Significance
This study describes the crystal structures of the full-length PriA DNA helicase, a multifunctional enzyme that mediates the essential process of restarting prematurely terminated DNA replication reactions in bacteria. Our findings reveal how PriA is able to recognize replication restart substrates through structure-specific DNA binding and interactions with the ssDNA-binding protein and how it exposes ssDNA that could be used to reload the replisome and reinitiate replication. These observations shed light on the physical mechanisms that allow cells to survive the common and potentially lethal problems posed by incomplete genome replication.
Keywords: X-ray crystal structure, single-molecule FRET
Abstract
Collisions between cellular DNA replication machinery (replisomes) and damaged DNA or immovable protein complexes can dissociate replisomes before the completion of replication. This potentially lethal problem is resolved by cellular “replication restart” reactions that recognize the structures of prematurely abandoned replication forks and mediate replisomal reloading. In bacteria, this essential activity is orchestrated by the PriA DNA helicase, which identifies replication forks via structure-specific DNA binding and interactions with fork-associated ssDNA-binding proteins (SSBs). However, the mechanisms by which PriA binds replication fork DNA and coordinates subsequent replication restart reactions have remained unclear due to the dearth of high-resolution structural information available for the protein. Here, we describe the crystal structures of full-length PriA and PriA bound to SSB. The structures reveal a modular arrangement for PriA in which several DNA-binding domains surround its helicase core in a manner that appears to be poised for binding to branched replication fork DNA structures while simultaneously allowing complex formation with SSB. PriA interaction with SSB is shown to modulate SSB/DNA complexes in a manner that exposes a potential replication initiation site. From these observations, a model emerges to explain how PriA links recognition of diverse replication forks to replication restart.
Replisomes frequently encounter obstructions, such as impassable DNA damage or frozen protein complexes, that can arrest replication and/or eject the replisome before completion of replication (1–4). Left unrepaired, these events lead to incomplete chromosome duplication, genomic instability, and cell death. All proliferating cells must contend with these clashes because replication blockades can arise from essential endogenous cellular processes. Replication restart, the process by which replisomes are reloaded onto abandoned replication forks, has been observed in both bacteria and eukaryotes. However, only the bacterial factors that mediate this process have been identified. The bacterial replication restart proteins thus serve as models for understanding the general mechanisms of replication restart in all cells (4).
Replication restart in bacteria is orchestrated by the multifunctional PriA DNA helicase (5, 6). The process is initiated by PriA binding to abandoned DNA replication forks or other appropriate replication reinitiation sites in a structure-specific manner. In this recognition step, PriA binds to duplex parental DNA and can accommodate either duplex or gapped leading- and lagging-strand DNA in branched replication fork structures (7–10). PriA’s ability to recognize multiple DNA structures appears to provide the flexibility needed for replication restart to be initiated on structures ranging from simple replication forks to D-loops that are produced by recombinational repair of dsDNA breaks (5, 6, 10). However, the structural mechanism by which PriA manages to bind to this array of substrates is unknown. In addition to structure-specific DNA binding, PriA takes advantage of a direct protein interaction with ssDNA-binding protein (SSB) tetramers to target its activity to replication forks. SSBs coat the lagging strand template when it is single-stranded and form a complex with PriA that stimulates PriA’s helicase activity (11–15). In this interaction, PriA binds to the extreme C terminus (Ct) of SSB, which is a known docking site for numerous genome maintenance proteins that process ssDNA within SSB/ssDNA nucleoprotein complexes (16). Once bound to an appropriate replication restart substrate, PriA remodels the lagging-strand arm to expose ssDNA by either unwinding DNA (if the lagging strand is duplex) or altering the SSB-bound template DNA (if the lagging strand is ssDNA) (17–19). The structural mechanisms underlying lagging-strand remodeling by PriA have not been well defined. PriA then recruits additional replication restart proteins (PriB and DnaT in Escherichia coli) to complete formation of the PriA/PriB/DnaT “primosome” complex that is competent to mediate the first step of replication restart: loading of the replicative helicase (DnaB in E. coli) onto the remodeled replication fork (20–24). Assembly of the full replisome is then mediated by DnaB activities and protein interactions.
Biochemical studies have identified a two-domain architecture for PriA that includes a DNA-binding domain (DBD; N-terminal ∼200 residues) and a helicase domain (HD; C-terminal ∼530 residues) (18, 21, 25) (Fig. 1A). However, despite decades of genetic and biochemical research, the lack of high-resolution structural information for full-length PriA has left the molecular mechanisms underlying abandoned replication fork recognition and processing unresolved. To better understand the roles and mechanisms of PriA in replication restart, we have determined the X-ray crystal structures of full-length PriA and the PriA/SSB-Ct complex and examined the biochemical consequences of PriA/SSB complex formation using a single-molecule approach. The structures reveal a modular arrangement for PriA in which its central helicase core is surrounded by an array of DNA-binding elements. These elements include two domains within the DBD [a 3′ DNA-binding domain (3′BD) and an unusual circularly permuted winged helix (WH) domain] and two domains within the HD [a Cys-rich region (CRR) that binds two Zn2+ ions and a C-terminal domain (CTD) with unanticipated similarity to the S10 subunit of the bacterial ribosome]. The positioning of these domains, coupled with their known or anticipated roles in DNA binding and replication fork remodeling, leads to a model that explains how PriA binds to branched DNA replication fork structures in a specific manner. SSB binds to PriA at a site formed at the interface of the 3′BD, CTD, and helicase core that appears to be positioned to allow simultaneous PriA binding to both DNA and SSB. PriA interaction with SSB through this site modulates the SSB/ssDNA complex in a manner that exposes ssDNA that could be used to reload DnaB onto abandoned replication forks. Together with the PriA DNA-binding model, these findings produce a molecular view of initial steps of PriA-mediated replication restart.
Fig. 1.
Structure of PriA DNA helicase. (A) Schematic diagram of PriA domain structure. (B) Crystal structure of KpPriA. Domains are colored as in A. ADP (red sticks) is bound within the helicase core, and two Zn2+ ions (gray spheres) are bound to the CRR. (C) Electrostatic surface features of KpPriA (blue, electropositive; red, electronegative; white, neutral). (D) Evolutionary conservation of KpPriA (a conservation scale, from variable to invariant among 150 PriA protein sequences, is shown below the structure).
Results and Discussion
PriA DNA Helicase X-Ray Crystal Structure Determination.
Several bacterial PriA proteins were initially selected as targets in our crystallographic effort to determine the structure of full-length PriA. Among these proteins, PriA from Klebsiella pneumoniae (KpPriA) crystallized in a form that diffracted X-rays to a resolution of 2.65 Å (Table S1). KpPriA shares 88% sequence identity with the prototypical E. coli PriA protein (EcPriA), and the two proteins display very similar DNA-binding and DNA-dependent ATPase activities with a synthetic forked DNA substrate (Fig. S1). Moreover, expression of the K. pneumoniae priA gene complements the filamentation and constitutive SOS phenotypes of priA- E. coli cells (Fig. S1). Thus, KpPriA and EcPriA are structural and functional homologs.
Single-wavelength anomalous diffraction data from a selenomethionine-substituted KpPriA crystal were used to calculate a 3.76-Å resolution experimental electron density map that allowed building of a partial model of the KpPriA structure. This partial structure was then used as a search model to solve the 2.65-Å resolution native KpPriA structure via molecular replacement (Table S1).
The KpPriA structure comprises six subdomains that cluster together (Fig. 1). Two of these subdomains (3′BD and WH) comprise the N-terminal DBD, and the remaining four (two lobes of the helicase core, CRR, and CTD) comprise the HD. The 3′BD, helicase core, CRR, and CTD interact with one another to form a shallow cup shape, whereas the WH domain projects away from the rest of KpPriA in a position that is stabilized by interactions with symmetrically related PriA proteins in the crystal lattice (Fig. 1B). Because the WH lacks direct contacts with the rest of KpPriA, its position is likely to be dynamic in the PriA monomer in solution, which may have led to conformational heterogeneity that prevented crystallization of other full-length PriA proteins. The electrostatic surface of PriA includes prominent basic patches presented by the 3′BD, WH, helicase, and CTD elements on its concave surface (Fig. 1C). A significant portion of the concave surface and more scattered patches on the convex surface are evolutionarily well conserved among PriA proteins (Fig. 1D).
PriA Structure Reveals Two Subdomains Within the DBD.
The structure of KpPriA showed that the DBD is subdivided into two DNA-binding elements. The first is the 3′BD, which has been shown previously to bind the 3′ end of the leading-strand arm of replication fork structures (26). This recognition is thought to help direct PriA replisomal reloading activity to appropriate DNA structures (25–28). The 3′BD structure from full-length KpPriA is strikingly similar to the previously determined structure of the isolated 3′BD from EcPriA (26) (rmsd = 1.8 Å for 90 common Cα atoms). Residues that are known to bind the 3′ end of nascent leading-strand DNA in EcPriA (26) form an exposed pocket in full-length KpPriA, making it available for leading strand recognition in the apo structure (Fig. 1). A leading-strand binding role for this site within full-length PriA is described further below in our model of the PriA/replication fork structure complex.
The second DBD element is a WH domain, which has an unusual circularly permuted topology that distinguishes it from the classic WH fold. Typical WH domains are defined by core helix–turn–helix folds with β-hairpin “wing” elements; these motifs bind the major and minor grooves of duplex DNA, respectively, in many WH domains (29) (Fig. S2). The helix–turn–helix fold is present within the PriA WH domain; however, the β-hairpin wing element has been split open and serves as the connection between the WH domain and the rest of PriA (Fig. 1B and Fig. S2). This β-hairpin splitting also alters the order of secondary structural elements in the PriA domain relative to other WH folds. This difference is accommodated by circular permutation in which the elements that serve as N- and C-terminal ends in most WH domains are directly linked (Fig. S2). Despite these differences, an EcPriA fragment comprising just the WH domain is able to bind partial duplex DNA in vitro, and a longer fragment that includes both the 3′BD and WH domains binds DNA replication fork structures with higher affinity than either domain alone, indicating that the two domains functionally cooperate (9). The most conserved surface of the WH domain presents a highly basic patch from the helix–turn–helix motif on the concave face of PriA that we speculate is important in replication fork DNA binding (below).
PriA HD Includes a Helicase Core and Unusual Zn2+-Binding Domain and CTD with Roles in DNA Binding and Unwinding.
The PriA HD is composed of a bilobed helicase core region that is buttressed by CRR and CTD elements (Figs. 1 and 2). The PriA helicase core shares significant similarity with other helicases, with canonical helicase motifs lining the interface between the two lobes. An ADP molecule is bound at this interface (Fig. 2A). Two features were identified within the helicase core that could provide important autoregulatory elements that restrict PriA ATPase activity in the absence of DNA. The first is a conserved aromatic-rich loop (ARL) in the N-terminal helicase lobe that extends from the helicase core to bind both the 3′BD and CTD (Fig. 2A). Similar ARLs in RecQ and PcrA DNA helicases bind directly to ssDNA and couple binding to structural changes that stimulate ATPase and helicase activities (30, 31). Mutations within this region of EcPriA block binding to D-loop DNA structures (25), which is consistent with the ARL having a direct role in DNA binding in PriA. The second feature is an extended helicase motif V that positions the side chain of Lys543 between the β-phosphate of ADP and the carboxyl group of Asp319 from helicase motif II (Fig. 2A). This carboxyl group coordinates an active site Mg2+ in other helicases. In the PcrA DNA helicase, a similar Lys modulates ATPase kinetics in a manner that appears to stabilize the ADP product complex (32). Both the ARL and Lys543 are highly conserved among PriA proteins (Fig. S1), and we predict that they help coordinate PriA DNA-binding and ATPase activities.
Fig. 2.
PriA domain features. (A) Core helicase motifs and the ARL (magenta, labeled) line the interface between the bilobed HD. An ADP molecule (red sticks) is shown. (B) PriA CRR binds two Zn2+ ions (gray spheres) and positions a β-hairpin that is proposed to act as a DNA unwinding wedge. (C) Close-up view of the CTD (yellow) with basic residues displayed as sticks. The surfaces of the 3′BD, helicase core, and CRR are rendered to highlight the extensive contacts made between the CTD and each domain. (D) Overlay of KpPriA CTD (yellow) with the S10 ribosomal subunit (gray, with rRNA shown in orange) (37). RNA-binding basic residues from S10 and similarly positioned residues from the KpPriA CTD are shown (sticks). (E) EcPriA CTD binding to fluorescently labeled dT28 (black), 10-bp duplex (blue), 38-bp duplex (green), and replication fork (red) DNA. Data are the mean of three independent experiments, with error bars representing 1 SD.
An unusual feature of the PriA HD is the presence of a CRR embedded within the C-terminal helicase lobe (Fig. 2B). This 50-residue insertion forms a structure on the surface of the helicase core in which two Zn2+ ions are coordinated by invariant Cys residues. A portion of the PriA CRR bears an unexpected structural similarity to the N-terminal domain of the Rpb9 subunit of eukaryotic RNA polymerase II [rmsd = 2.3 Å for 39 common Cα atoms (33)], although the analogous Rpb9 domain only binds a single Zn2+ ion. The functional significance of this structural similarity is not known. Previous biochemical experiments have shown that sequence changes to Zn2+-binding Cys residues in the EcPriA CRR can eliminate helicase, but not ATPase, activity and can block assembly of PriB onto DNA-bound PriA (24, 34), implicating the CRR in multiple functions in PriA. Interestingly, a β-hairpin within the CRR is in a similar position relative to the PriA helicase core to “wedge” elements that are critical for DNA strand separation in other related helicases (Fig. S3). Given the position of the CRR and noted defects of EcPriA CRR Cys variants in DNA unwinding, the β-hairpin within the CRR is an excellent candidate to function as a DNA unwinding wedge in PriA. In this activity, the helicase core would directionally translocate on an ssDNA tail of a partial duplex DNA and pull the duplex into the β-hairpin wedge, which would split the duplex into two strands. Given the 3′-5′ translocase and helicase activity of PriA and its preference for unwinding the lagging strand of replication fork structures (11–13, 35, 36), this model suggests that lagging strand template ssDNA would be engaged by the helicase core, whereas the duplex portion of the lagging strand would be unwound by the CRR. This arrangement could create a PriA/DNA structure onto which PriB can dock during primosome assembly and may be important for creating ssDNA that acts as a loading site for DnaB.
The final element of the HD is the CTD, which forms a central core in PriA that directly contacts all domains but the WH (Fig. 2C). The CTD is unexpectedly similar to the S10 ribosomal subunit (rmsd = 2.2 Å for 68 common Cα atoms), which binds branched rRNA within the bacterial ribosome (37) (Fig. 2D). Several basic residues within the CTD project toward the HD near where ssDNA is predicted to bind and are positioned similar to RNA-binding residues in S10, highlighting their possible roles in DNA binding in PriA. To test the DNA-binding properties of the PriA CTD, a recombinant EcPriA CTD was constructed based on the KpPriA structure and tested for its ability to bind to fluorescein-labeled DNA structures. The isolated CTD was able to bind to a variety of DNA structures in vitro, including ssDNA, duplex DNA, and a replication fork mimic (duplex DNA with 3′ and 5′ ssDNA tails) (Fig. 2E), consistent with roles for the CTD in DNA binding in the context of full-length PriA.
Structure and Function of the PriA/SSB Complex.
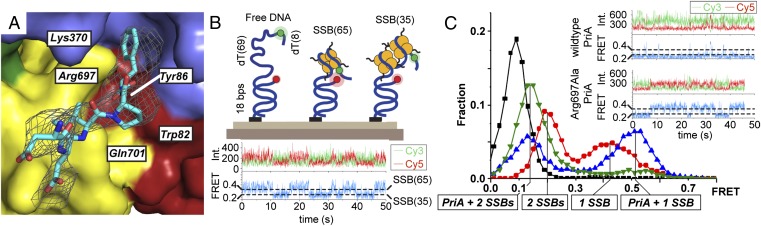
In addition to structure-specific DNA binding, PriA interacts with the SSB C terminus (Ct) at replication forks (13–15). To identify the SSB-Ct binding site on PriA, we determined the 4.1-Å resolution structure of KpPriA bound to an SSB-Ct peptide (Table S1). Although the resolution of the structure was modest, clear electron density for the SSB-Ct peptide was observed in the Fo-Fc maps that permitted fitting of the peptide (Fig. 3A). The SSB-Ct binds at an evolutionarily conserved site formed at the junction of the CTD, helicase core, and 3′BD that is on the opposite face of PriA relative to the DNA-binding surface (Fig. 1). This partitioning would allow PriA to contact DNA and SSB simultaneously. The PriA SSB-Ct binding site shares similarity with binding pockets characterized in other SSB-interacting proteins, including the presence of a prominent basic residue (Arg697) near the α-carboxyl group of the C-terminal–most residue of the SSB-Ct (Fig. 3A). In other proteins, sequence changes at the α-carboxyl interaction position dramatically destabilize their interactions with SSB (38–41).
Fig. 3.
PriA/SSB complex structure and function. (A) Fo-Fc omit electron density (contoured at 1.7-σ in the image) identifies the SSB-Ct peptide-binding site on PriA. Invariant PriA residues are labeled. (B, Upper) Schematic of the smFRET assay that distinguishes free DNA, SSB65-bound DNA, and SSB35-bound DNA (44). (B, Lower) Trace of an individual DNA molecule transitioning between SSB65- and SSB35-binding modes. Cy3 (green), Cy5 (red), and FRET (blue) intensities fluctuate over time due to mode interconversion. (C) smFRET-efficiency histograms of DNA alone (black), DNA with 20 nM SSB (red), 20 nM SSB with 1 μM PriA (green), and 20 nM SSB with 1 μM Arg697Ala PriA (blue). (Inset) Representative single-molecule time traces are shown for PriA and Arg697Ala PriA experiments. Int., intensity.
Because SSB binding to ssDNA blocks reloading of DnaB (42), we used a single-molecule FRET (smFRET) assay to test whether direct binding of PriA to SSB could modulate SSB/DNA complexes in a manner that exposes a potential ssDNA replisome reloading site. E. coli SSB binds ssDNA in either a highly cooperative mode in which 35 nucleotides are bound per tetramer (SSB35) or a less cooperative mode that binds 65 nucleotides per tetramer (SSB65) (43); these modes can be distinguished in our assay by their differing FRET efficiencies between the fluorescence donor/acceptor pair (Cy3/Cy5) on the DNA substrate (44) (Fig. 3B). When a single SSB tetramer is bound in the SSB65 mode, Cy3/Cy5 is in close proximity (FRET efficiency of ∼0.4), but when two SSB tetramers are bound to the DNA in the SSB35 mode, Cy3/Cy5 is further apart (FRET efficiency of ∼0.2) (44, 45) (Fig. 3B). In the absence of SSB, the labeled DNA molecule is unconstrained, which leads to a very low FRET efficiency (efficiency of ∼0.1; Fig. 3). Our experimental conditions, which include excess free SSB, allowed spontaneous interconversion between the SSB35 and SSB65 modes (Fig. 3B, Lower).
Interestingly, the addition of EcPriA had two effects on SSB/DNA complexes in the smFRET assay: PriA strongly stabilized the SSB35 binding mode over the SSB65 mode, and PriA modestly reduced the SSB35 mode FRET efficiency (Fig. 3C). These changes are consistent with PriA exposing ssDNA (due to the SSB65-to-SSB35 transition) and with PriA binding to the newly exposed ssDNA, which result in the slightly lower FRET efficiency state. In a simple model of this PriA/SSB/DNA ternary complex, PriA could associate at the single strand/duplex junction in the smFRET substrate and slide the two SSB tetramers to the end of the ssDNA tail. Interestingly, an Arg697Ala EcPriA variant, in which a key SSB-binding residue is neutralized, fails to alter the SSB35/SSB65 distribution (Fig. 3C). This variant retains DNA-binding and unwinding activities and can still alter the FRET efficiencies of both SSB modes, suggesting that the observed modulation of SSB/DNA-binding modes is dependent upon a functional PriA/SSB interaction and not due to differential DNA-binding abilities of the PriA variant (Fig. 3C and Figs. S4 and S5). These data indicate that PriA complex formation with SSB induces structural changes in the SSB/DNA complex that expose ssDNA, which can be captured by PriA. A recent study showed that the PriC protein, which initiates a parallel replication restart pathway in E. coli and a small number of closely related bacterial species, also preferentially stabilizes the SSB35 mode (46). These findings suggest that SSB-binding mode remodeling could be a general requirement for DNA replication restart.
Model for PriA-Mediated Replication Restart.
A model emerges from our data to explain how PriA can recognize diverse DNA replication fork structures (Fig. 4). The arrangement of DBDs within the PriA structure complements the positions of the three DNA arms of branched replication fork and D-loop structures, which could facilitate structure-specific DNA binding by PriA. Within this array of domains, we propose that the 3′BD would recognize the 3′ end of the leading strand as has been observed previously (26), whereas the HD would preferentially bind to lagging-strand DNA. Lagging-strand binding by the HD in this model is consistent with PriA’s noted 3′-5′ translocase and helicase activities and its preference for unwinding the nascent lagging strand of replication fork structures (11–13, 35, 36). This arrangement places the unreplicated parental DNA in an ideal position to be bound by the highly basic surface presented by the helix–turn–helix fold in the PriA WH domain (Figs. 1C and 4). The isolated EcPriA WH domain has been shown to bind partial duplex DNA (9), further supporting its suggested role in parental DNA binding. Finally, given the ability of the CTD to bind to a variety of DNA structures (Fig. 2E), it could contribute to binding ssDNA, duplex DNA, or branched DNA that is presented by the helicase core and/or the 3′BD. Our binding model thus explains how the structure of PriA is specifically adapted to recognize appropriate substrates for replication restart.
Fig. 4.
PriA replication restart initiation models. PriA recognizes abandoned DNA replication forks with either duplex (Upper) or SSB-bound ssDNA (Lower) lagging strands and processes these to expose ssDNA necessary for full primosome assembly and reloading of the replicative helicase. Nascent DNA strands are colored gray. Replisomal reassembly proceeds spontaneously after replicative helicase loading.
Following recognition of an abandoned replication fork, PriA must alter the lagging strand to expose an ssDNA site for reloading the replicative helicase. Depending upon whether the lagging strand is duplex or single-stranded, this step requires DNA unwinding or remodeling of SSB-coated ssDNA, respectively. As described above, unwinding of the lagging strand could be accomplished by translocation of the core HD along the lagging-strand template DNA, with the CRR acting as a wedge to unwind duplex DNA (Fig. 4, Upper). For SSB/DNA remodeling, the smFRET studies presented in Fig. 3 support a model in which PriA acts by directly interacting with SSB bound to the lagging strand. Formation of the PriA/SSB complex exposes ssDNA from SSB to which PriA can then bind, leading to formation of a PriA/SSB/DNA ternary complex where PriA holds ssDNA in preparation for reloading of the replicative DnaB helicase. Additionally, PriA translocase activity could possibly “push” SSB along the lagging-strand template to expose additional ssDNA to which primosome components (PriB and DnaT) can bind and that will ultimately serve as a binding site for DnaB. A similar “pushing” mechanism in which SSB is promoted along DNA by a directional enzyme has been demonstrated for the RecA recombinase in vitro (47).
Taken together, these models provide insights into the molecular mechanisms that govern the initial steps of replication restart in bacteria. Through a combination of its structure-specific DNA-binding and unwinding properties and its ability to manipulate SSB/DNA complexes, PriA appears to have adapted to function on a variety of abandoned DNA replication fork structures that could form in response to diverse challenges during replication in cells. We speculate that these mechanistic features allow PriA to act selectively on bona fide sites for replication restart while avoiding the potential problems that would arise from nonselective replication restart in other loci throughout the genome. Similar core biochemical activities may form minimal functional requirements that could aid in the identification of eukaryotic replication restart factors.
Materials and Methods
Detailed experimental procedures used in this study can be found in SI Materials and Methods and in Tables S2 and S3. A summary of the experimental procedures follows.
Structure Determination.
KpPriA and selenomethionine-substituted KpPriA were crystallized using the hanging-drop vapor diffusion method, and the structure of apo KpPriA was determined by a combination of single-wavelength anomalous dispersion and molecular replacement phasing. The structure of the KpPriA/SSB-Ct peptide complex was determined by molecular replacement using the apo KpPriA structure as a search model. Coordinate and structure factor files for apo KpPriA and the KpPriA/SSB-Ct peptide complex crystal structures are available at the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB ID codes: 4NL4 and 4NL8, respectively).
DNA-Binding Experiments.
Equilibrium DNA binding by PriA proteins or domains was measured as protein-dependent changes in fluorescence anisotropy of fluorescein-labeled DNA.
smFRET Experiments.
Partial duplex DNA with donor (Cy3) and acceptor (Cy5) labels was immobilized on a quartz slide and first bound by SSB and subsequently by EcPriA or an EcPriA variant. Total internal fluorescence microscope-measured time-resolved FRET of individual molecules and FRET efficiency histograms were generated from >5,000 molecules each.
Supplementary Material
Acknowledgments
We thank Kenneth Satyshur and staff at the Advanced Photon Source (Life Science Collaborative Access Team) for assistance with data collection. We thank James Berger, Matthew Lopper, and members of the J.L.K. laboratory for manuscript review. This work was supported by National Institutes of Health (NIH) Grants GM098885 (to J.L.K. and S.J.S.) and GM065367 (to T.H.) and by National Science Foundation Grant 0822613 (to T.H.). T.H. is an employee of the Howard Hughes Medical Institute. B.B. and S.R.W. were supported, in part, by an NIH training grant in Molecular Biosciences (Grant GM07215).
Footnotes
Conflict of interest statement: J.L.K. is a cofounder of Replisoma, Inc.
This article is a PNAS Direct Submission.
Data deposition: The structure-factor and atomic coordinate files for PriA and the PriA/SSB complex have been submitted to the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB ID codes 4NL4 and 4NL8).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318001111/-/DCSupplemental.
References
- 1.Cox MM, et al. The importance of repairing stalled replication forks. Nature. 2000;404(6773):37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 2.Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006;7(12):932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- 3.Merrikh H, Zhang Y, Grossman AD, Wang JD. Replication-transcription conflicts in bacteria. Nat Rev Microbiol. 2012;10(7):449–458. doi: 10.1038/nrmicro2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeeles JT, Poli J, Marians KJ, Pasero P. Rescuing stalled or damaged replication forks. Cold Spring Harb Perspect Biol. 2013;5(5):a012815. doi: 10.1101/cshperspect.a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gabbai CB, Marians KJ. Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival. DNA Repair (Amst) 2010;9(3):202–209. doi: 10.1016/j.dnarep.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masai H, Tanaka T, Kohda D. Stalled replication forks: Making ends meet for recognition and stabilization. Bioessays. 2010;32(8):687–697. doi: 10.1002/bies.200900196. [DOI] [PubMed] [Google Scholar]
- 7.McGlynn P, Al-Deib AA, Liu J, Marians KJ, Lloyd RG. The DNA replication protein PriA and the recombination protein RecG bind D-loops. J Mol Biol. 1997;270(2):212–221. doi: 10.1006/jmbi.1997.1120. [DOI] [PubMed] [Google Scholar]
- 8.Nurse P, Liu J, Marians KJ. Two modes of PriA binding to DNA. J Biol Chem. 1999;274(35):25026–25032. doi: 10.1074/jbc.274.35.25026. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka T, Mizukoshi T, Sasaki K, Kohda D, Masai H. Escherichia coli PriA protein, two modes of DNA binding and activation of ATP hydrolysis. J Biol Chem. 2007;282(27):19917–19927. doi: 10.1074/jbc.M701848200. [DOI] [PubMed] [Google Scholar]
- 10.Manhart CM, McHenry CS. The PriA replication restart protein blocks replicase access prior to helicase assembly and directs template specificity through its ATPase activity. J Biol Chem. 2013;288(6):3989–3999. doi: 10.1074/jbc.M112.435966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee MS, Marians KJ. Escherichia coli replication factor Y, a component of the primosome, can act as a DNA helicase. Proc Natl Acad Sci USA. 1987;84(23):8345–8349. doi: 10.1073/pnas.84.23.8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lasken RS, Kornberg A. The primosomal protein n′ of Escherichia coli is a DNA helicase. J Biol Chem. 1988;263(12):5512–5518. [PubMed] [Google Scholar]
- 13.Cadman CJ, McGlynn P. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 2004;32(21):6378–6387. doi: 10.1093/nar/gkh980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lecointe F, et al. Anticipating chromosomal replication fork arrest: SSB targets repair DNA helicases to active forks. EMBO J. 2007;26(19):4239–4251. doi: 10.1038/sj.emboj.7601848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozlov AG, Jezewska MJ, Bujalowski W, Lohman TM. Binding specificity of Escherichia coli single-stranded DNA binding protein for the chi subunit of DNA pol III holoenzyme and PriA helicase. Biochemistry. 2010;49(17):3555–3566. doi: 10.1021/bi100069s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol. 2008;43(5):289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones JM, Nakai H. Escherichia coli PriA helicase: Fork binding orients the helicase to unwind the lagging strand side of arrested replication forks. J Mol Biol. 2001;312(5):935–947. doi: 10.1006/jmbi.2001.4930. [DOI] [PubMed] [Google Scholar]
- 18.Chen HW, North SH, Nakai H. Properties of the PriA helicase domain and its role in binding PriA to specific DNA structures. J Biol Chem. 2004;279(37):38503–38512. doi: 10.1074/jbc.M404769200. [DOI] [PubMed] [Google Scholar]
- 19.Shlomai J, Kornberg A. A prepriming DNA replication enzyme of Escherichia coli. II. Actions of protein n′: A sequence-specific, DNA-dependent ATPase. J Biol Chem. 1980;255(14):6794–6798. [PubMed] [Google Scholar]
- 20.Low RL, Shlomai J, Kornberg A. Protein n, a primosomal DNA replication protein of Escherichia coli. Purification and characterization. J Biol Chem. 1982;257(11):6242–6250. [PubMed] [Google Scholar]
- 21.Lopper M, Boonsombat R, Sandler SJ, Keck JL. A hand-off mechanism for primosome assembly in replication restart. Mol Cell. 2007;26(6):781–793. doi: 10.1016/j.molcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng JY, Marians KJ. The ordered assembly of the phiX174-type primosome. I. Isolation and identification of intermediate protein-DNA complexes. J Biol Chem. 1996;271(26):15642–15648. doi: 10.1074/jbc.271.26.15642. [DOI] [PubMed] [Google Scholar]
- 23.Ng JY, Marians KJ. The ordered assembly of the phiX174-type primosome. II. Preservation of primosome composition from assembly through replication. J Biol Chem. 1996;271(26):15649–15655. doi: 10.1074/jbc.271.26.15649. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Nurse P, Marians KJ. The ordered assembly of the phiX174-type primosome. III. PriB facilitates complex formation between PriA and DnaT. J Biol Chem. 1996;271(26):15656–15661. doi: 10.1074/jbc.271.26.15656. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka T, et al. DNA binding of PriA protein requires cooperation of the N-terminal D-loop/arrested-fork binding and C-terminal helicase domains. J Biol Chem. 2002;277(41):38062–38071. doi: 10.1074/jbc.M204397200. [DOI] [PubMed] [Google Scholar]
- 26.Sasaki K, et al. Structural basis of the 3′-end recognition of a leading strand in stalled replication forks by PriA. EMBO J. 2007;26(10):2584–2593. doi: 10.1038/sj.emboj.7601697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka T, Masai H. Stabilization of a stalled replication fork by concerted actions of two helicases. J Biol Chem. 2006;281(6):3484–3493. doi: 10.1074/jbc.M510979200. [DOI] [PubMed] [Google Scholar]
- 28.Mizukoshi T, Tanaka T, Arai K, Kohda D, Masai H. A critical role of the 3′ terminus of nascent DNA chains in recognition of stalled replication forks. J Biol Chem. 2003;278(43):42234–42239. doi: 10.1074/jbc.C300285200. [DOI] [PubMed] [Google Scholar]
- 29.Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10(1):110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 30.Dillingham MS, Soultanas P, Wigley DB. Site-directed mutagenesis of motif III in PcrA helicase reveals a role in coupling ATP hydrolysis to strand separation. Nucleic Acids Res. 1999;27(16):3310–3317. doi: 10.1093/nar/27.16.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zittel MC, Keck JL. Coupling DNA-binding and ATP hydrolysis in Escherichia coli RecQ: Role of a highly conserved aromatic-rich sequence. Nucleic Acids Res. 2005;33(22):6982–6991. doi: 10.1093/nar/gki999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soultanas P, Dillingham MS, Velankar SS, Wigley DB. DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase. J Mol Biol. 1999;290(1):137–148. doi: 10.1006/jmbi.1999.2873. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Bushnell DA, Silva DA, Huang X, Kornberg RD. Initiation complex structure and promoter proofreading. Science. 2011;333(6042):633–637. doi: 10.1126/science.1206629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zavitz KH, Marians KJ. Helicase-deficient cysteine to glycine substitution mutants of Escherichia coli replication protein PriA retain single-stranded DNA-dependent ATPase activity. Zn2+ stimulation of mutant PriA helicase and primosome assembly activities. J Biol Chem. 1993;268(6):4337–4346. [PubMed] [Google Scholar]
- 35.Heller RC, Marians KJ. Unwinding of the nascent lagging strand by Rep and PriA enables the direct restart of stalled replication forks. J Biol Chem. 2005;280(40):34143–34151. doi: 10.1074/jbc.M507224200. [DOI] [PubMed] [Google Scholar]
- 36.Lee MS, Marians KJ. Differential ATP requirements distinguish the DNA translocation and DNA unwinding activities of the Escherichia coli PRI A protein. J Biol Chem. 1990;265(28):17078–17083. [PubMed] [Google Scholar]
- 37.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484(7393):256–259. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 38.Lu D, Keck JL. Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I. Proc Natl Acad Sci USA. 2008;105(27):9169–9174. doi: 10.1073/pnas.0800741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marceau AH, et al. Structure of the SSB-DNA polymerase III interface and its role in DNA replication. EMBO J. 2011;30(20):4236–4247. doi: 10.1038/emboj.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryzhikov M, Koroleva O, Postnov D, Tran A, Korolev S. Mechanism of RecO recruitment to DNA by single-stranded DNA binding protein. Nucleic Acids Res. 2011;39(14):6305–6314. doi: 10.1093/nar/gkr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inoue J, et al. A mechanism for single-stranded DNA-binding protein (SSB) displacement from single-stranded DNA upon SSB-RecO interaction. J Biol Chem. 2011;286(8):6720–6732. doi: 10.1074/jbc.M110.164210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LeBowitz JH, McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J Biol Chem. 1986;261(10):4738–4748. [PubMed] [Google Scholar]
- 43.Bujalowski W, Lohman TM. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry. 1986;25(24):7799–7802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- 44.Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J Mol Biol. 2007;369(5):1244–1257. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou R, et al. SSB functions as a sliding platform that migrates on DNA via reptation. Cell. 2011;146(2):222–232. doi: 10.1016/j.cell.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wessel SR, et al. PriC-mediated DNA replication restart requires PriC complex formation with the single-stranded DNA-binding protein. J Biol Chem. 2013;288(24):17569–17578. doi: 10.1074/jbc.M113.478156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461(7267):1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.