Significance
ADP/ATP carriers are archetypal members of the mitochondrial carrier family of transport proteins, which are thought to operate by a common but unresolved mechanism. Members of this family play key roles in many aspects of cell physiology and are implicated in several severe human diseases. Here, we present the structures of Aac2p and Aac3p, ADP/ATP carriers from Saccharomyces cerevisiae, determined by X-ray crystallography. Together with mutagenesis and functional assays, the structures support an alternating-access transport mechanism involving domain-based motions, where salt-bridge networks act as gates, providing access to a central substrate-binding site.
Keywords: membrane protein, cardiolipin binding, X-ray crystallography, serine kinks, adenine nucleotide translocase
Abstract
The mitochondrial ADP/ATP carrier imports ADP from the cytosol and exports ATP from the mitochondrial matrix. The carrier cycles by an unresolved mechanism between the cytoplasmic state, in which the carrier accepts ADP from the cytoplasm, and the matrix state, in which it accepts ATP from the mitochondrial matrix. Here we present the structures of the yeast ADP/ATP carriers Aac2p and Aac3p in the cytoplasmic state. The carriers have three domains and are closed at the matrix side by three interdomain salt-bridge interactions, one of which is braced by a glutamine residue. Glutamine braces are conserved in mitochondrial carriers and contribute to an energy barrier, preventing the conversion to the matrix state unless substrate binding occurs. At the cytoplasmic side a second salt-bridge network forms during the transport cycle, as demonstrated by functional analysis of mutants with charge-reversed networks. Analyses of the domain structures and properties of the interdomain interfaces indicate that interconversion between states involves movement of the even-numbered α-helices across the surfaces of the odd-numbered α-helices by rotation of the domains. The odd-numbered α-helices have an L-shape, with proline or serine residues at the kinks, which functions as a lever-arm, coupling the substrate-induced disruption of the matrix network to the formation of the cytoplasmic network. The simultaneous movement of three domains around a central translocation pathway constitutes a unique mechanism among transport proteins. These findings provide a structural description of transport by mitochondrial carrier proteins, consistent with an alternating-access mechanism.
Mitochondrial carriers are a family of proteins that transport a diverse range of nucleotides, amino acids, inorganic ions, fatty acids, keto acids, and cofactors across the inner mitochondrial membrane (1). The carriers link the biochemical pathways in the cytoplasm with those in the mitochondrial matrix, thereby playing key roles in many aspects of cell physiology. There are many rare, but severe, human diseases associated with defective mitochondrial carriers (2).
The ADP/ATP carriers are archetypal members of the mitochondrial carrier family (3). ADP/ATP carriers play the essential role of importing ADP into the mitochondrial matrix, where it can be phosphorylated by ATP synthase, and of exporting newly synthesized ATP into the cytosol, replenishing the cell with metabolic energy. ADP/ATP carriers have been intensively studied, because of their high natural abundance and the availability of specific inhibitors, which lock the carrier in two distinct states. The atractylosides, such as carboxyatractyloside (CATR) (4, 5), lock the carrier in the cytosolic state (c-state) with the substrate-binding site accessible to the intermembrane space, which is confluent with the cytosol. Bongkrekic acid (6) locks the carrier in the matrix state (m-state), with the substrate-binding site accessible to the mitochondrial matrix.
Saccharomyces cerevisiae contains three isoforms of the ADP/ATP carrier, AAC1 (7), AAC2 (8), and AAC3 (9). Aac2p is the principal ADP/ATP carrier expressed in aerobically growing yeast, whereas Aac1p is only expressed at low levels (10, 11). Aac3p is expressed almost exclusively under anaerobic growth conditions (9), where it is thought to transport ATP produced by glycolysis into the mitochondrion (12). Our current understanding of the function of mitochondrial carriers depends largely upon studies of Aac2p (3, 10, 11, 13–15).
Mitochondrial carriers consist of three tandem related sequences, each proposed to be folded into two transmembrane α-helices linked by an extrinsic region (16), and each containing the signature motif Px[DE]xx[KR] (17). The projection structure of atractyloside-inhibited Aac3p in a lipid environment revealed a monomer composed of six transmembrane α-helices surrounding a central translocation pathway, arranged with threefold pseudosymmetry (18). The atomic structure of the bovine ADP/ATP carrier (AAC1) in complex with CATR (19, 20) also showed three domains, each consisting of an odd-numbered transmembrane helix, a loop including a short matrix helix, and an even-numbered transmembrane helix. Proline residues in the highly conserved Px[DE]xx[KR] signature motif are found at sharp kinks in the odd-numbered helices. The charged residues of the motif form salt bridges on the matrix side, closing the central cavity to the matrix. These salt bridges constitute the matrix salt-bridge network.
The transport mechanism of mitochondrial carriers is unresolved. Previous proposals for the mechanism were based on a functional dimer (19, 21, 22), but subsequent work has shown that carriers are functionally monomeric (23, 24). The first step toward a mechanism consistent with a monomeric carrier was the identification of a single substrate-binding site in the central cavity, as shown by distance and chemical constraints (25), molecular dynamics simulations (26, 27), sequence analysis (28), and mutagenesis (29). Sequence analysis of the three homologous repeats found in members of the mitochondrial carrier family highlighted a conserved motif [YF][DE]xx[KR] on the cytoplasmic side, which has been suggested to form a salt-bridge network when the carrier is in the m-state (28). However, there is no experimental evidence that this network forms, and not all residues of this network have been modeled in the c-state, because density for the extreme C-terminal region is missing in the bovine AAC1 structures (19, 20). Unfortunately, the m-state is highly unstable in detergent solution, preventing its structural characterization.
Here, we have determined the atomic structures of CATR-inhibited Aac2p and Aac3p by X-ray crystallography. Each isoform was recombinantly expressed and crystallized in two different crystal forms. The structures reveal differences between the yeast and bovine orthologs and new features important for the mechanism. Based on structural and functional analyses, we provide evidence that mitochondrial carriers function via a domain-based alternating-access transport mechanism.
Results
Overall Structures of Aac2p and Aac3p.
Structures of Aac2p were determined using data to 2.5-Å and 3.2-Å resolution from a C2221 and P212121 crystal, respectively. Structures of Aac3p were solved using data collected to 3.4-Å and 3.2-Å resolution from a P212121 and P21 crystal of Aac3p, respectively. Data collection and refinement statistics are shown in SI Appendix, Table S1. Aac2p and Aac3p share 52% sequence identity with bovine AAC1, and 90% sequence identity with each other. The ability to compare structures determined in different crystal forms allows the significance of similarities and differences in the structures to be assessed.
The yeast ADP/ATP carriers consist of six transmembrane α-helices (H1–H6), which are tilted by about 45° with respect to the membrane plane (Fig. 1 A and B). The odd-numbered transmembrane helices are kinked at proline or serine residues (discussed later), so that the carrier forms a barrel-like structure. The central cavity is open to the cytoplasmic side of the membrane, but closed to the mitochondrial matrix. There are three short α-helices on the matrix side (h12, h34, and h56) that are positioned at the surface of the membrane. Aac2p and Aac3p show threefold pseudosymmetry, in agreement with earlier observations (16, 18). The overall fold is similar to that of the bovine ADP/ATP carrier (19) (SI Appendix, Table S2). Comparing all structures reveals differences in the positions of helices, predominantly on the cytoplasmic side, especially for H1 and H6 (SI Appendix, Fig. S1). Differences in these regions are also seen between copies related by noncrystallographic symmetry, and are reflected in higher-temperature factors for these regions, indicative of the dynamic nature of the molecule. Electron density maps for all structures show clear density for the inhibitor CATR, bound in the central cavity, with similar binding interactions to the bovine AAC1 structures (19, 20) (SI Appendix, Note S1, Fig. S2, and Table S3). Several of the yeast ADP/ATP carrier crystal forms reveal the presence of crystallographic dimers, but no consistent dimerization interface is found, confirming that carriers are structurally monomeric (SI Appendix, Note S2, and Fig. S3).
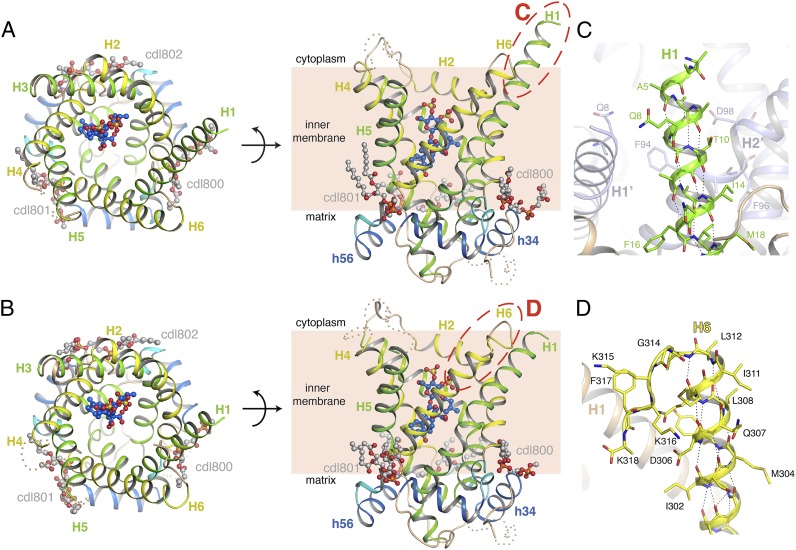
Fig. 1.
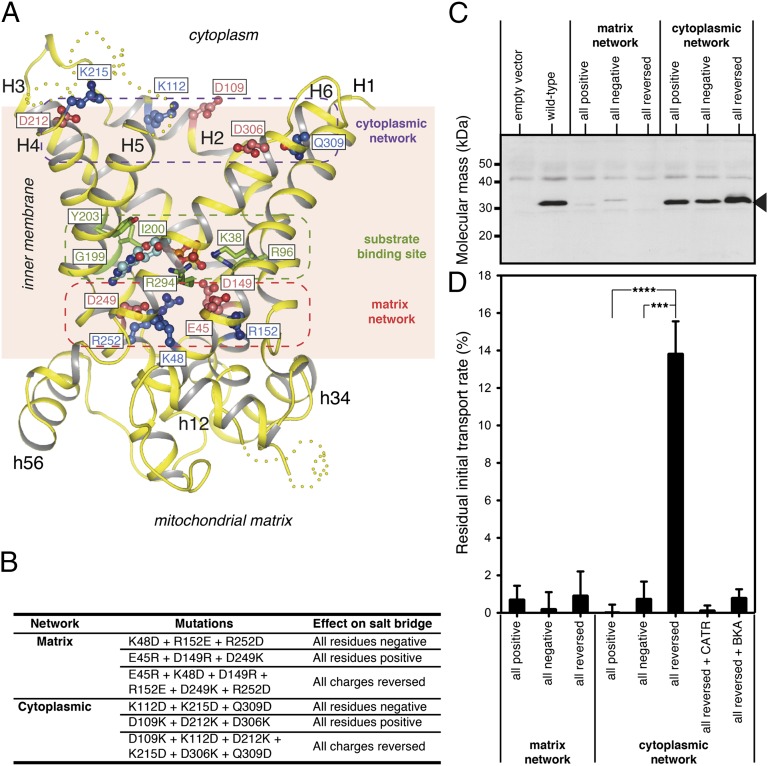
Architecture of the yeast ADP/ATP carriers. (A) Aac3p (chain B of the P21 crystal), viewed from the cytoplasm (Left) and from the membrane (Right). (B) Equivalent views of Aac2p (chain A of the P212121 crystal). Odd-numbered helices are shown in green, even-numbered helices in yellow, matrix helices in blue, and linker helices in cyan. Cardiolipin and CATR molecules are shown in ball-and-stick representation, with gray-colored carbons for cardiolipin and blue-colored carbons for CATR. The cytoplasmic loop between H4 and H5 and part of the loop between H3 and matrix helix h34 are missing, and are indicated as wheat-colored dots, following the position of these elements in the bovine carrier. Red-dashed ovals show the location of the close-ups in C and D. (C) The N-terminal region of Aac3p (chain B of the P21 crystal). H1 is in green, and the rest of the structure in wheat. A symmetry-related molecule is shown in pale blue, and residues making crystal contacts to the N-terminal region are shown as sticks. (D) The C-terminal region of Aac2p (chain A of the P212121 crystal). The C-terminal region is highlighted in yellow. In C and D, hydrogen bonds are shown by black-dotted lines. Side-chains for some residues in the N- and C-terminal regions have poor density, and have therefore been modeled to the Cβ atom.
Sequences of yeast ADP/ATP carriers show that they have a significantly extended N-terminal region compared with the mammalian orthologs. The P21 Aac3p crystal shows that H1 extends almost to the N terminus, protruding about 8 Å from the cytoplasmic surface of the protein (Fig. 1 A and C). This part is ordered because it is sandwiched between H1 and H2 of a neighboring molecule, whereas there is no density for it in the other crystal forms. Compared with AAC1, the yeast carriers have an extra turn of α-helix on the matrix end of H1 (SI Appendix, Fig. S4).
The P212121 crystal of Aac2p reveals the position of the C-terminal region, which is complete in molecule A (Fig. 1D). H6 shows regular α-helical hydrogen bonding up to residue Phe313, but then adopts a sharp turn at Gly314 (highly conserved among fungal and plant ADP/ATP carriers) and surprisingly folds back into the cavity. There are few interactions between the C terminus and the rest of the protein, although Phe317 (or Tyr in other fungal and plant ADP/ATP carriers) forms hydrophobic contacts with Met29 from H1, and an anion-π interaction with Asp26 (30), both conserved residues in fungal and plant ADP/ATP carriers. The density for molecule B in the Aac2p P212121 crystal and the molecules in the Aac3p crystal forms are consistent with the C-terminal region adopting the same fold.
Apart from the N-terminal region, there are no obvious structural differences between Aac2p and Aac3p. Both carriers can fully complement growth of the S. cerevisiae WB12 strain on nonfermentable carbon sources, indicating that the transport rates are not growth limiting. These results indicate that different isoforms exist to allow differential responses at the level of gene expression to changes in aerobic conditions.
Cardiolipin–Protein Interactions.
Cardiolipin is known to bind tightly to ADP/ATP carriers (31). Electron density maps reveal the presence of three cardiolipin molecules bound to the yeast carriers, in a pocket between the matrix helices and the matrix end of the even-numbered transmembrane helices (Fig. 1 A and B). The cardiolipins lie in a similar position to those modeled in the bovine AAC1 structures (19, 20). The electron density is strong for the phosphate moieties and glycerol backbone, but is present for only part of the acyl chains.
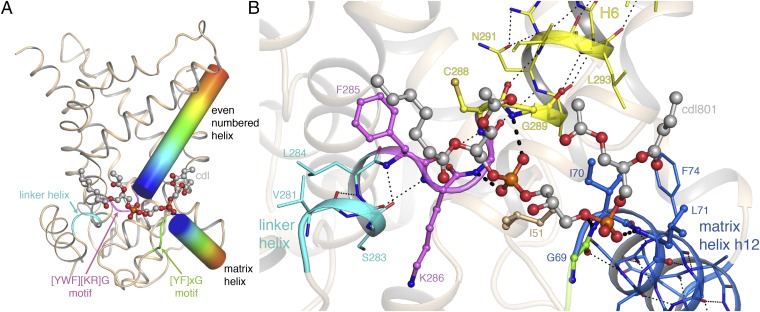
The 21 new observations of cardiolipin-binding sites make it possible to identify the common factors mediating lipid-protein interactions (SI Appendix, Table S4). Two highly conserved glycine residues lie close to the cardiolipin phosphate moieties, and play a key role in forming the binding-site. One residue is part of a highly conserved [YWF][KR]G motif, which is crucial for function (32) and is responsible for the break between the linker and even-numbered helices, and the offset in their positions. The other glycine is part of a conserved [YF]xG motif at the N-terminal ends of the matrix helices in the majority of carrier sequences. The negatively charged phosphate groups of the cardiolipin molecules are positioned near the N-terminal ends of the helices. Here, they interact with the positively charged end of the helix dipoles (Fig. 2A) and are able to form hydrogen bonds to the exposed amide groups (Fig. 2B and SI Appendix, Table S4).
Fig. 2.
Cardiolipin binding to Aac2p. (A) Schematic representation of the key elements of the cardiolipin-binding sites. The even-numbered and matrix helices for one binding site are shown as cylinders, with a rainbow color scheme (N terminus, blue; C terminus, red), the linker helix is shown in cyan. (B) Detailed view of the binding site for cardiolipin (Cdl801 in chain A of the P212121 crystal form). Residues in the linker helix, even-numbered helix, and matrix helix are shown with cyan, yellow, and blue carbon atoms, respectively. Hydrogen bonds within these helices are shown as thin black-dotted lines. Residues in the [YWF][KR]G and [YF]xG motifs are shown in violet and green, respectively. Residues contacting cardiolipin are shown with spheres. Hydrogen-bond interactions between protein and cardiolipin are shown as thick black-dashed lines. Electron density is present for only part of the cardiolipin acyl chains, and they have therefore been modeled with truncated acyl chains.
Cardiolipin binding was originally thought to be electrostatic in nature and mediated by lysine residues (31); however, the positively charged residues of the [YWF][KR]G motif (Lys286 in Fig. 2B) are not involved in cardiolipin binding, with their side-chains oriented away from the lipid in all of the cardiolipin-binding sites examined, with one exception (SI Appendix, Table S4). The conserved aromatic residues of the motif are involved in hydrophobic contacts with the acyl chain. There is no conserved binding site for the cardiolipin acyl chains, and they are observed to adopt a range of conformations, making hydrophobic contacts with the protein.
The Serine Kink in H3 Mimics Proline.
The structure of bovine AAC1 showed that the proline residues in the mitochondrial carrier signature motifs (Px[DE]xx[KR]) lie at sharp kinks in the odd-numbered transmembrane helices (19). The structure around the signature motifs on H1 and H5 of the fungal ADP/ATP carriers shows the disruption of conventional α-helical hydrogen bonding at the proline, because of the loss of a backbone amide (Fig. 3A). The disruption, combined with steric restrictions at the proline, allows the kink in the helix. These prolines are highly conserved in the mitochondrial carrier family (28), but the proline on H3 is substituted by serine in over 40% of ADP/ATP carrier sequences (14), including Aac2p and Aac3p. This substitution is also found on H1 of yeast Ymr166c, a carrier of unknown function. Serine residues are frequently found near kinks in transmembrane helices because the side-chain can hydrogen bond to the backbone carbonyl of the i-3, i-4, or i+4 residue (33–35).
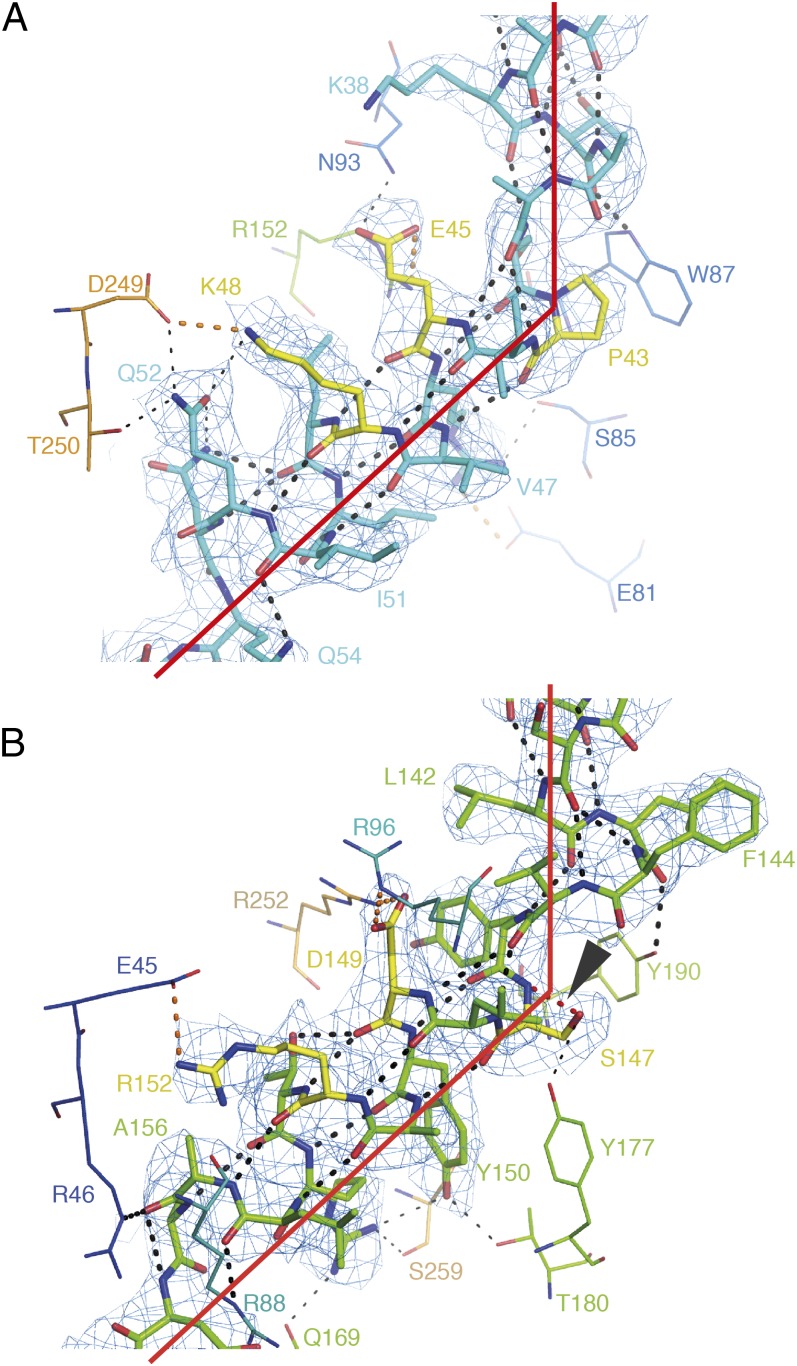
Fig. 3.
Proline and serine residues in the signature motifs of Aac2p. (A) The signature motif of H1 (cyan) and (B) of H3 (green) of Aac2p (chain A of P212121). Residues interacting with the side-chains are shown in line representation. Residues of the signature motif are shown in yellow. Hydrogen bonds involving backbone atoms are shown as thick black-dotted lines, hydrogen bonds not involving backbone atoms are shown as thin black-dotted lines. Salt bridges are shown as orange-dotted lines. The density, shown as a blue mesh, is a 2mFo-DFc map, contoured at 1 σ, and displayed within 2 Å of the atoms. In B, the unusual hydrogen bond from the side-chain of Ser147 to its own backbone amide is highlighted in red dots, and indicated by an arrowhead.
The serine substitution in Aac2p and Aac3p leaves the kink in H3 intact, with the 50° kink angle being similar to that observed for the proline-containing odd-numbered helices (Fig. 3B). In both cases, a network of interactions between amino acid side-chains helps to stabilize the extreme kink (Fig. 3). Unexpectedly, the structures of Aac2p and Aac3p show that the serine kink differs from those observed in other structures. Analysis of the optimal hydrogen-bonding arrangements (36) shows that the side-chain of Ser147 makes an unusual hydrogen bond to its own backbone amide group, mimicking the structure of proline. This arrangement is rare (37), but not without precedence (38, 39). The side-chain of Ser147 also forms a hydrogen bond with the side-chain of Tyr177 on h34, a highly conserved amino acid among ADP/ATP carriers with serine in the signature motif on H3.
Glutamine Residues Brace the Salt Bridges of the Matrix Network.
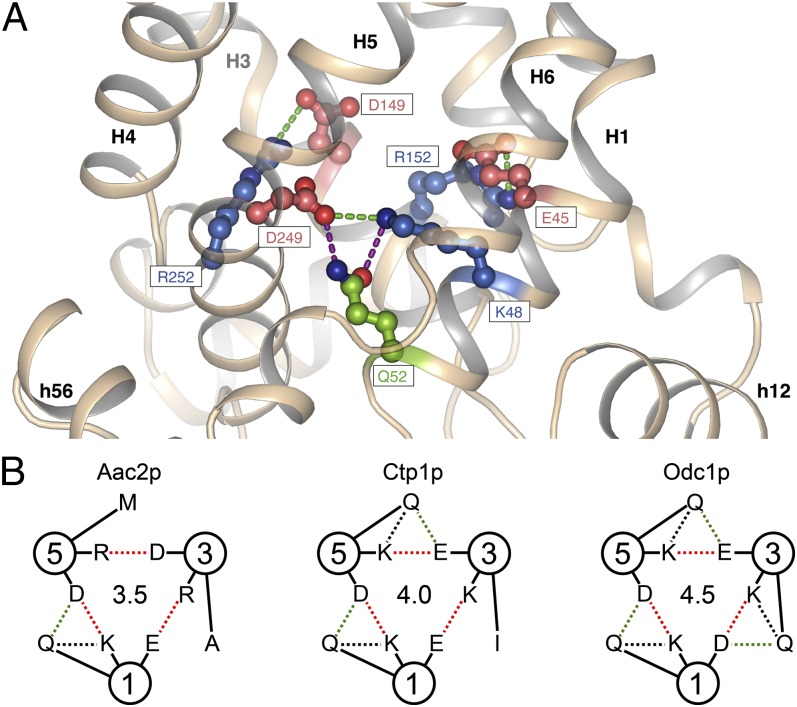
The salt-bridge network at the matrix side of the cavity is formed by the charged residues of the signature motif Px[DE]xx[KR] (15, 19). In all yeast-carrier structures, a conserved glutamine residue is found on the matrix side of the salt bridge between domains 1 and 3 (H1–H5), which forms hydrogen bonds with both charged residues (Fig. 4A). One hydrogen bond is between residues of the same domain (intradomain, H1–H1) and one between residues of domains 1 and 3 (interdomain, H1–H5). In this configuration the residue forms a brace, helping to hold the salt-bridge residues in place. Highly conserved glutamine residues can be found in an equivalent position in all three domains of mitochondrial carriers (SI Appendix, Fig. S5). Aac2p and Aac3p have only one brace, whereas the mitochondrial citrate carrier Ctp1 (40) and the oxodicarboxylate carrier Odc1p (41) are predicted to have two and three braces, respectively (Fig. 4B).
Fig. 4.
Glutamine residues brace salt bridge links of the matrix network. (A) Close-up of the bonding arrangement of glutamine with the salt bridge of the matrix network that links domains 1 and 3 of Aac2p. (B) Interactions of glutamine residues with salt-bridge residues of the matrix network of Aac2p and the yeast citrate carrier Ctp1 and oxodicarboxylate carrier Odc1p. The circles represent the odd-numbered α-helices H1, H3, and H5, with the residues indicated in the one-letter amino acid code. The interdomain and intradomain polar interactions with glutamine are shown in green- and black-dashed lines, respectively, whereas the salt bridges are shown in red. The central numbers give an estimate of the strength of interaction network in terms of number of salt bridges, assuming that hydrogen bonds have about half the interaction energy of salt bridges.
Experimental Evidence for Formation of a Cytoplasmic Salt-Bridge Network During the Transport Cycle.
Analysis of the three homologous sequence repeats found throughout the carrier family highlighted a conserved motif [YF][DE]xx[KR] on the cytoplasmic side, which could form a salt-bridge network when the carrier is in the m-state (28). The atomic models of the bovine ADP/ATP carrier lack one of the residues of this putative cytoplasmic salt-bridge network because it is in the extreme C-terminal region, which is missing from the models. The C-terminal region of the yeast ADP/ATP carriers is fully resolved (Fig. 1D), allowing all residues of the proposed cytoplasmic network to be modeled. Both the matrix and putative cytoplasmic networks are at the water–membrane interface, consistent with their role as gates in an alternating-access mechanism (Fig. 5A). The residues of the proposed cytoplasmic network are positioned such that they could interact upon closure of the cavity at the cytoplasmic side.
Fig. 5.
The cytoplasmic salt-bridge network forms during the transport cycle. (A) Lateral view of the structure of the yeast ADP/ATP carrier Aac2p with the positively charged (blue) and negatively charged (red) residues of the matrix and cytoplasmic salt-bridge networks indicated. Shown also are the contact points of the substrate-binding site (green) interacting with ADP (light blue) docked in the site. (B) Mutations introduced into the matrix and cytoplasmic networks to change the residues to all positively charged, all negatively charged, and fully reversed matrix or cytoplasmic salt-bridge networks. (C) Western blot of lactococcal membranes expressing wild-type and mutant Aac2p carriers. An antibody raised against Neurospora crassa ADP/ATP carrier was used and the band for Aac2p is indicated by an arrowhead. Approximately 10 μg total protein was loaded per lane. (D) Residual ADP uptake rate corrected for background of mutant Aac2p carriers compared with the wild-type rate in fused lactococcal membranes. The specific initial uptake rate of wild-type Aac2p (100%) was 0.788 nmol⋅min−1⋅mg−1 of protein. For active transporters the residual rates were also determined in the presence of CATR and bongkrekic acid (BKA). The initial transport rates were determined in the linear part of the uptake curve. The data are represented by the average plus SD of three independent experiments. Student t test P values are indicated: ***P ≤ 0.001 and ****P ≤ 0.0001.
To test the hypothesis that the cytoplasmic salt bridge forms during the transport cycle, residues of the matrix and proposed cytoplasmic network were mutated to create a network with only positively charged residues, only negatively charged residues, or with residues that have the opposite charge (Fig. 5B). The mutant carriers with the altered networks were expressed in the cytoplasmic membrane of Lactococcus lactis. The mutants of the matrix network did not express (Fig. 5C), indicating a crucial role in the biogenesis of the carrier, and no significant transport activity could be measured (Fig. 5D). In contrast, mutants of the putative cytoplasmic network expressed to approximately wild-type levels (Fig. 5C). Mutant carriers with an altered cytoplasmic network in which the six residues were either all positively or all negatively charged (three mutations each) were not active, as the same charge may oppose their interaction. However, when further mutations were introduced to interchange the residues of the cytoplasmic network (six mutations in total), the transport activity was restored to ∼14% of the wild-type rate (Fig. 5D). The transport activity of the mutant with the reversed network was fully sensitive to CATR and bongkrekic acid, like the wild-type, demonstrating that the activity was specific and that the c- and m-states were still capable of binding inhibitors. These results are consistent with a transport mechanism in which salt bridges of the cytoplasmic network form through the interaction of positively and negatively charged residues.
Intradomain Interactions Predict Domain Rigidity.
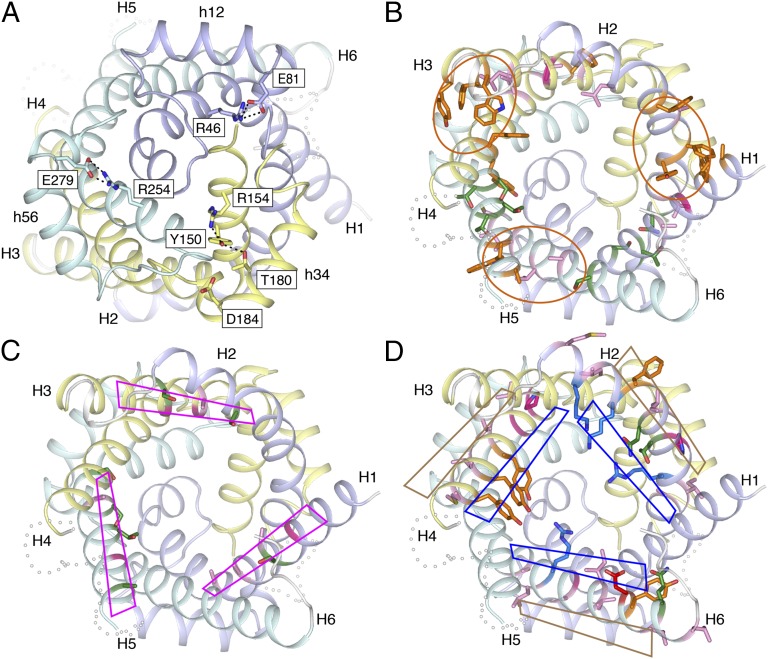
Three observations suggest that ADP/ATP carriers may function by domain motions. First, considering the bovine AAC1, Aac2p, and higher-resolution Aac3p structures, there are significantly more intradomain polar interactions than interdomain interactions (P value 0.02, paired two-tailed t test) (SI Appendix, Table S5). More interactions are found on the matrix side of the carrier compared with the cytoplasmic side, indicating that these interactions stabilize the c-state. Second, interactions link the odd and matrix helices of a domain together. One of these intradomain interactions is between an arginine residue in H1 and H5 and a glutamate residue of a highly conserved EG motif at the C-terminal end of matrix helices h12 and h56, respectively (Fig. 6A). In bovine AAC1, only the intradomain interaction in domain 3 is observed (20). In domain 2 of the yeast carriers, the glutamate residue on matrix helix h34 is replaced by aspartate, whose shorter side-chain cannot bridge the distance to the arginine on H3. In Aac2p, the arginine (Arg154) is linked to Thr180 in the matrix helix via a hydrogen-bond network involving Tyr150, compensating for the loss of interactions with Asp184. On the basis of sequence conservation, these intradomain interactions are likely to be important for carriers in general. Third, at the cytoplasmic side aromatic residues dominate the intradomain interhelical interface (Fig. 6B): five in the H1–H2 interface, five in the H3–H4 interface, and three in the H5–H6 interface. Many of these residues are conserved in all three domains of mitochondrial carriers, consistent with their importance for function (32, 42, 43). The residues are bulky, with few rotamers, allowing them to form a rigid interface. Clusters of small residues are found on the matrix side of the intradomain interfaces. Some of these clusters mediate intradomain interactions; for example, in Aac2p, Asn90, Thr39, and Ser42 comprise a hydrogen bond network in the H1–H2 intradomain interface, and a hydrophobic cluster is found in the H3–H4 intradomain interface (Ala139, Leu142, Phe193, and Val197). Together, these three observations are consistent with the helices of each domain moving together as a rigid-body.
Fig. 6.
Conserved properties of the intra- and interdomain interfaces suggest a domain-based mechanism of transport. (A) Intradomain hydrogen bonds and salt bridges link the matrix-side of the odd-numbered helices to the matrix helices. The networks involve negatively charged residues of the EG motif on the matrix helices. View from the matrix side of the membrane. (B) Many of the residues in the intradomain interface are aromatic or form hydrophobic or polar clusters. The aromatic clusters are highlighted by orange ovals. Side-chains for Phe23 and Phe231 are not modeled because of poor density. (C) Residues of the odd-numbered α-helices close to the interdomain interface have no or small side-chains, and include the conserved GxxxG motif. The interfaces are highlighted by magenta rhomboids. The side-chain of Ser230 is not modeled. (D) Residues of the even-numbered α-helices close to the interdomain interface are either hydrophobic and face the membrane, or are hydrophilic and face the cavity. The hydrophobic and hydrophilic faces are shown as brown and blue rhomboids, respectively. In A–D, domain 1 is colored light blue, domain 2 pale yellow, and domain 3 pale cyan. For B–D, the carrier is viewed from the cytoplasmic side and polar, aliphatic, aromatic, structural, positively charged, and negatively charged residues are shown in green, pink, orange, magenta, blue, and red, respectively.
Residues in the Interdomain Interface Predict Domain Motion.
The surface of the interdomain interfaces on the odd-numbered α-helices consists almost entirely of amino acid residues with no or small side-chains, except for the cytoplasmic ends (Fig. 6C). This includes the well-conserved symmetrical sequence motif GxxxG, where the glycines are positioned on the same face of the helix separated by one turn, which has been shown to be important for function (42). These motifs are commonly found in membrane proteins in regions of interhelical close contact (44). On the even-numbered α-helices, hydrophobic residues face the membrane and hydrophilic residues face the cavity, neither pointing toward the odd-numbered α-helices (Fig. 6D). The only exceptions are at the N-terminal end, where Ala92, Pro195 and Ala290 (Aac2p) point directly into the interface, but they have fairly small side-chains. These properties are consistent with a dynamic interdomain interface.
Discussion
In general, transporters of ions and small solutes have been proposed to function according to an alternating-access mechanism (3, 45). Key requirements for this mechanism are a central substrate-binding site and two flanking gates that regulate access to either side of the membrane. For mitochondrial carriers, no structural mechanism has been defined. The availability of atomic models of mitochondrial carriers from different species, and in different crystal forms, allows us to identify the molecular features that govern transport by an alternating-access mechanism.
The matrix salt-bridge network is part of the gate that closes the cavity to the matrix side of the membrane in the c-state (15, 19). The charged residues of the matrix salt-bridge network form hydrogen bonds to glutamine residues, which provide additional stabilization by forming a brace (Fig. 4A). The number of these braces differs between mitochondrial carriers of different function (Fig. 4B). The high degree of conservation of the glutamine residues in equivalent positions in the three domains indicates an important role for these interactions in the transport mechanism. It has been suggested previously that the interactions of the network provide an energy barrier that has to be overcome by substrate binding in order for the substrate to be translocated (28). The interdomain braces strengthen the interaction network and increase the magnitude of the energy barrier. The yeast ADP/ATP carriers have one brace and three salt bridges in the extended matrix network, providing a significant energy barrier that prevents conversion to the m-state in the absence of substrate. A single substrate-binding site has been identified in the central cavity, which corresponds to the middle of the membrane (25, 29). There is consensus on the residues involved in ADP binding to the ADP/ATP carrier (25–28). The adenine moiety is bound in a hydrophobic pocket, which in Aac2p consists of Gly199, Ile200, and Tyr203, and the major interaction is an aromatic stacking arrangement (Fig. 5A). The two phosphates, carrying three negative charges, are most likely to be bound by three positively charged residues Arg96, Arg294, Lys38 (28), and possibly also Arg253 (26, 27). Thus, the interaction energy of substrate binding matches that of the extended matrix salt-bridge network, allowing conversion to the m-state only when substrate is bound, enforcing a strict equimolar exchange mechanism (3, 28). The citrate (Ctp1) and oxodicarboxylate (Odc1) carrier might have two and three braces, respectively, indicating that the interaction energy of the extended network differs between carriers. Here the interaction energies could also balance if interactions with negatively charged residues involved in proton coupling are also considered (46). The extra helical turn on the matrix side of H1 in the yeast ADP/ATP carriers causes H1 to end close to the position of the central threefold pseudosymmetry axis (SI Appendix, Fig. S4), increasing the thickness of the matrix gate by ∼3 Å.
Here, experimental evidence for formation of the cytoplasmic salt-bridge network during transport is presented. This network forms part of the cytoplasmic gate when the carrier is open to the matrix side. The new structures reveal the position of the C-terminal region of the carriers, allowing all of the residues involved in the cytoplasmic salt-bridge network to be modeled. The C-terminal region folds back into the central cavity, and may form an additional component of the cytoplasmic gate at the cytoplasmic side of the network. Consistent with this role, large fluorescence changes are observed upon ATP binding in an Aac2p mutant that has a single tryptophan at the position of Phe317 (13). The thicker gates flanking the central substrate-binding site in the yeast ADP/ATP carriers have been reduced in the mammalian counterparts, which could reflect adaptations to challenges in different environments.
We have investigated the roles of the tripartite structural features in a transport mechanism in which each domain functions as a unit in the transport cycle. There are significantly more polar interactions within domains than between domains. Conserved hydrogen-bond networks link the odd and matrix helices of the domain together. There are significantly more polar interactions on the matrix side of the carrier than on the cytoplasmic side, where bulky aromatic residues lie between the helices of the domain. Taken together, these observations support the notion that the domains function as rigid-bodies in the transport cycle. The observation that the transmembrane helices are at a 45° tilt implies that the domain motions are within the membrane plane, otherwise domains would clash.
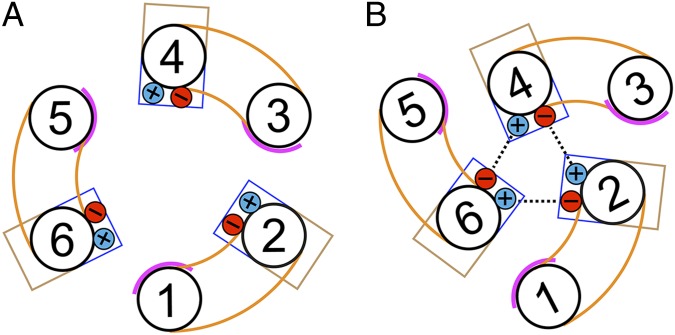
At the interdomain interface, the odd-numbered α-helices only have residues with small side-chains, whereas the even-numbered α-helices have residues that point either into the membrane or into the cavity. This arrangement would allow the even-numbered α-helices to move across the surface of the odd-numbered α-helices (Fig. 7). In this way, the hydrophobic residues on the even-numbered α-helices remain facing the membrane, whereas the hydrophilic residues keep facing the cavity. Once the even-numbered α-helices have moved into the center of the carrier, the positively and negatively charged residues of the cytoplasmic network engage, closing the carrier on the cytoplasmic side. For this mechanism to work, it is required that the axes of rotation are around the odd-numbered α-helices. The L-shape of the odd-numbered α-helices, because of sharp kinks at proline or serine residues (Fig. 3), plays a key role in this mechanism. Serine acts as a proline mimic, explaining why Ser147 can be mutated to Pro without dramatically changing the transport kinetics of the carrier (14). Proline or a proline mimic allows regular hydrogen bonding to be broken at the kink. Analysis of mutants at the proline position suggests that maintenance of the kink is crucial for transport (42). The L-shape is maintained by a network of interactions between amino acid side-chains, predominantly on the matrix side of the kink. The L-shaped arrangement could function as a lever, coupling outward motion at the matrix side of the odd-numbered α-helices during disruption of the matrix network to an inward motion of the cytoplasmic end of the even-numbered α-helices, allowing formation of the cytoplasmic network. The domain motions must be simultaneous, otherwise clashes and gaps in the protein structure would occur. A simultaneous motion of the three domains around a central translocation pathway is unique among transport proteins.
Fig. 7.
Proposed domain motions in the transport cycle. (A and B) Schematic representation of the helical arrangement at the cytoplasmic side in the c-state and m-state, respectively. The domain structures are shown in orange. Pink lines indicate the smooth surface of the odd-numbered α-helices in the interdomain interface. Blue and brown boxes represent hydrophilic and hydrophobic residues on the even-numbered α-helices at the interdomain interface, which face either the cavity or the membrane, respectively. The positive and negatively charged residues of the cytoplasmic salt-bridge network are shown in blue and red, respectively. The formation of the cytoplasmic salt-bridge network is shown as dashed lines.
The tightly bound cardiolipin molecules are important but not essential for function (47–49). The cardiolipins are positioned with their phosphate groups at the N-terminal ends of the even-numbered and matrix α-helices, acting as a cap. The phosphate moieities of cardiolipin provide hydrogen-bonding partners to the amide groups at the ends of the helices. Without these hydrogen-bond acceptors, polar amide groups would be exposed in a hydrophobic environment, which would be energetically unfavorable. The position of the cardiolipin phosphate groups also allows an electrostatic interaction with the helix dipoles, stabilizing binding of cardiolipin, which is likely to carry two negative charges at physiological pH (50). When modeled as a macrodipole in a low dielectric medium, a helix dipole provides the effect of isolated half-unit charges, positive at the N terminus and negative at the C terminus (51, 52). Theoretical models confirm a relatively large helix dipole effect for transmembrane helices, where one end is exposed to solvent and one within the lipid bilayer (53), as is observed for the even-numbered and matrix helices. Hydrogen bonding, together with electrostatic interactions with the helix dipoles, explains the tight binding of cardiolipin to ADP/ATP carriers.
Mitochondrial ADP/ATP carriers are one of the most highly expressed proteins in the inner membrane, which has a high protein density. The dimeric structure of cardiolipin links the even-numbered helix from one domain with the matrix helix from a neighboring domain. We propose that this interface is mobile, and thus cardiolipin may help protect a dynamic region of the carrier. Cardiolipin could allow close packing of protein monomers by preventing interactions with other proteins, which could impede conformational changes. Cardiolipin could therefore act as “grease” at a dynamic interface.
In conclusion, the structures combined with functional analysis of mutant carriers have allowed us to identify the molecular features involved in the interconversion between c- and m-states, consistent with an alternating-access mechanism. We have determined the structures of two mitochondrial ADP/ATP carriers by using recombinant, rather than native, proteins, which opens the way for structural studies of other mitochondrial carriers.
Methods
The AAC2 and AAC3 genes were each cloned into a modified pYES3 vector, with an N-terminal His tag and Factor Xa cleavage site upstream of the carrier gene. Recombinant Aac2p and Aac3p proteins were expressed in S. cerevisiae strain WB12. Mitochondria were prepared from disrupted cells, and protein solubilized with undecyl-β-d-maltoside. Proteins were purified by Ni Sepharose affinity chromatography, and exchanged into 5-cyclohexyl-1-pentyl-β-d-maltoside (Aac2p) or n-decyl-β-d-maltoside (Aac3p) while bound to the Ni Sepharose resin. Following Factor Xa cleavage and concentration, protein was crystallized using vapor diffusion techniques. Both proteins were crystallized in two different crystal forms, and data collection was performed at the European Synchrotron Radiation Facility beamline ID23-2. The structures were solved by molecular replacement, initially using the coordinates of the bovine ADP/ATP carrier (P21212 crystal form, PDB ID code 1OKC) as a search model. Functional assays were performed on protein expressed in Lactococcus lactis under the control of a nisin A-inducible promoter. Following expression, membranes were isolated and fused with liposomes, and transport rates determined by measuring uptake of 14C-labeled ADP. Detailed materials and methods can be found in SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Dr. Manfred Burghammer and Dr. David Flot, at the European Synchrotron Radiation Facility (Grenoble, France) beamlines ID13 and ID23-2, for excellent technical support with crystal screening and data collection; Dr. Shane Palmer for yeast fermenter runs; Dr. Yang Lee for insightful discussions; Drs. Jade Li, Andrew Leslie, and Garib Murshudov for advice with crystallographic analysis; and Dr. Richard Henderson and Prof. Sir John Walker for comments on the manuscript. Additional crystal screening and data collection were performed on beamline I24 at the Diamond Light Source, Didcot, UK, and at beamline X06SA at the Swiss Light Source, Villigen, Switzerland. This work was funded by the Medical Research Council and the European Community’s Seventh Framework Programme FP7/2007-2013 under Grant agreement HEALTH-F4-2007-201924, European Drug Initiative on Channels and Transporters Consortium.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates, structure factors, and Fourier map coefficients for the Aac2p C2221 and P212121 crystal forms, and for the Aac3p P21 and P212121 crystal forms have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4C9G, 4C9H, 4C9Q, and 4C9J, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320692111/-/DCSupplemental.
References
- 1.Palmieri F. The mitochondrial transporter family SLC25: Identification, properties and physiopathology. Mol Aspects Med. 2013;34(2–3):465–484. doi: 10.1016/j.mam.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Palmieri F. Diseases caused by defects of mitochondrial carriers: A review. Biochim Biophys Acta. 2008;1777(7–8):564–578. doi: 10.1016/j.bbabio.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Klingenberg M. The ADP and ATP transport in mitochondria and its carrier. Biochim Biophys Acta. 2008;1778(10):1978–2021. doi: 10.1016/j.bbamem.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 4.Duee ED, Vignais PV. [Exchange between extra- and intramitochondrial adenine nucleotides] Biochim Biophys Acta. 1965;107(1):184–188. doi: 10.1016/0304-4165(65)90419-8. French. [DOI] [PubMed] [Google Scholar]
- 5.Vignais PV, Vignais PM, Defaye G. Adenosine diphosphate translocation in mitochondria. Nature of the receptor site for carboxyatractyloside (gummiferin) Biochemistry. 1973;12(8):1508–1519. doi: 10.1021/bi00732a007. [DOI] [PubMed] [Google Scholar]
- 6.Henderson PJ, Lardy HA. Bongkrekic acid. An inhibitor of the adenine nucleotide translocase of mitochondria. J Biol Chem. 1970;245(6):1319–1326. [PubMed] [Google Scholar]
- 7.O’Malley K, Pratt P, Robertson J, Lilly M, Douglas MG. Selection of the nuclear gene for the mitochondrial adenine nucleotide translocator by genetic complementation of the op1 mutation in yeast. J Biol Chem. 1982;257(4):2097–2103. [PubMed] [Google Scholar]
- 8.Lawson JE, Douglas MG. Separate genes encode functionally equivalent ADP/ATP carrier proteins in Saccharomyces cerevisiae. Isolation and analysis of AAC2. J Biol Chem. 1988;263(29):14812–14818. [PubMed] [Google Scholar]
- 9.Kolarov J, Kolarova N, Nelson N. A third ADP/ATP translocator gene in yeast. J Biol Chem. 1990;265(21):12711–12716. [PubMed] [Google Scholar]
- 10.Lawson JE, Gawaz M, Klingenberg M, Douglas MG. Structure-function studies of adenine nucleotide transport in mitochondria. I. Construction and genetic analysis of yeast mutants encoding the ADP/ATP carrier protein of mitochondria. J Biol Chem. 1990;265(24):14195–14201. [PubMed] [Google Scholar]
- 11.Gawaz M, Douglas MG, Klingenberg M. Structure-function studies of adenine nucleotide transport in mitochondria. II. Biochemical analysis of distinct AAC1 and AAC2 proteins in yeast. J Biol Chem. 1990;265(24):14202–14208. [PubMed] [Google Scholar]
- 12.Visser W, et al. Involvement of mitochondria in the assimilatory metabolism of anaerobic Saccharomyces cerevisiae cultures. Microbiology. 1994;140(Pt 11):3039–3046. doi: 10.1099/13500872-140-11-3039. [DOI] [PubMed] [Google Scholar]
- 13.Clémençon B, et al. Structure-function relationships of the C-terminal end of the Saccharomyces cerevisiae ADP/ATP carrier isoform 2. J Biol Chem. 2008;283(17):11218–11225. doi: 10.1074/jbc.M709565200. [DOI] [PubMed] [Google Scholar]
- 14.Babot M, Blancard C, Pelosi L, Lauquin GJM, Trézéguet V. The transmembrane prolines of the mitochondrial ADP/ATP carrier are involved in nucleotide binding and transport and its biogenesis. J Biol Chem. 2012;287(13):10368–10378. doi: 10.1074/jbc.M111.320697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson DR. The yeast ADP/ATP carrier. Mutagenesis and second-site revertants. Biochim Biophys Acta. 1996;1275(1–2):133–137. doi: 10.1016/0005-2728(96)00063-1. [DOI] [PubMed] [Google Scholar]
- 16.Saraste M, Walker JE. Internal sequence repeats and the path of polypeptide in mitochondrial ADP/ATP translocase. FEBS Lett. 1982;144(2):250–254. doi: 10.1016/0014-5793(82)80648-0. [DOI] [PubMed] [Google Scholar]
- 17.Nelson DR, Felix CM, Swanson JM. Highly conserved charge-pair networks in the mitochondrial carrier family. J Mol Biol. 1998;277(2):285–308. doi: 10.1006/jmbi.1997.1594. [DOI] [PubMed] [Google Scholar]
- 18.Kunji ER, Harding M. Projection structure of the atractyloside-inhibited mitochondrial ADP/ATP carrier of Saccharomyces cerevisiae. J Biol Chem. 2003;278(39):36985–36988. doi: 10.1074/jbc.C300304200. [DOI] [PubMed] [Google Scholar]
- 19.Pebay-Peyroula E, et al. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426(6962):39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- 20.Nury H, et al. Structural basis for lipid-mediated interactions between mitochondrial ADP/ATP carrier monomers. FEBS Lett. 2005;579(27):6031–6036. doi: 10.1016/j.febslet.2005.09.061. [DOI] [PubMed] [Google Scholar]
- 21.Huang SG, Odoy S, Klingenberg M. Chimers of two fused ADP/ATP carrier monomers indicate a single channel for ADP/ATP transport. Arch Biochem Biophys. 2001;394(1):67–75. doi: 10.1006/abbi.2001.2520. [DOI] [PubMed] [Google Scholar]
- 22.Palmieri F, Indiveri C, Bisaccia F, Krämer R. Functional properties of purified and reconstituted mitochondrial metabolite carriers. J Bioenerg Biomembr. 1993;25(5):525–535. doi: 10.1007/BF01108409. [DOI] [PubMed] [Google Scholar]
- 23.Bamber L, Harding M, Monné M, Slotboom DJ, Kunji ER. The yeast mitochondrial ADP/ATP carrier functions as a monomer in mitochondrial membranes. Proc Natl Acad Sci USA. 2007;104(26):10830–10834. doi: 10.1073/pnas.0703969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunji ER, Crichton PG. Mitochondrial carriers function as monomers. Biochim Biophys Acta. 2010;1797(6-7):817–831. doi: 10.1016/j.bbabio.2010.03.023. [DOI] [PubMed] [Google Scholar]
- 25.Robinson AJ, Kunji ER. Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc Natl Acad Sci USA. 2006;103(8):2617–2622. doi: 10.1073/pnas.0509994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dehez F, Pebay-Peyroula E, Chipot C. Binding of ADP in the mitochondrial ADP/ATP carrier is driven by an electrostatic funnel. J Am Chem Soc. 2008;130(38):12725–12733. doi: 10.1021/ja8033087. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Tajkhorshid E. Electrostatic funneling of substrate in mitochondrial inner membrane carriers. Proc Natl Acad Sci USA. 2008;105(28):9598–9603. doi: 10.1073/pnas.0801786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson AJ, Overy C, Kunji ER. The mechanism of transport by mitochondrial carriers based on analysis of symmetry. Proc Natl Acad Sci USA. 2008;105(46):17766–17771. doi: 10.1073/pnas.0809580105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monné M, et al. Substrate specificity of the two mitochondrial ornithine carriers can be swapped by single mutation in substrate binding site. J Biol Chem. 2012;287(11):7925–7934. doi: 10.1074/jbc.M111.324855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philip V, et al. A survey of aspartate-phenylalanine and glutamate-phenylalanine interactions in the protein data bank: Searching for anion-π pairs. Biochemistry. 2011;50(14):2939–2950. doi: 10.1021/bi200066k. [DOI] [PubMed] [Google Scholar]
- 31.Beyer K, Klingenberg M. ADP/ATP carrier protein from beef heart mitochondria has high amounts of tightly bound cardiolipin, as revealed by 31P nuclear magnetic resonance. Biochemistry. 1985;24(15):3821–3826. doi: 10.1021/bi00336a001. [DOI] [PubMed] [Google Scholar]
- 32.Cappello AR, et al. Functional and structural role of amino acid residues in the even-numbered transmembrane alpha-helices of the bovine mitochondrial oxoglutarate carrier. J Mol Biol. 2006;363(1):51–62. doi: 10.1016/j.jmb.2006.08.041. [DOI] [PubMed] [Google Scholar]
- 33.Ballesteros JA, Deupi X, Olivella M, Haaksma EE, Pardo L. Serine and threonine residues bend alpha-helices in the χ(1) = g(-) conformation. Biophys J. 2000;79(5):2754–2760. doi: 10.1016/S0006-3495(00)76514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray TM, Matthews BW. Intrahelical hydrogen bonding of serine, threonine and cysteine residues within alpha-helices and its relevance to membrane-bound proteins. J Mol Biol. 1984;175(1):75–81. doi: 10.1016/0022-2836(84)90446-7. [DOI] [PubMed] [Google Scholar]
- 35.Hall SE, Roberts K, Vaidehi N. Position of helical kinks in membrane protein crystal structures and the accuracy of computational prediction. J Mol Graph Model. 2009;27(8):944–950. doi: 10.1016/j.jmgm.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Hooft RW, Sander C, Vriend G. Positioning hydrogen atoms by optimizing hydrogen-bond networks in protein structures. Proteins. 1996;26(4):363–376. doi: 10.1002/(SICI)1097-0134(199612)26:4<363::AID-PROT1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 37.Eswar N, Ramakrishnan C. Deterministic features of side-chain main-chain hydrogen bonds in globular protein structures. Protein Eng. 2000;13(4):227–238. doi: 10.1093/protein/13.4.227. [DOI] [PubMed] [Google Scholar]
- 38.Hegde SS, et al. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science. 2005;308(5727):1480–1483. doi: 10.1126/science.1110699. [DOI] [PubMed] [Google Scholar]
- 39.Buchko GW, et al. Characterization of two potentially universal turn motifs that shape the repeated five-residues fold—Crystal structure of a lumenal pentapeptide repeat protein from Cyanothece 51142. Protein Sci. 2006;15(11):2579–2595. doi: 10.1110/ps.062407506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaplan RS, Mayor JA, Gremse DA, Wood DO. High level expression and characterization of the mitochondrial citrate transport protein from the yeast Saccharomyces cerevisiae. J Biol Chem. 1995;270(8):4108–4114. doi: 10.1074/jbc.270.8.4108. [DOI] [PubMed] [Google Scholar]
- 41.Palmieri L, et al. Identification in Saccharomyces cerevisiae of two isoforms of a novel mitochondrial transporter for 2-oxoadipate and 2-oxoglutarate. J Biol Chem. 2001;276(3):1916–1922. doi: 10.1074/jbc.M004332200. [DOI] [PubMed] [Google Scholar]
- 42.Cappello AR, et al. Functional and structural role of amino acid residues in the odd-numbered transmembrane alpha-helices of the bovine mitochondrial oxoglutarate carrier. J Mol Biol. 2007;369(2):400–412. doi: 10.1016/j.jmb.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 43.Miniero DV, et al. Functional and structural role of amino acid residues in the matrix alpha-helices, termini and cytosolic loops of the bovine mitochondrial oxoglutarate carrier. Biochim Biophys Acta. 2011;1807(3):302–310. doi: 10.1016/j.bbabio.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 44.Russ WP, Engelman DM. The GxxxG motif: A framework for transmembrane helix-helix association. J Mol Biol. 2000;296(3):911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 45.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211(5052):969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 46.Kunji ER, Robinson AJ. Coupling of proton and substrate translocation in the transport cycle of mitochondrial carriers. Curr Opin Struct Biol. 2010;20(4):440–447. doi: 10.1016/j.sbi.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 47.Brandolin G, et al. Kinetic, binding and ultrastructural properties of the beef heart adenine nucleotide carrier protein after incorporation into phospholipid vesicles. Biochim Biophys Acta. 1980;592(3):592–614. doi: 10.1016/0005-2728(80)90103-6. [DOI] [PubMed] [Google Scholar]
- 48.Jiang F, Rizavi HS, Greenberg ML. Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non-fermentable carbon sources. Mol Microbiol. 1997;26(3):481–491. doi: 10.1046/j.1365-2958.1997.5841950.x. [DOI] [PubMed] [Google Scholar]
- 49.Krämer R, Klingenberg M. Enhancement of reconstituted ADP, ATP exchange activity by phosphatidylethanolamine and by anionic phospholipids. FEBS Lett. 1980;119(2):257–260. doi: 10.1016/0014-5793(80)80266-3. [DOI] [PubMed] [Google Scholar]
- 50.Olofsson G, Sparr E. Ionization constants pKa of cardiolipin. PLoS ONE. 2013;8(9):e73040. doi: 10.1371/journal.pone.0073040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hol WG. The role of the alpha-helix dipole in protein function and structure. Prog Biophys Mol Biol. 1985;45(3):149–195. doi: 10.1016/0079-6107(85)90001-x. [DOI] [PubMed] [Google Scholar]
- 52.Hol WG, van Duijnen PT, Berendsen HJ. The alpha-helix dipole and the properties of proteins. Nature. 1978;273(5662):443–446. doi: 10.1038/273443a0. [DOI] [PubMed] [Google Scholar]
- 53.Sengupta D, Behera RN, Smith JC, Ullmann GM. The alpha helix dipole: Screened out? Structure. 2005;13(6):849–855. doi: 10.1016/j.str.2005.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.