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005. p. xxviii. [Google Scholar]
- 22.Smibert P, et al. A Drosophila genetic screen yields allelic series of core microRNA biogenesis factors and reveals post-developmental roles for microRNAs. RNA. 2011;17(11):1997–2010. doi: 10.1261/rna.2983511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y, Han J, Yeom KH, Jin H, Kim VN. Drosha in primary microRNA processing. Cold Spring Harb Symp Quant Biol. 2006;71:51–57. doi: 10.1101/sqb.2006.71.041. [DOI] [PubMed] [Google Scholar]
- 24.Pressman S, Reinke CA, Wang X, Carthew RW. A Systematic Genetic Screen to Dissect the MicroRNA Pathway in Drosophila. G3 (Bethesda) 2012;2(4):437–448. doi: 10.1534/g3.112.002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bellemer C, et al. Microprocessor dynamics and interactions at endogenous imprinted C19MC microRNA genes. J Cell Sci. 2012;125(Pt 11):2709–2720. doi: 10.1242/jcs.100354. [DOI] [PubMed] [Google Scholar]
- 26.Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: Sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126(18):4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- 27.Wu YC, Chen CH, Mercer A, Sokol NS. Let-7-complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Dev Cell. 2012;23(1):202–209. doi: 10.1016/j.devcel.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin R, et al. A Drosophila pasha mutant distinguishes the canonical microRNA and mirtron pathways. Mol Cell Biol. 2009;29(3):861–870. doi: 10.1128/MCB.01524-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117(1):69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 30.Azzam G, Smibert P, Lai EC, Liu JL. Drosophila Argonaute 1 and its miRNA biogenesis partners are required for oocyte formation and germline cell division. Dev Biol. 2012;365(2):384–394. doi: 10.1016/j.ydbio.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu HH, et al. A complete developmental sequence of a Drosophila neuronal lineage as revealed by twin-spot MARCM. PLoS Biol. 2010;8(8):E1000461. doi: 10.1371/journal.pbio.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weng R, Cohen SM. Drosophila miR-124 regulates neuroblast proliferation through its target anachronism. Development. 2012;139(8):1427–1434. doi: 10.1242/dev.075143. [DOI] [PubMed] [Google Scholar]
- 33.Xu B, Hsu PK, Stark KL, Karayiorgou M, Gogos JA. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell. 2013;152(1-2):262–275. doi: 10.1016/j.cell.2012.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lester HA, Karschin A. Gain of function mutants: Ion channels and G protein-coupled receptors. Annu Rev Neurosci. 2000;23:89–125. doi: 10.1146/annurev.neuro.23.1.89. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.