Significance
Many chemicals are valuable because they bind to other molecules. Chemical theory cannot directly design “binders.” However, we might recreate in the laboratory the Darwinian processes that nature uses to create binders. This in vitro evolution uses nucleic acids as binders, libraries of DNA/RNA to survive a selection challenge before they can have “children” (systematic evolution of ligands by exponential enrichment, SELEX). Unfortunately, with only four nucleotides, natural DNA/RNA often yields only poor binders, perhaps because they are built from only four building blocks. Synthetic biology has increased the number of DNA/RNA building blocks, with tools to sequence, PCR amplify, and clone artificially expanded genetic information systems (AEGISs). We report here the first example of a SELEX using AEGIS, producing a molecule that binds to cancer cells.
Keywords: synthetic biology, next generation sequencing, nucleic acids, cancer cell SELEX
Abstract
Artificially expanded genetic information systems (AEGISs) are unnatural forms of DNA that increase the number of independently replicating nucleotide building blocks. To do this, AEGIS pairs are joined by different arrangements of hydrogen bond donor and acceptor groups, all while retaining their Watson–Crick geometries. We report here a unique case where AEGIS DNA has been used to execute a systematic evolution of ligands by exponential enrichment (SELEX) experiment. This AEGIS–SELEX was designed to create AEGIS oligonucleotides that bind to a line of breast cancer cells. AEGIS–SELEX delivered an AEGIS aptamer (ZAP-2012) built from six different kinds of nucleotides (the standard G, A, C, and T, and the AEGIS nonstandard P and Z nucleotides, the last having a nitro functionality not found in standard DNA). ZAP-2012 has a dissociation constant of 30 nM against these cells. The affinity is diminished or lost when Z or P (or both) is replaced by standard nucleotides and compares well with affinities of standard GACT aptamers selected against cell lines using standard SELEX. The success of AEGIS–SELEX relies on various innovations, including (i) the ability to synthesize GACTZP libraries, (ii) polymerases that PCR amplify GACTZP DNA with little loss of the AEGIS nonstandard nucleotides, and (iii) technologies to deep sequence GACTZP DNA survivors. These results take the next step toward expanding the power and utility of SELEX and offer an AEGIS–SELEX that could possibly generate receptors, ligands, and catalysts having sequence diversities nearer to that displayed by proteins.
A quarter century ago, various laboratories independently sought to apply in vitro selection, commonly called “SELEX” (systematic evolution of ligands by exponential enrichment), to create DNA and RNA (collectively “xNA”) “aptamers”, xNA molecules that bind specifically to many different types of targets (1–3). SELEX follows a simple recipe that includes (i) making a library of nucleic acid molecules; (ii) placing the library in contact with the target to separate molecules in the library that bind from those that do not; (iii) amplifying “survivors” by PCR; and (iv) after a sufficient number of cycles of selection, recovering individual aptamers that are sequenced, resynthesized, and characterized as molecular entities having unambiguous molecular structures.
SELEX has had substantial success, with xNA aptamers now known for many targets, including small molecules, carbohydrates, peptides, and various kinds of cancer cells (4–7). Nevertheless, considerable effort has been devoted to improving SELEX (8–10). Improvements were initially sought by adding functionalized side chains to one or more of the standard nucleotides used in the SELEX library (11–13). More recently, Perrin and coworkers have had notable success obtaining catalytic DNAzymes using heavily functionalized DNA libraries (14). Following a slightly different rationale, SomaLogic has obtained slow off-rate modified aptamers (SOMAmers) by appending hydrophobic side chains (e.g., benzyl, naphthyl, tryptamino, and isobutyl) to nucleobases (15). In one set of SOMAmers targeted against ∼800 different human proteins, affinities in the 0.1 pM–1 μM range are reported (16).
However, another approach to increase the power of aptamers is to increase the number of independently replicating nucleotides in the xNA library. For example, Hirao and coworkers recently added a fifth nucleotide to aptamers against IFN-gamma and VEGF-165 (17). Adding nucleotides to xNA libraries offers the possibility of obtaining higher information density in aptamer sequences, a richer variety of folds, more control over folding interactions, and xNA libraries having sequences and functional diversity more like proteins.
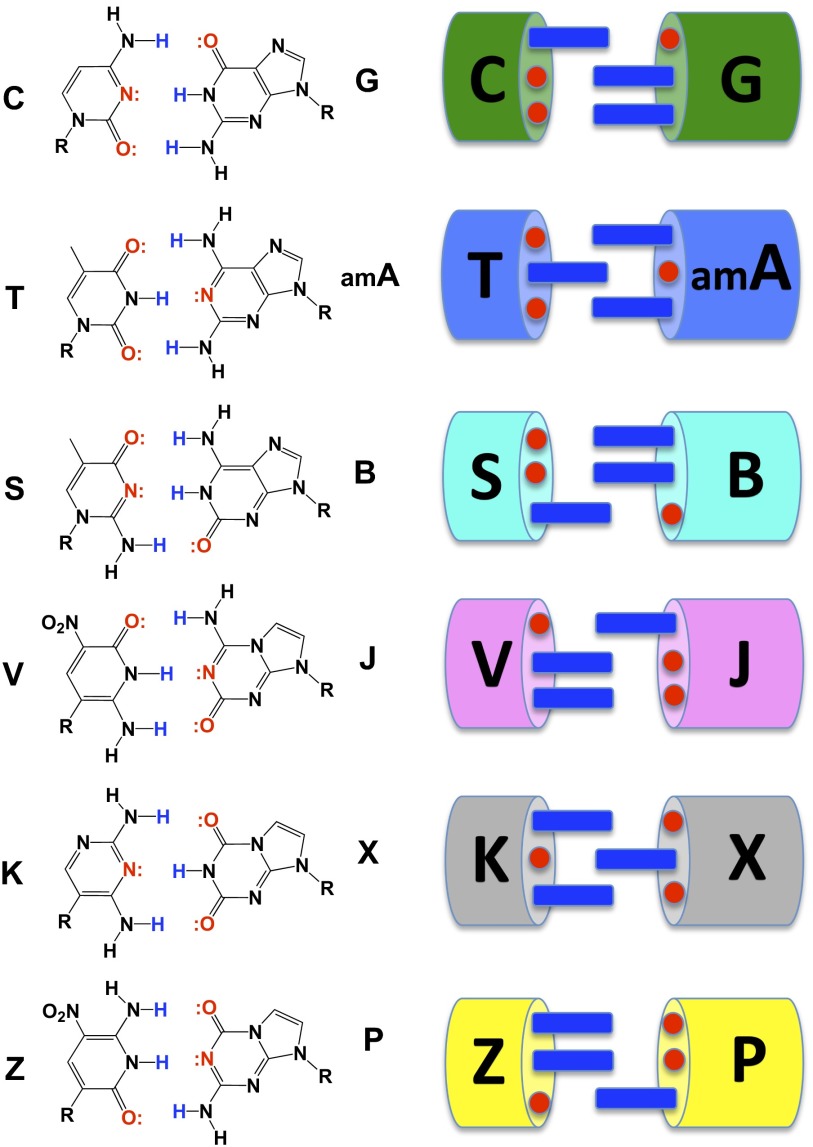
Another approach starts with the recognition that the two nucleobase pairs found in natural xNA [G:C and A:T(U)] do not exploit all possible hydrogen bonding patterns (Fig. 1). Therefore, rearranging hydrogen bond donor and acceptor groups on the nucleobases can increase the number of independently replicable nucleosides in xNA from 4 to 12 (forming six nucleobase pairs) (18). In this artificially expanded genetic information system (AEGIS), the 12 different nucleotide “letters” pair via six distinguishable hydrogen bonding patterns to give a system that can pair, be copied, and evolve like natural DNA, but with higher information density and more functional groups (19). This laid the grounds for the work reported here, an “AEGIS–SELEX.”
Fig. 1.
(Left) Watson–Crick pairing follows two rules of complementarity: (i) size complementarity (large purines pair with small pyrimidines) and (ii) hydrogen bonding complementarity [hydrogen bond donors (blue) pair with hydrogen bond acceptors (red)]. Rearranging hydrogen bond donors and acceptors on the bases gives an artificially expanded genetic information system (AEGIS). An xNA biopolymer having functionalized AEGIS components may allow SELEX to yield protein-like aptamers better than the standard DNA and RNA biopolymers. The example of AEGIS–SELEX reported here adds the Z:P pair, shown at the bottom of the molecular structures. (Right) The images show the general idea behind AEGIS in cartoon form.
We report here a unique example of AEGIS–SELEX. This AEGIS–SELEX exploits two additional nucleotides [2-amino-8-(1′-β-d-2-deoxyribofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)one, trivially called P, and 6-amino-5-nitro-3-(1′-β-d-2′-deoxyribofuranosyl)-2(1H)-pyridone, trivially called Z] (20–23). GACTZP AEGIS–SELEX is based on the following features of a molecular biology for “second generation” AEGIS that have been developed over the past few years in these laboratories:
-
(i)
Methods to synthesize libraries containing 1012–1014 different AEGIS oligonucleotides (20).
- (ii)
-
(iii)
Procedures to sequence AEGIS DNA molecules that emerge after multiple rounds of selection (22).
In this AEGIS–SELEX effort, we chose an especially challenging target, a line of breast cancer cells (MDA-MB-231) (25). In addition to exploiting the latest whole-cell SELEX technology that has been developed with standard GACT SELEX, this target has considerable medical interest (26).
Results and Discussion
GACTZP Aptamer Selection Scheme and AEGIS–SELEX Progression.
AEGIS–SELEX began with the solid-phase synthesis of a GACTZP DNA library having two primer binding sequences (16 nt each) flanking a 20-nt random region (N1N2…N20). Each of the 20 randomized sites was synthesized to have all six (GACTZP) nucleotide phosphoramidites in equal amounts (SI Appendix, Table S1). In subsequent analysis, the library was digested and the nucleotide fragments were quantitated by HPLC to show that Z and P were present in the random region. The ratio of all six nucleotides was T/G/A/C/Z/P ∼ 1.5/1.2/1.0/1.0/1.0/0.5. We did not think it necessary to resynthesize the library to get ratios closer to a 1:1:1:1:1:1 mixture, although this might be done in future work using unequal amounts of phosphoramidites based on these data.
A sample (5 nmol) of the GACTZP library was then subjected to sequential binding and elution from the line of breast cancer cells, MDA-MB-231. The pool of DNA survivors was collected after each round of selection and amplified by six-letter GACTZP PCR with a mixture of nucleotide triphosphates (dGTP, dATP, dCTP, dTTP, dZTP, and dPTP, SI Appendix, Table S2) using Hot Start Taq DNA polymerase (TaKaRa). The product was recovered by binding to solid-phase streptavidin, followed by elution with NaOH. The resulting single-stranded DNA was subjected to the next round of selection (Fig. 2). No negative selection was incorporated into this procedure.
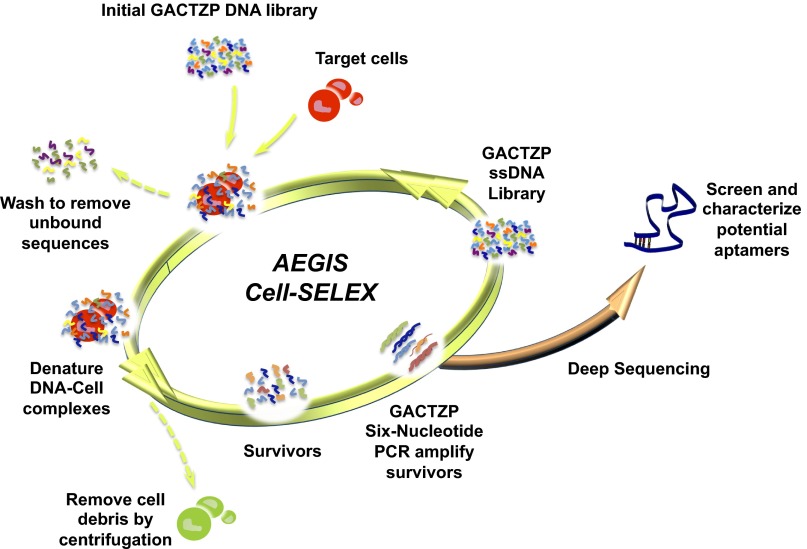
Fig. 2.
Schematic of the AEGIS cell–SELEX used here. A GACTZP DNA library, consisting of randomized sequences and primer sites, was first incubated with the target cells. Unbound sequences were then removed by washing. Bound sequences having affinity to the target cells were collected. “Survivors” in the enriched collection were amplified using a FITC-labeled 5′-primer and a biotinylated 3′-primer by GACTZP six-nucleotide PCR (22). Enriched FITC-conjugated single-stranded DNA obtained from PCR products was used for the next round of selection. The binding affinity of survivors from 9th round up to 12th round of selection was monitored by flow cytometer. The survivors from 12th round selection were subjected to deep sequencing.
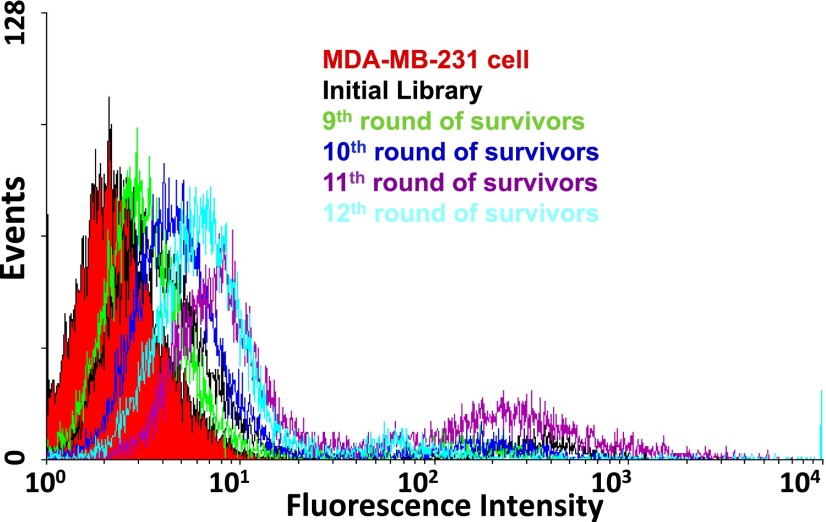
To increase the selection pressure in later rounds of AEGIS–SELEX, the number of cells and incubation times were gradually reduced, and the volume of washing buffer and the number of washes was increased. Starting after nine rounds of selection, the progress of the AEGIS–SELEX experiment was monitored by flow cytometry to measure the binding of the total library to the target cells. The amount of surviving GACTZP total DNA bound to MDA-MB-231 increased from 9th round to 11th round (Fig. 3), but not further after 11th round. Therefore, the AEGIS–SELEX was stopped at 12th round, and the survivors were prepared for deep sequencing.
Fig. 3.
Monitoring the progress of GACTZP AEGIS Cell–SELEX using flow cytometer. Vertical axis indicates the number of cells counted having the fluorescence intensity indicated by the horizontal axis, with the increase in intensity from 9th round indicating the increased number of fluorescein-labeled aptamers bound per cell. Binding assays showed the enrichment of breast cancer cell (MDA-MB-231) binders with an optimum obtained after 11th round, and no further improvement after the 12th round.
Deep Sequencing GACTZP DNA Survivors Using Next Generation Sequencing Technology.
Deep sequencing was done following the “conversion” strategy previously reported (22). The enriched GACTZP DNA survivors recovered after 12 rounds of AEGIS–SELEX were divided into two equal portions. These were separately converted by barcoded copying into standard DNA using two conversion protocols (SI Appendix, Table S3). In the first protocol, sites holding Z and P nucleotides in the GACTZP survivors were converted predominantly into sites holding C and G nucleotides, respectively; less than 15% were other nucleotides. Under the second conversion protocol, sites holding Z were converted to sites holding a mixture of C and T, with their ratio lying between 60:40 and 40:60, depending on the sequence surrounding that site. Sites holding P are converted to a mixture of G and A with roughly the same range of ratios, again depending on the sequence context surrounding that site.
Following conversion, two barcoded samples were combined and submitted for Ion Torrent “next generation” sequencing at the University of Florida DNA sequencing Core facility (Interdisciplinary Center for Biotechnology Research at the University of Florida). Reads that did not contain exactly matched barcodes and/or forward and reverse priming sequences were discarded. To minimize miscalling, any sequence present as fewer than 80 copies in the whole library was removed from the analysis. The remaining reads (357,574 in total) were then clustered using software custom designed at the Foundation for Applied Molecular Evolution, which ignored differing barcodes during the clustering and accepted single-step changes within sequence reads. Clustered sequences were then separated by barcode, with variable sites being compared between each barcode. The clustered sequences obtained under the first conversion conditions (Z to C and P to G) serve as reference for the clustered sequences obtained under the second conversion conditions. Sites where C and T were found in approximately equal amounts after conversion under the second conditions were assigned as Z in their “parent.” Sites where G and A were found in approximately equal amounts after conversion under the second conditions were assigned as P in their parent (SI Appendix, Table S4).
The inferred ancestral sequences were aligned to identify dominant candidate aptamers. One dominant aptamer sequence was represented by 101,224 independent reads and constituted ∼30% of the survivors (SI Appendix, Table S4). This aptamer was named ZAP-2012 (Z And P, 20 nucleotide random region, 12 cycles of selection), and its sequence was inferred to be:
5′-TCCCGAGTGACGCAGC-CCCCGGZGGGATTPATCGGT-GGACACGGTGGCTGAC-3′. ZAP-2012 contains a single Z at the position of 23 and a single P at the position of 30 (Z23-P30). The second and third most abundant sequences were about 5% and 3% of the population.
Determine the Binding Affinity of the Dominant Aptamer ZAP-2012 and Variants.
The ZAP-2012 aptamer was then resynthesized in a form carrying a 5′-biotin by solid phase synthesis (ABI 394 DNA) from standard phosphoramidites (Glen Research) and dZ and dP phosphoramidites (Firebird Biomolecular Sciences). Analogous molecules lacking Z (C23-P30 and T23-P30), lacking P (Z23-G30 and Z23-A30), or lacking both Z and P (C23-G30, T23-G30, T23-A30, and C23-A30) were also synthesized with a 5′-biotin (SI Appendix, Table S5).
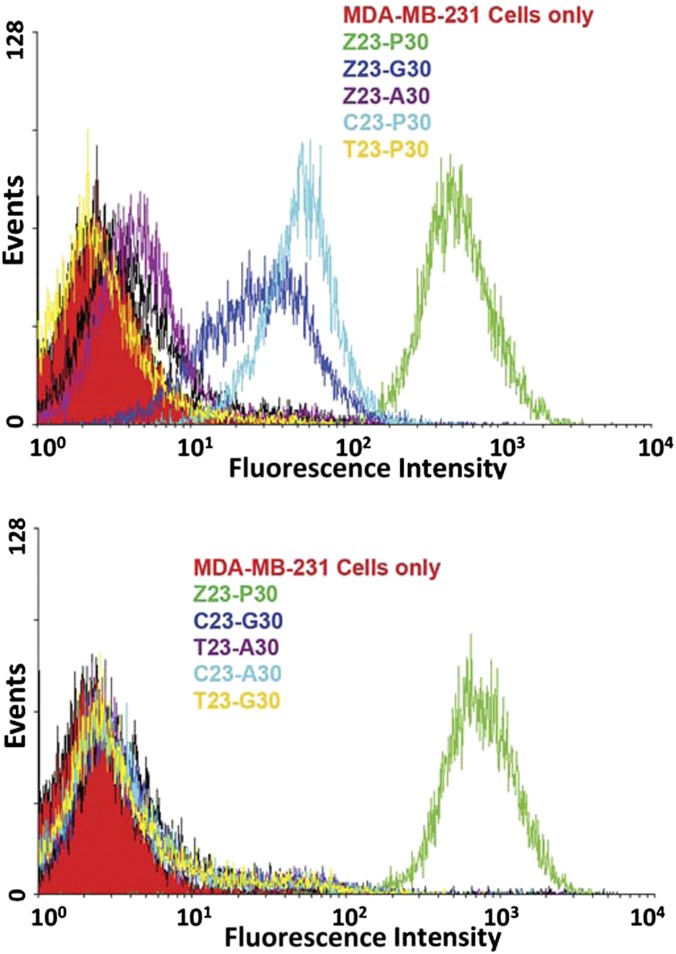
These were each used at 250 nM in a flow cytometry assay (labeling with streptavidin-PE-Cy5.5) to test their binding to the MDA-MB-231 target cells (Fig. 4). The original nonbinding library was used as a negative control. The ZAP-2012 sequence bound strongly; its mutant forms gave either reduced binding (C23-P30, Z23-G30, and Z23-A30) or no binding at all (T23-P30, and all sequences lacking both Z and P). These studies illustrated that the Z and P nucleotides in ZAP-2012 significantly contribute to the binding affinity.
Fig. 4.
Flow cytometry measurement of the binding of chemically resynthesized ZAP-2012 aptamer (5′-biotin-TCCCGAGTGACGCAGC-CCCCGGZGGGATTPATCGGT-GGACACGGTGGCTGAC-3′, the random region is underlined) obtained using AEGIS Cell–SELEX to MDA-MB-231 breast cancer cells (green curve). The vertical axis indicates the number of cells counted having the fluorescence (from biotin-bound Cy5.5-conjugated streptavidin) intensity indicated by the horizontal axis. Also shown (other colors) are binding data for chemically resynthesized ZAP-2012 mutants. (Upper) Where the Z and P are separately replaced with standard nucleotides, binding affinity is reduced. (Lower) Where the Z and P are both replaced with standard nucleotides, binding affinity is lost.
We also resynthesized and tested the binding affinity of secondary survivors in the population, contributing from 5.0 to 0.4% of the total population of survivors. Among these 12 sequences, 3 had no Z and P; the remainder had either a single Z (and no P), a single P (and no Z), or one of each (SI Appendix, Table S6). Compared with the binding signal of the ZAP-2012, all 12 secondary candidates gave negligible binding signals (SI Appendix, Figs. S1 and S2). To rule out the misassignment of the Z and P in these candidates, we replaced Z by either C or T, and replaced P by either G or A, to produce all possible fully standard sequence analogs, to exclude the possibility that these might be the “true” aptamer arising from the selection (variants indicated by blue color in SI Appendix, Table S6). Again, all these candidate sequences gave no binding signals at 250 nM (SI Appendix, Fig. S1). This shows that both Z and P are required for ZAP-2012 to bind.
Determination of Dissociation Constant of Aptamer ZAP-2012.
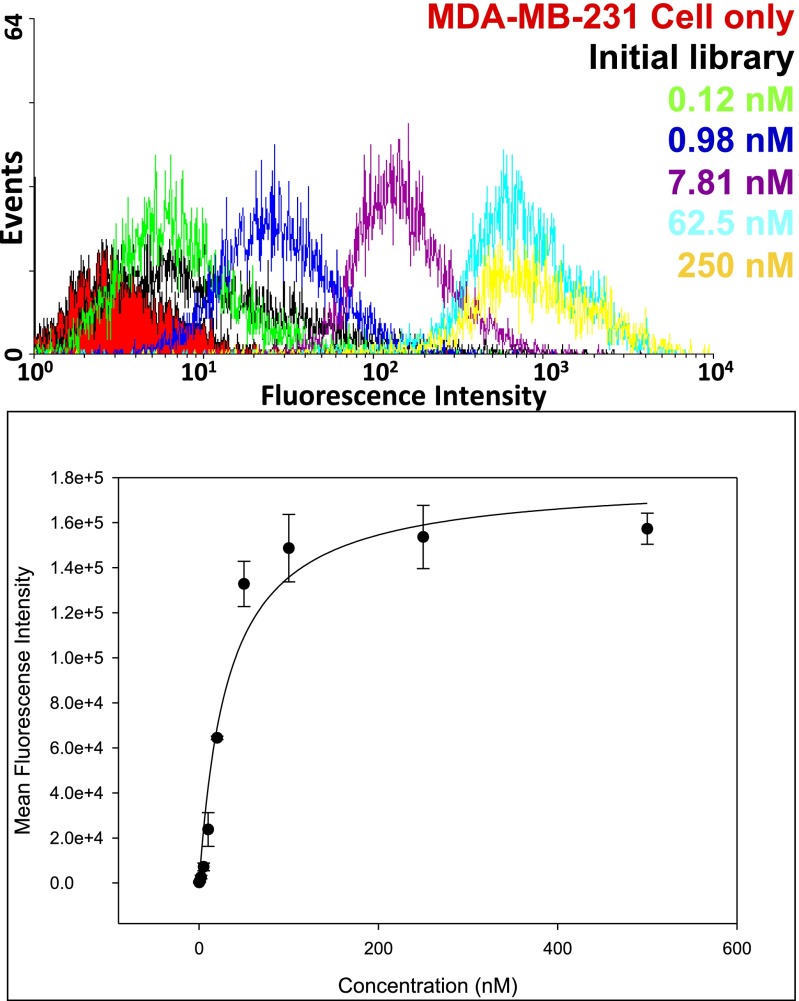
We then estimated the dissociation constant (Kdiss) of the ZAP-2012 aptamer against breast cancer cells (MDA-MB-231) using serial dilutions (0.1 nM–500 nM final concentration) (Fig. 5). The biotin-labeled unselected library was used as a negative control to assess background binding. All binding assays were done in triplet. The mean fluorescence intensity of the unselected library was subtracted from that of the corresponding aptamer with the target cells to determine the specific binding of the labeled aptamer. The apparent Kdiss was obtained by fitting the intensity of binding versus the concentration of the aptamers to the equation Y = Bmax X/(Kdiss + X); Y is the mean fluorescence intensity, at the concentration of aptamer = X in nanomoles; Bmax is maximal binding, using SigmaPlot. From these data, ZAP-2012 (Z23-P30) bound to the cells (MDA-MB-231) with an apparent Kdiss = 30 ± 1 nM (Fig. 5). If the Z in Z23-P30 is replaced by C to give C23-P30, the Kdiss increases to 160 ± 31 nM. If the P in Z23-P30 is replaced by G to give Z23-G30, the Kdiss increases to 442 ± 130 nM. All other mutant forms lacking Z (T23-P30), lacking P (Z23-A30), or lacking both Z and P (C23-G30, T23-A30, C23-A30, and T23-G30) gave almost no binding (the Kdiss increases to >1 µM, Fig. 4).
Fig. 5.
Estimation of the dissociation constant (Kdiss) for the ZAP-2012 aptamer. (Upper) Binding signals of diluted aptamers (0.1–500 nM). (Lower) Binding curve, Kdiss = 30 ± 1 nM.
Discussion
These results show that AEGIS–SELEX works in cell-targeted SELEX. This, in turn, requires the performance of each step in the molecular biology of the GACTZP system. These steps include (i) library synthesis without substantial loss of diversity, (ii) selection of nucleic acid analogs that incorporate AEGIS components by their binding to cells, (iii) six-nucleotide PCR without substantial loss of the Z:P pair, and (iv) downstream deep sequencing.
Although we have completed only one AEGIS–SELEX experiment, some brief comments might be made to compare its results to our experience with standard GACT SELEX. Here, AEGIS–SELEX generated a nanomolar aptamer against this cell target after 12 rounds of selection. Typically, 15–20 rounds of standard GACT SELEX are required to get similar affinities against cell targets. For example, 18 rounds of standard SELEX were required to get aptamers having Kdiss values ranging from 45 nM (best) to 208 nM (worst) against lung cancer cells (27). Between 18 and 20 rounds of standard GACT were required to get aptamers that bind with Kdiss values ranging from 302 to 0.7 nM for colon cancer cells (28). The tightest binding aptamer in this series is the best against any cell target obtained by Tan and coworkers. It evidently binds to protein tyrosine kinase 7, a protein present on the surface of most cancer cells (7). Here, it should be noted that the length of the randomized regions was variable between these different SELEX experiments, typically between 35 and 45 nucleotides (compared with the 20 nucleotides used here). Whereas this might be significant to the outcome, variation in sequence length has not been observed to generally affect the affinity of the aptamers generated from standard libraries.
The rapid appearance of aptamers with nanomolar affinities may be attributable to the absence of a negative selection step in this experiment. However, it is also consistent with the hypothesis that AEGIS libraries are richer reservoirs of nanomolar affinity aptamers than standard xNA libraries, perhaps because they have richer diversity.
In this context, it is interesting to note that, given the experimentally confirmed diversity of the original GACTZP, we might expect an “average” aptamer having a 20-nucleotide random region to contain about 3 Zs and 1.5 Ps. However, ZAP-2012 contains just one Z and one P. This slight depletion has many possible explanations, of course. However, as one hypothesis, the retention of Z:P pairs may be modestly disadvantaged during the six-letter GACTZP PCR (22). Thus, the final composition of surviving aptamers would reflect both selection for aptamers containing Z and P (for their contribution to binding) and counterselection against aptamers containing Z and P (as these might be slightly more difficult to PCR amplify).
This balance has precedent in standard GACT SELEX. Here the G–C richness in survivor populations may be selected for (in the binding step), as it supports certain kinds of folds. However, G–C richness is also believed to be selected against (in the PCR step), as G-rich sequences are difficult to PCR amplify. More work is necessary to understand this balance for AEGIS–SELEX, as it is for standard SELEX.
Regardless of these comments relevant to AEGIS–SELEX in general, the Z and P AEGIS components are clearly necessary for ZAP-2012 itself to function; removing them decreases or eliminates binding. Interestingly, the ZAP-2012 aptamer also gave very intense signal labeling of the MDA-MB-231 cells, ∼10-fold higher than with standard aptamers. Further, the 30 nM affinity of the ZAP-2012 aptamer sits in the middle of the values obtained for aptamers generally targeted against cells. However, we do not know whether or not this binding is specific to cancer cells (that is, transformed cell lines that have been immortalized in culture); we have not done a control with breast cells that have not been immortalized.
This is, of course, just one of many efforts to improve the performance of SELEX by altering the matrix upon which selection operates, as noted in the introduction (11–17). Accordingly, it is important to note the differences between what we have done here and what is being done in other laboratories.
First, as noted in the introduction, others (and we ourselves, ref. 12) have previously attempted to simply add functionality to the four standard nucleotides. AEGIS–SELEX is different, in that it adds independently replicable nucleotide letters to the SELEX library. Whereas the nitro group on Z is certainly a functionality not known in natural DNA, and may contribute substantially to the binding interaction between ZAP-2012 and the MDA-MB-231 cells, the primary advance of the work reported here is that it is increasing the number of independently replicating nucleotides. This opens the door to an AEGIS–SELEX supported by a full 12 nucleotides, some functionalized, others not.
Other groups have also attempted to add nucleotides to PCR (29–32). Especially persuasive work has been reported by Hirao and coworkers, who recently incorporated Ds (a steric complement to the nonstandard nucleobase Px) into DNA aptamers that bind to VEGF and IFN-gamma, with dramatically improved results (17). These results were obtained not with a full SELEX, but rather by placing nonstandard nucleotides at predetermined positions in a random sequence. This expedient was necessary because the Ds:Px pair, being more radically different in structure than the second generation AEGIS pairs that we have developed, is difficult to amplify and sequence in multiple and consecutive sequences (32).
In this respect, we cannot overemphasize the value of the sequencing technology that was applied to this work. Without it, we would have been constrained to simply adding single nonstandard nucleotides at predetermined sites. However, AEGIS–SELEX clearly would be improved by (i) polymerases with still higher levels of fidelity and efficiency, so that less selection against Z:P pairs occurs during PCR amplification; (ii) a technology for directly sequencing Z:P pairs; and, more expansively, (iii) strains of Escherichia coli cells that can propagate plasmids containing GACTZP DNA. All are being developed in these laboratories.
Materials and Methods
GACTZP DNA libraries and aptamers containing Z and P were synthesized and purified by methods described in SI Appendix. AEGIS–SELEX procedures, deep sequencing of GACTZP DNA survivors, and analysis of potential aptamer candidates are also described in SI Appendix. Detailed materials and methods can be found in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We are indebted to the Defense Threat Reduction Agency (HDTRA1-13-1-0004) and the National Aeronautics and Space Administration (NNX10AT28G) for long-standing support of basic research in the S.A.B. laboratory in the area of nucleic acid chemistry. The W.T. laboratory is indebted to the National Institutes of Health (R21CA122648, GM079359, and CA133086), the National Key Scientific Program of China (2011CB911000), and the China National Instrumentation Program 2011YQ03012412 for support of this work.
Footnotes
Conflict of interest statement: S.A.B., Z.Y., and the Foundation for Applied Molecular Evolution hold patents and patent applications for various of the AEGIS compounds and in vitro selection with expanded genetic alphabets.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311778111/-/DCSupplemental.
References
- 1.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 2.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344(6265):467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 3.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 4.Famulok M. Oligonucleotide aptamers that recognize small molecules. Curr Opin Struct Biol. 1999;9(3):324–329. doi: 10.1016/S0959-440X(99)80043-8. [DOI] [PubMed] [Google Scholar]
- 5.Sun W, Du L, Li M. Aptamer-based carbohydrate recognition. Curr Pharm Des. 2010;16(20):2269–2278. doi: 10.2174/138161210791792877. [DOI] [PubMed] [Google Scholar]
- 6.Gopinath SCB. Methods developed for SELEX. Anal Bioanal Chem. 2007;387(1):171–182. doi: 10.1007/s00216-006-0826-2. [DOI] [PubMed] [Google Scholar]
- 7.Shangguan D, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci USA. 2006;103(32):11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Proske D, Blank M, Buhmann R, Resch A. Aptamers—basic research, drug development, and clinical applications. Appl Microbiol Biotechnol. 2005;69(4):367–374. doi: 10.1007/s00253-005-0193-5. [DOI] [PubMed] [Google Scholar]
- 9.Li N, et al. Technical and biological issues relevant to cell typing with aptamers. J Proteome Res. 2009;8(5):2438–2448. doi: 10.1021/pr801048z. [DOI] [PubMed] [Google Scholar]
- 10.Sefah K, Shangguan D, Xiong X, O’Donoghue MB, Tan W. Development of DNA aptamers using Cell-SELEX. Nat Protoc. 2010;5(6):1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 11.Tarasow TM, Eaton BE. Dressed for success: Realizing the catalytic potential of RNA. Biopolymers. 1998;48:29–37. [Google Scholar]
- 12.Battersby TR, et al. Quantitative analysis of receptors for adenosine nucleotides obtained via in vitro selection from a library incorporating a cationic nucleotide analog. J Am Chem Soc. 1999;121(42):9781–9789. doi: 10.1021/ja9816436. [DOI] [PubMed] [Google Scholar]
- 13.Tolle F, Mayer G. Dressed for success. Applying chemistry to modulate aptamer functionality. Chem Sci. 2013;4:60–67. [Google Scholar]
- 14.Hollenstein M, Hipolito CJ, Lam CH, Perrin DM. Toward the combinatorial selection of chemically modified DNAzyme RNase A mimics active against all-RNA substrates. ACS Comb Sci. 2013;15(4):174–182. doi: 10.1021/co3001378. [DOI] [PubMed] [Google Scholar]
- 15.Gold L, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE. 2010;5(12):e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraemer S, et al. From SOMAmer-based biomarker discovery to diagnostic and clinical applications: A SOMAmer-based, streamlined multiplex proteomic assay. PLoS ONE. 2011;6(10):e26332. doi: 10.1371/journal.pone.0026332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimoto M, Yamashige R, Matsunaga K-I, Yokoyama S, Hirao I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat Biotechnol. 2013;31(5):453–457. doi: 10.1038/nbt.2556. [DOI] [PubMed] [Google Scholar]
- 18.Benner SA. Understanding nucleic acids using synthetic chemistry. Acc Chem Res. 2004;37(10):784–797. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]
- 19.Benner SA, Yang Z, Chen F. Synthetic biology, tinkering biology, and artificial biology. What are we learning? CR (East Lansing, Mich) 2010;14:372–387. doi: 10.1016/j.crci.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang ZY, Hutter D, Sheng PP, Sismour AM, Benner SA. Artificially expanded genetic information system: A new base pair with an alternative hydrogen bonding pattern. Nucleic Acids Res. 2006;34(21):6095–6101. doi: 10.1093/nar/gkl633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang ZY, Sismour AM, Sheng PP, Puskar NL, Benner SA. Enzymatic incorporation of a third nucleobase pair. Nucleic Acids Res. 2007;35(13):4238–4249. doi: 10.1093/nar/gkm395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z, Chen F, Alvarado JB, Benner SA. Amplification, mutation, and sequencing of a six-letter synthetic genetic system. J Am Chem Soc. 2011;133(38):15105–15112. doi: 10.1021/ja204910n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen F, et al. Recognition of an expanded genetic alphabet by type-II restriction endonucleases and their application to analyze polymerase fidelity. Nucleic Acids Res. 2011;39(9):3949–3961. doi: 10.1093/nar/gkq1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z, Chen F, Chamberlin SG, Benner SA. Expanded genetic alphabets in the polymerase chain reaction. Angew Chem Int Ed Engl. 2010;49(1):177–180. doi: 10.1002/anie.200905173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang K, et al. A novel aptamer developed for breast cancer cell internalization. ChemMedChem. 2012;7(1):79–84. doi: 10.1002/cmdc.201100457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sefah K, et al. Cell-based selection provides novel molecular probes for cancer stem cells. Int J Cancer. 2013;132(11):2578–2588. doi: 10.1002/ijc.27936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiménez E, et al. Generation of lung adenocarcinoma DNA aptamers for cancer studies. PLoS ONE. 2012;7(10):e46222. doi: 10.1371/journal.pone.0046222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sefah K, et al. DNA aptamers as molecular probes for colorectal cancer study. PLoS ONE. 2010;5(12):e14269. doi: 10.1371/journal.pone.0014269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delaney JC, et al. High-fidelity in vivo replication of DNA base shape mimics without Watson-Crick hydrogen bonds. Proc Natl Acad Sci USA. 2003;100(8):4469–4473. doi: 10.1073/pnas.0837277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henry AA, Romesberg FE. Beyond A, C, G and T: Augmenting nature’s alphabet. Curr Opin Chem Biol. 2003;7(6):727–733. doi: 10.1016/j.cbpa.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 31.Hirao I, et al. An unnatural hydrophobic base pair system: Site-specific incorporation of nucleotide analogs into DNA and RNA. Nat Methods. 2006;3(9):729–735. doi: 10.1038/nmeth915. [DOI] [PubMed] [Google Scholar]
- 32.Kimoto M, Kawai R, Mitsui T, Yokoyama S, Hirao I. An unnatural base pair system for efficient PCR amplification and functionalization of DNA molecules. Nucleic Acids Res. 2009;37(2):e14. doi: 10.1093/nar/gkn956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.