Significance
Cyclin D2 is a cell cycle regulator with spatially restricted expression. Loss and gain of function in animal models also revealed a role in cell differentiation, but the mechanisms underlying this are incompletely understood. The cardiogenic transcription factor GATA4 is an upstream regulator of cyclin D2. We show that GATA4 and cyclinD2 are part of a forward reinforcing loop in which cyclin D2 feeds back to enhance GATA4 activity through direct interaction. Mutations in GATA4 that abrogate cyclin D2 interactions are associated with human congenital heart disease. The results unravel a unique transcriptional role of cyclin D2 that may underlie its cell specificity. The finding that cyclin D2 is a cardiogenic GATA4 cofactor may be exploitable therapeutically for heart repair.
Keywords: GATA transcription factors, heart development, heart repair
Abstract
The G1 cyclins play a pivotal role in regulation of cell differentiation and proliferation. The mechanisms underlying their cell-specific roles are incompletely understood. Here, we show that a G1 cyclin, cyclin D2 (CycD2), enhances the activity of transcription factor GATA4, a key regulator of cardiomyocyte growth and differentiation. GATA4 recruits CycD2 to its target promoters, and their interaction results in synergistic activation of GATA-dependent transcription. This effect is specific to CycD2 because CycD1 is unable to potentiate activity of GATA4 and is CDK-independent. GATA4 physically interacts with CycD2 through a discreet N-terminal activation domain that is essential for the cardiogenic activity of GATA4. Human mutations in this domain that are linked to congenital heart disease interfere with CycD2-GATA4 synergy. Cardiogenesis assays in Xenopus embryos indicate that CycD2 enhances the cardiogenic function of GATA4. Together, our data uncover a role for CycD2 as a cardiogenic coactivator of GATA4 and suggest a paradigm for cell-specific effects of cyclin Ds.
D-type cyclins [Cyclin D1 (CycD1), CycD2, and CycD3] and their partners, the cyclin-dependent kinases (CDKs), are important regulators of the cell cycle (1), connecting the cell cycle machinery with the extracellular environment. In response to mitogenic signals, cyclin Ds associate with CDK4 and CDK6, phosphorylate the tumor suppressor retinoblastoma protein Rb, along with the family members p107 and p130, and, together with cyclin E/A-CDK2 complexes, control the G1/S-phase transition in the cell cycle (1, 2). In addition to the Rb protein family, cyclin D–CDK4 complex was found to phosphorylate other transcription factors involved in cell proliferation/arrest, such as the transcription factor SMAD3 and the myb like transcription factor, DMP1 (3, 4). Moreover, Cyclin Ds have been shown to interact with transcription coactivators/corepressors such as CBP, p300, P/CAF and HDACs (5–8) and a repressor domain distinct from the CDK regulatory domain has been identified on CycD1 (9). As such, Cyclin Ds play important roles as CDK-dependent and -independent transcriptional modulators. Most transcription studies so far have focused on CycD1 but some of the reported interactions appear to be specific to a particular Cyclin D family member (10). In other cases, interaction of different Cyclin Ds with the same transcription factor, as in the case of the peroxisome proliferator-activated receptor (PPAR)γ, leads to opposing outcomes (11, 12). Although the mechanisms underlying these differential effects remain uncertain, the findings, together with the distinct expression pattern of the three Cyclin Ds (1), have raised intriguing questions regarding cell-specific functions of Cyclin Ds in normal development and disease.
In addition to cell cycle regulation, Cyclin Ds participate in several other cellular processes, including hormone regulation (13, 14) and cell differentiation; for example, CycD1 (but not CycD2) promotes neurogenesis in a cell cycle-independent manner (15), whereas CycD3 appears to play a unique function in lymphocyte development (16) and adipogenesis (12). In all cases, gain and loss of Cyclin Ds lead to profound changes in gene expression through transcriptional mechanisms that remain incompletely understood. The production of transgenic mice homozygous for one or more Cyclin D allele has confirmed the requirement for specific Cyclin Ds in organ development (17). CycD2 appears to play an important role in the embryonic and postnatal heart, where it has been shown to be a direct target of the cardiogenic transcription factor GATA4 (18, 19). CycD2 has been linked to cardiomyocyte regeneration (20), hypertrophy (21), and stress response (22).
In the present study, we show that CycD2 but not CycD1 is a potent coactivator of GATA4, a critical cardiac regulator. CycD2 physically interacts with GATA4, is recruited to GATA4 target promoters, and enhances GATA4-dependent transcription. Moreover, CycD2 potentiates GATA4-dependent cardiogenesis but does not affect other cell fates induced by GATA4, including endoderm. The data provide a mechanism that may underlie, at least in part, cell specificity of Cyclin Ds and a rationale for targeting cell cycle proteins for cardiac repair.
Results
CycD2 Is a GATA4 Transcriptional Coactivator.
CycD2 is a direct GATA4 target in the heart (18, 19), where it is coexpressed with GATA4 in fetal and neonate cardiomyocytes. In the mouse heart, CycD2 transcripts have been reported as early as embryonic day (E) 9 in the developing ventricles (23). Importantly, CycD2 transcripts were detected by PCR in E7.5 embryos and were localized in the primitive streak and in mesodermal cells migrating from it; these cells were not positive for CycD1 (24). We carried out immunohistochemical analysis of CycD2 and GATA4 using consecutive embryonic tissue sections. As shown in Fig. S1, CycD2 and GATA4 colocalized in the mesothelial cells of the future cardiogenic plate at E7.5 and within numerous myocardial and cushion cells in E9.5 and later-stage embryos. We extended our analysis to Xenopus, where, in addition to the previously described prominent CNS expression (25), we detected CycD2 in the developing heart of stage 30 Xenopus embryos, around the onset of cardiomyocyte differentiation (Fig. S1).
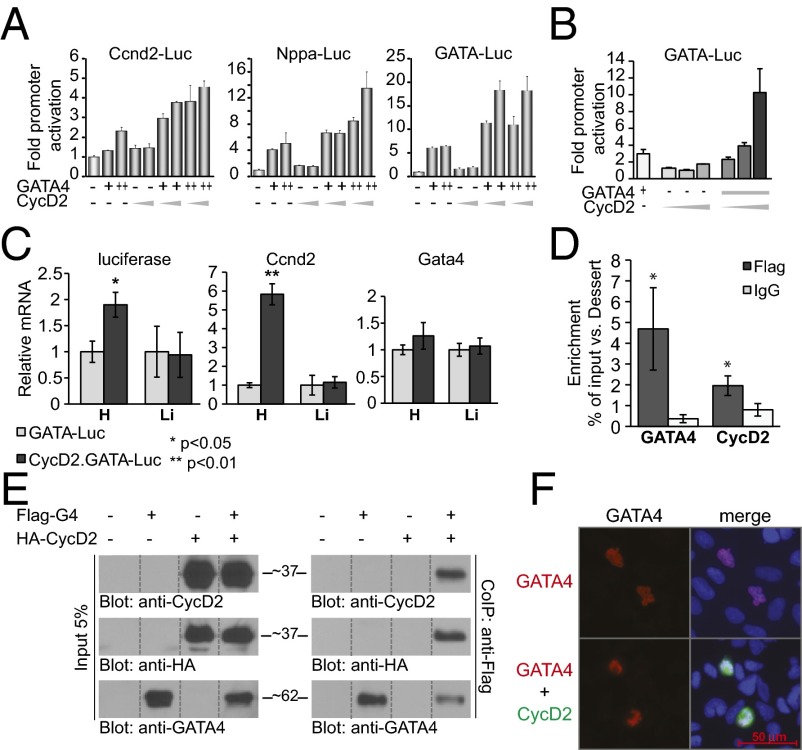
We tested whether CycD2 affects GATA4 activity. Cotransfection of CycD2 and GATA4 in National Institutes of Health (NIH) 3T3 cells showed that CycD2 enhances GATA4 activity on its target promoters, including Nppa and CycD2 (ccnd2) (Fig. 1A, Left and Center); CycD2 enhancement of GATA4 transcription was also observed on the minimal GATA-dependent promoter, suggesting that GATA-binding sites were sufficient to support synergy (Fig. 1A, Right). This effect was also observed in HL1 cells, which are mouse atrial cardiomyocytes (Fig. 1B). To test whether in vivo CycD2 can potentiate GATA4 activity, GATA-Luc transgenic mice were crossed with mice overexpressing CycD2 specifically in the heart (under the control of the α-MHC promoter). Quantitative PCR analysis confirmed that ccnd2 transcripts are specifically up-regulated in the heart but have no effect on GATA4 encoding mRNA levels (Fig. 1C), consistent with our published results (18). Up-regulation of ccnd2 resulted in a twofold higher GATA-dependent luciferase transcription specifically in the heart of double transgenics but not in other organs exhibiting GATA activity like liver (Fig. 1C). Interestingly, increased GATA activity was not associated with increased phosphorylation of S105, the major MAPK target residue on GATA4 (Fig. S2). To further confirm that CycD2 associates with GATA-binding sites on GATA4 target promoters, we performed chromatin immunoprecipitation (ChIP) assays on the endogenous ccnd2 gene using cell lines with stable overexpression of Flag-CycD2. As shown in Fig. 1D, CycD2 was enriched twofold on the −99 GATA site relative to the Dessert gene (used as internal control). GATA4 was also enriched at this site, consistent with our previous results (18).
Fig. 1.
GATA4 and Cyclin D interaction. (A and B) Transient transactivation of CycD2 (ccnd2), Nppa, and the minimal GATA promoter by GATA4 and CycD2 in NIH 3T3 cells (A) and in HL-1 atrial cardiomyocytes (B); 250 and 500 ng of GATA4 expression vector and 3 and 4 μg of CycD2 expression vector were used in the case of NIH cells and 25 ng of GATA4 and 0.5, 1, and 3 μg of CycD2 expression vectors were used in the case of HL-1 cells. (C) Transcript levels of luciferase, ccnd2, and Gata4 in the heart and liver of GATA-Luc transgenic mice crossed with HA-ccnd2 mice. Note the increase of luciferase mRNA levels of CycD2.GATA-Luc mice relative to those of GATA-Luc mice specifically in the heart (Left). (Center) Control to show the overexpression of CycD2 in the hearts of CycD2.GATA-Luc mice but not in the liver. (Right) No effect of CycD2 overexpression on Gata4 mRNA levels. The results are shown as means ± SEM (n = 3). *P < 0.05; **P < 0.01. (D) Enrichment of CycD2 and GATA4 on CycD2 promoter, as revealed by ChIP. Dessert is used as a negative gene. IgG is a negative control. Flag-GATA4 and Flag-CycD2 C2C12 stable cell lines were used. The results are means ± SEM (n = 3). *P < 0.05 vs. IgG. (E) CycD2 coimmunoprecipitates with GATA4 in vivo. Nuclear extracts from 293T cells transfected with Flag-GATA4 and/or HA-CycD2 expression vectors were immunoprecipitated using an anti-Flag antibody, separated on 8% (vol/vol) SDS/PAGE, transferred to poly(vinylidene difluoride) (PVDF) membranes, and subjected to immunoblotting using anti-HA, anti-CycD2, or anti-GATA4 antibodies. The results here are from one representative experiment of three. For consistency, the relevant lanes were spliced from the same blot image and assembled in the order shown. (F) Immunocytochemical analysis of 293T cells transiently transfected with 100 ng of Flag-GATA4 with or without 1 μg of HA-CycD2. Red is GATA4 staining; green is CycD2; blue marks nuclei (Hoechst). Note cotransfection of GATA4 with CycD2 does not affect GATA4 nuclear levels.
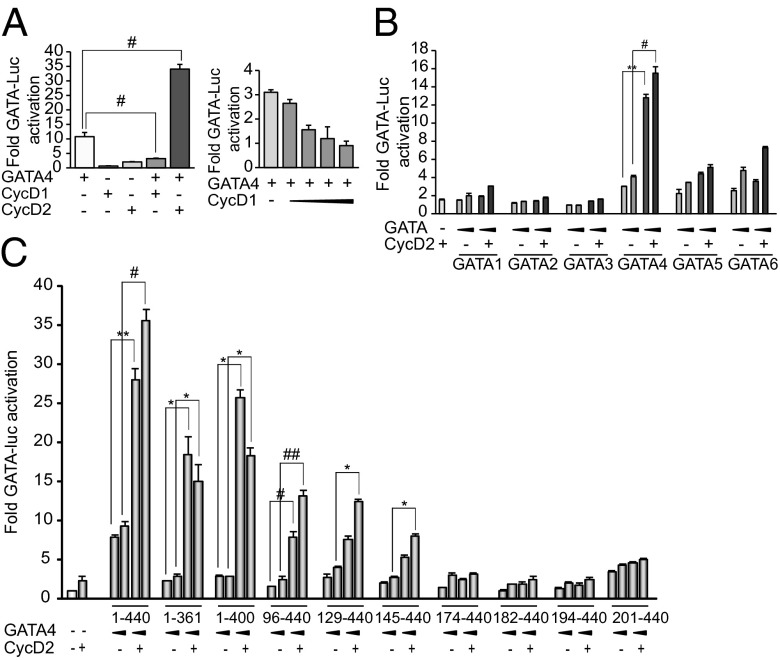
These results suggested that GATA4 could recruit CycD2 to target promoters. To test this possibility, we performed coimmunoprecipitation assays using cells expressing Flag-GATA4 and HA-CycD2 alone or in combination. As shown in Fig. 1E, CycD2 coimmunoprecipitated with GATA4. Of note, CycD2 did not increase levels of nuclear GATA4 nor did it result in detectable changes in subcellular distribution (Fig. 1E, Left, and Fig. 1F). Next, we tested whether enhancement of GATA4 transcriptional activity could also be observed with the related CycD1. Cotransfection of GATA4 with CycD1 showed that CycD1 is unable to enhance GATA4-dependent transcription (Fig. 2A); in fact, CycD1 inhibited GATA4 activity in a dose-dependent manner (Fig. 2A, Right). This result is consistent with a recent report showing that cardiac-specific CycD1 overexpression in mice reduces GATA4 protein levels and inhibits cardiomyocyte differentiation (26). The specificity of CycD2/GATA interaction to GATA4 was also checked by testing the ability of CycD2 to synergize with the hematopoietic GATA family members GATA1, -2 , and -3, as well as the other two cardiac members, GATA5 and GATA6. Fig. 2B shows that CycD2 produced no transcriptional enhancement when combined with GATA1–3; a modest but reproducible enhancement of the activity of the other cardiac GATA proteins was detected. Thus, CycD2 appears to enhance GATA4 activity in preference over other GATA family members.
Fig. 2.
Specificity of GATA4-CycD2 interaction. (A, Left) Transient transactivation of GATA-dependent promoter by GATA4 and CycD1 or CycD2 in NIH 3T3 cells; 500 ng of GATA4 and 4 μg of CycD1 or CycD2 expression vectors were used. Note that CycD1 does not synergize but rather inhibits GATA4 activity. #P < 0.005 vs. GATA4. (Right) Transient transactivation of GATA-dependent promoter by GATA4 and CycD1 in NIH 3T3 cells; 10 ng of GATA4 and 100, 200, 500, and 3,000 ng of CycD1 were used. Note that CycD1 inhibits GATA4 activity in a dose-dependent manner. (B) Transient transactivation of GATA-dependent promoter by GATA1, -2, -3, -4, -5, or -6 and CycD2 in NIH 3T3 cells; 250 and 500 ng of GATA and 4 μg of CycD2 expression vectors were used in the cases of GATA1, -2, -3, -4, and -6; 15 and 25 ng of GATA and 1 μg of CycD2 expression vectors were used in the case of GATA5. Note a statistically significant synergy of CycD2 with GATA4. Modest but reproducible synergy was seen in case of GATA5 and 6. No synergy was seen with the hematopoietic GATA members even at different doses. **P < 0.01; #P < 0.005 vs. GATA4. (C) Structure–function analysis of GATA4/CycD2 activation of the GATA-dependent promoter. Transient transfections were carried out in NIH 3T3 cells using the indicated GATA4 and/or CycD2 expression vectors and the GATA-Luc reporter; 250 and 500 ng of GATA4 and 4 μg of CycD2 were used. The results shown are those of one representative experiment of three carried out in duplicates with the SD of the mean. *P < 0.05; **P < 0.01; #P < 0.005; ##P < 0.0001 vs. GATA4.
A Discrete GATA4 N-Terminal Domain Is Required for CycD2 Synergy.
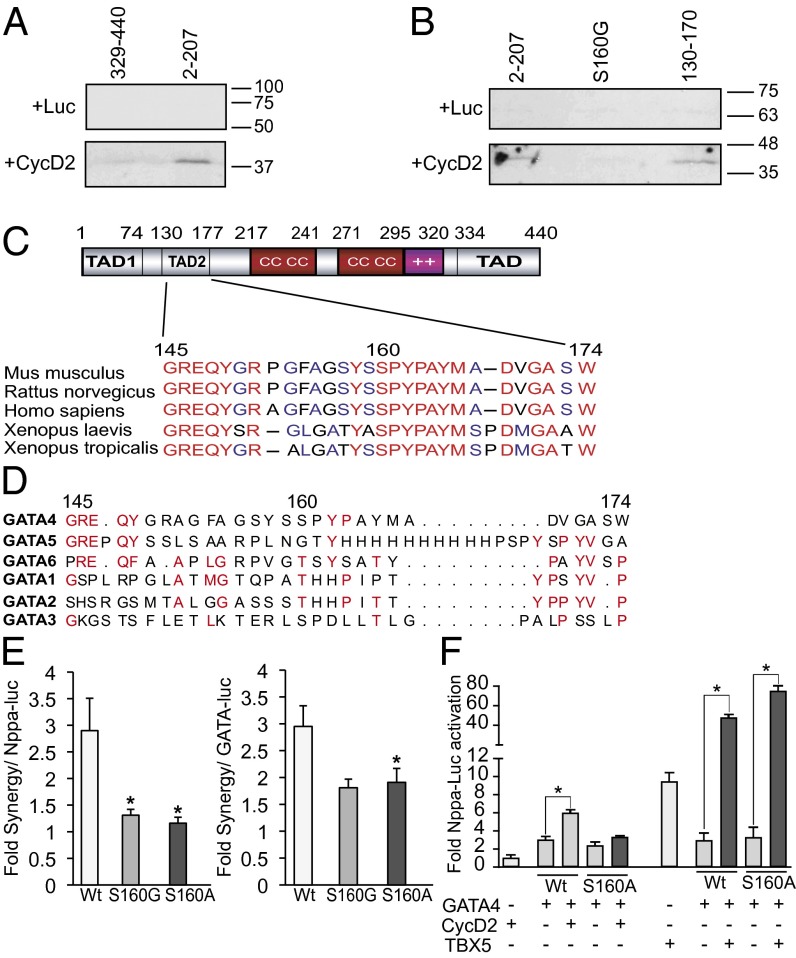
To determine which region of GATA4 is required for CycD2 interaction, we carried out a structure–function analysis of GATA4. Mutant proteins that harbored various C-terminal or N-terminal deletions were used. All mutants were tested for their ability to be expressed in the nucleus at similar levels and bind DNA using Western blot analysis and electrophoretic mobility-shift assays (Fig. S3). As expected, N- and C-terminal GATA4 deletions reduced transcriptional activity. When the same constructs were tested for synergy with CycD2, removal of the C-terminal region had minimal effect on the synergy. Removal of the first 145 aa reduced but did not abrogate the synergy. However, removal of the first 174 aa abolished the synergy, suggesting that amino acids 145–174 on the GATA4 protein are critical for interaction with CycD2 (Fig. 2C). To determine whether the GATA4 domain required for synergy is involved in physical interaction with CycD2, in vitro pull-down assays were performed with GST- produced N- and C-terminal GATA4 fusion proteins and in vitro-translated CycD2. Fig. 3A shows that CycD2 is able to physically interact with the N- but not C-terminal region of GATA4. Moreover, a GATA4 fragment containing amino acids 130–170 was sufficient for physical interaction with CycD2 (Fig. 3B).
Fig. 3.
CycD2 physically interacts with N-terminal GATA4 in vitro. (A) In vitro-translated radiolabeled CycD2 protein (or luciferase protein as a negative control) was incubated with glutathione Sepharose beads containing GST–N-terminal GATA4 or GST–C-terminal GATA4 fusion proteins. The bound proteins were then resolved by SDS/PAGE and revealed by autoradiography. Note that CycD2 binds to N-terminal GATA4 (second lane). The experiment is one representative of two. (B) Amino acids 130–170 are sufficient to interact with CycD2, and S160 is required for this interaction. In vitro-translated radiolabeled CycD2 protein were incubated with glutathione Sepharose beads containing GST alone, GST–N-terminal region 2–207 of GATA4, the same GST–N-terminal part of GATA4 harboring the S160G mutation or GST–130–170 GATA4 fusion proteins. The bound proteins were then resolved by SDS/PAGE and revealed by autoradiography. In vitro-translated luciferase protein was used as a negative control. Note that CycD2 binds to amino acids 2–207 of GATA4 and 130–170 but not S160G mutant. The experiment is one representative of two. (C) Schematic representation of GATA4 protein. Note that the 145- to 174-aa region, and particularly amino acid S160, is highly conserved among the mouse, rat, human, and Xenopus. (D) Alignment of the CycD2-interacting domain (145–174) of human GATA4 with the other GATA members as indicated. Note that this region is highly divergent among the different GATA members. (E) Fold synergy of CycD2 with WT (Wt) GATA4 or the indicated S160 GATA4 mutants on Nppa promoter (Left) or GATA-dependent promoter (Right). Note the reduced fold of synergy with both S160G and S160A. *P < 0.05 vs. Wt. (F) Transient transactivation of Nppa promoter by WT or S160A GATA4 with or without either CycD2 or TBX5; 250 ng of GATA4 was used with 4 μg of CycD2; 5 ng of GATA4 was used with 50 ng of TBX5. Note the S160 mutation attenuates synergy with CycD2 but not TBX5. *P < 0.05 vs. GATA4.
The 145- to 174-aa region of GATA4 lies within the second N-terminal transactivation domain and is conserved across GATA4 proteins from different species from zebrafish to human but not in other members of the GATA family (Fig. 3 C and D). Two conserved amino acids in this region, S160 and P163, have been reported mutated in humans with congenital heart defects (27–30). We tested the effect of S160 mutation on interaction with CycD2. As shown in Fig. 3B, mutation S160G greatly reduced physical interaction of GATA4 with CycD2. We produced GATA4 proteins with mutations on S160 and P163. As shown in Fig. S4, the mutant proteins retained their ability to be expressed and bind DNA as well as wild-type (WT) GATA4. However, their baseline activity, as well as their synergy with CycD2 but not with Tbx5, were significantly attenuated (Fig. 3 E and F and Fig. S4).
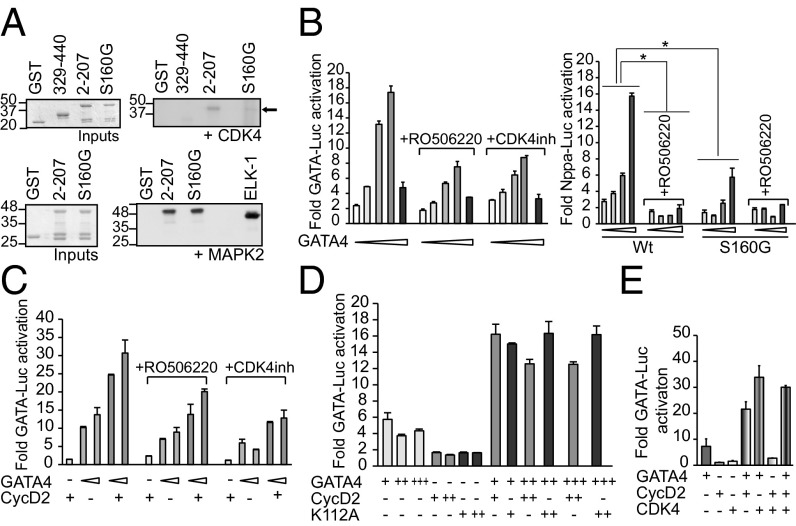
Bioinformatics analysis using the Motif Scan Web site (http://scansite.mit.edu/motifscan_seq.phtml) suggested that S160 may be a putative CDK phosphorylation site. We tested whether S160 can be phosphorylated by CDK4 using in vitro kinase assay; when active CDK4 kinase was incubated with N- and C-terminal GST GATA4 fusion proteins phosphorylation of the N- but not C-terminal GATA4 domain was detected. Incubation of CDK4 kinase with GST–N-terminal GATA4 harboring the S160G mutation greatly reduced this phosphorylation, suggesting that S160 is a major CDK4 phosphorylation site on GATA4 protein (Fig. 4A). The same mutation had no impact on MAPK phosphorylation of the 2–207 GATA4 domain, consistent with previous studies that found S105 to be the major MAPK target in the N-terminal domain of GATA4 (31). Interestingly, GATA4 S105 phosphorylation level was similar in the hearts of WT mice and in those overexpressing CycD2 (18), suggesting that CycD2-dependent phosphorylation takes place at distinct residues (Fig. S2). Next, we tested the effect of CDK4 inhibition on GATA4 activity and synergy with CycD2. Luciferase assays performed on NIH 3T3 cells cotransfected with increasing doses of GATA4 protein in presence of pharmacologic CDK4 inhibitors showed reduced GATA4 activation of its target promoters (Fig. 4B). Interestingly, CDK4 inhibition had a more modest effect on the S160 mutant than on WT GATA4, consistent with this residue being an important CDK4 target (Fig. 4B). These results suggest that CDK4 kinase activity is required for maximal GATA4 transcriptional activity. We checked whether CDK4 activity interferes with GATA4-CycD2 synergy. As shown in Fig. 4C, the CDK4 inhibitors decreased GATA4 activation of the GATA-Luc reporter but did not interfere with CycD2 synergy, with the fold enhancement decreasing modestly from 2.5 to 2 in the presence of either inhibitor. This suggested that CycD2 was acting as a CDK4-independent coactivator. Cyclin Ds were shown to interact with their catalytic partners CDK4/6 through a lysine residue in the cyclin box (32). To check whether interaction of CycD2 with GATA4 was indeed independent of the CDK4-binding site on CycD2, we generated a K112A CycD2 mutant. When cotransfected with GATA4, this mutant was able to enhance GATA4 activation of a GATA-dependent promoter in a manner similar to WT CycD2 (Fig. 4D). Consistent with this result, the presence of both CDK4 and CycD2 with GATA4 did not produce greater activation of the GATA-Luc reporter than either GATA4-CDK4 or GATA4-CycD2 (Fig. 4E).
Fig. 4.
Phosphorylation of GATA4 at S160 is required for its transcriptional activity. (A) In vitro CDK4 or MAPK phosphorylation of GST-GATA4 fusion proteins. Active CDK4 kinase was able to phosphorylate N-terminal GST-GATA4 (2–207) fusion protein (black arrow) but not GST alone, nor C-terminal GST-GATA4 (329–440) nor N-terminal GATA4 harboring the S160G mutation. MAPK phosphorylation of N-terminal GATA4 was not affected by S160 mutation. The experiment is one representative of three. (B) Inhibition of CDK4 reduces GATA4 activity. (Left) NIH 3T3 cells were transiently transfected with the GATA-dependent promoter and increasing doses of GATA4 expression vector (5, 10, 50, 100, and 500 ng) with or without treatment with CDK4 inhibitor; 2 μM the indicated CDK4 inhibitor was used. The cells were treated the following day after transfection and kept for 18 h. (Right) Effect of the CDK4 inhibitor RO506220 on S160G GATA4 mutant. NIH 3T3 cells were transiently transfected with Nppa-Luc and increasing doses of the indicated GATA4 expression vector (10, 50, 100, and 250 ng) with or without treatment with CDK4 inhibitor as above. Note S160G activity is affected less prominently by CDK4 inhibitor RO506220 than its WT. *P < 0.05 vs. Wt. (C) Inhibition of CDK4 does not affect GATA4/Cyclin D twofold synergy. NIH 3T3 cells were transiently transfected with GATA-Luc, 50 or 100 ng of GATA4, and/or 1 μg of CycD2 expression vectors with or without treatment with CDK4 inhibitors as above. (D) CycD2/GATA4 synergy does not require CDK-binding site. NIH 3T3 cells were transiently transfected with GATA-Luc, GATA4, and/or CycD2 expression vectors. K112A CycD2 mutant was prepared by PCR-mediated mutagenesis; 50, 250, and 500 ng of GATA4 expression vector and 1 and 4 μg of CycD2 and K112A expression vectors were used. (E) Lack of triple synergy between CycD2, CDK4, and GATA4. NIH 3T3 cells with transiently transfected with GATA-Luc, CDK4 (50 ng), and/or GATA4 (25 ng) and/or CycD2 (50 ng) expression vectors. Note that fold synergy of GATA4/CycD2 does not increase with addition of CDK4.
CycD2 Enhances Cardiogenic Activity of GATA4.
The CycD2 interaction domain overlaps with a region of GATA4 that was recently found to be essential for its cardiogenic activity (33), and mutations therein that decrease synergy with CycD2 have been associated with congenital heart disease (Fig. 3). This raised the possibility that CycD2 might cooperate with GATA4 in cardiogenesis.
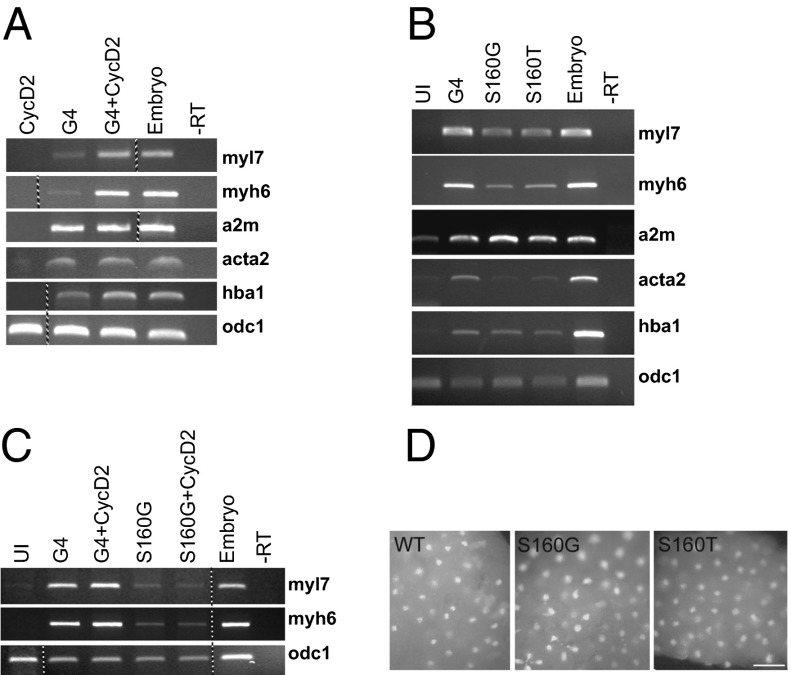
To check the consequences of CycD2/GATA4 interaction on cardiogenesis, we examined the ability of CycD2 to alter the activity of GATA4 in ectodermal (animal cap) explants from Xenopus embryos. To enhance detection of anticipated positive effect of CycD2 on activity of GATA4, we used suboptimal amount of GATA4 encoding mRNA (300 pg; optimal range is 400–1,000 pg) (34), together with a range of CycD2 encoding mRNA concentrations from 10 to 500 pg to determine that the optimal dose of CycD2 mRNA is 100 pg. As shown in Fig. 5A, CycD2 mRNA specifically stimulated the cardiogenic activity of GATA4, as evident by the induction of the cardiomyocyte-specific markers myosin light chain 7 (myl7) and myosin heavy chain 6 (myh6) but not the endodermal marker endodermin (a2m), the smooth muscle actin marker (acta2), or globin, the hematopoietic marker (hba1). Moreover, and consistent with its reduced transcriptional activity, GATA4 S160 mutations that reduce CycD2 synergy (S160G) or are associated with human congenital heart defects (S160T) decreased GATA4-dependent induction of cardiomyocytes (Fig. 5B) but not of markers of endoderm and blood. In addition, the S160G mutation abrogated the ability of CycD2 to potentiate GATA4 cardiogenesis (Fig. 5C). This was not attributable to changes in protein levels given that nuclear expression of the mutants tested was similar to that of WT GATA4 in the explants (Fig. 5D). Collectively, these results indicate that CycD2 can act as a coactivator of GATA4 in cardiogenesis.
Fig. 5.
CycD2 potentiates cardiogenic activity of GATA4. (A) CycD2 stimulates cardiogenic activity of the rat GATA4. Animal cap explants injected with 100 pg of CycD2 mRNA, 300 pg of Gata4 mRNA (G4), or the two combined were analyzed for expression of indicated markers at stage 34. Myl7 and myh6 are exclusively expressed in cardiac myocytes, endodermin (a2m) is an endodermal marker, smooth muscle actin (acta2) marks smooth muscle, and globin (hba1) is a marker of blood. Note that stimulation of GATA4 activity by CycD2 is restricted to induction of cardiac but not other cell fates. (B) S160 is important for cardiogenic activity of GATA4; 400 pg of indicated GATA4 constructs were injected. Animal explants were cultured as above, and expression of indicated markers were determined by RT-PCR. (C) S160 is required for stimulation of GATA4 activity by CycD2; 100 pg of CycD2 mRNA and 300 pg of the indicated GATA4 constructs were injected per embryo. Expression of myl7, myh6, and odc1 were determined by RT-PCR. The dotted lines between lanes in A and C indicate that the image shown was assembled either from two gels run in parallel or derives from the same gel after splicing out unnecessary lanes. (D) Immunofluorescence assay to determine the nuclear localization of the indicated GATA4 constructs in stage 6–7 embryos. Anti-GATA4 antibody was used. (Scale bar: 0.1 mm.)
Discussion
Transcription factor GATA4 is a critical regulator of heart development and homeostasis, where it has multiple essential and nonredundant roles in cell growth, survival, and differentiation. Moreover, GATA4 is a cardiogenic factor that can activate the genetic program required to convert stem and precursor cells to the cardiogenic lineage (34–36). CycD2 is a direct GATA4 transcription target in the heart, and its cardiomyocytes up-regulation in transgenic mice enhances postischemic heart repair and rescues GATA4 haploinsufficiency (18, 20). Here, we show that CycD2 but not CycD1 enhances transcriptional activity of GATA4 and potentiates GATA4-dependent cardiomyogenesis. Moreover, CycD2 interacts physically with a GATA4 N-terminal activation domain that overlaps with a recently identified GATA4 cardiogenic domain (33). We find that mutations within this domain that reduce CycD2-GATA4 synergy are associated with decreased GATA4 cardiogenic activity and with human congenial heart disease. Together, the results suggest the existence of a positive GATA4-CycD2 feedback loop in cardiomyocytes that may be exploited for expansion of cardiomyogenic progenitors for heart repair therapy.
CycD2 effect on GATA4 activity was not reproduced by CycD1, which in fact inhibited GATA4 transcriptional activity. This latter result is consistent with a recently published study suggesting that CycD1 causes GATA4 degradation through a jumonji pathway (26). However, unlike the finding of Nakajima et al. (26), that CDK4 inhibits GATA4 activity, we find that CDK4 enhances GATA4 activity. These discrepancies may reflect stage- and region-specific effects of GATA4 through differential interactions with particular Cyclin D members. Such differential effects of Cyclin Ds on cell fate determination have been observed in neurogenesis where CycD1 can inhibit differentiation by preventing G1 lengthening (37) and interfering with Neuro D (38); yet, CycD1 can also promote neurogenesis in a cell cycle independent manner (15). The differential effects of CycD1 and -D2 on GATA4 are in line with the different effects of the two family members when overexpressed in transgenic hearts (20) and their distinct interaction with other cell cycle regulators (39) including in cardiomyocytes (40). Importantly, the finding that CycD2 cooperates with GATA4 in cardiogenesis is consistent with the findings that up-regulated CycD2 expression in postnatal hearts is associated with increased resistance to drug induced cardiac dysfunction (18), as well as enhanced post ischemic repair (20). This role in cardiomyocytes is reminiscent of the dual role of CycD2 in pancreatic and neurogenic progenitor expansion and differentiation (41, 42).
The mechanism by which CycD2 enhances GATA4 activity does not appear to require CDK4 although a CDK4 phosphorylation site lies within the CycD2 interaction domain on GATA4. First, mutation of the CDK4 interaction domain on CycD2 has no effect on synergy; second, pharmacologic inhibition of CDK4 decreases basal GATA4 activity but has only minimal effect on synergy with CycD2. No effect of CycD2 on GATA4 mRNA or protein levels and subcellular distribution were detected, suggesting that CycD2 affects GATA4 transcriptional activity likely through protein–protein interactions.
Although the role of CycD2 in differentiation and homeostasis is supported by loss- and gain-of-function in model animals (22, 41, 42), the mechanisms underlying these effects are incompletely understood. Unlike CycD1, whose emerging role as a transcriptional regulator has received significant attention lately (reviewed in ref. 43), little is known regarding CycD2 function in transcription outside regulation of the Rb pathway. The only well-characterized CycD2 interactors are the CDKs, in contrast to CycD1 and -D3, for which several interacting partners have been reported. For instance, CycD1 has been shown to interact with the histone acetyl transferase p300/CREB-binding protein-associated protein (P/CAF), thus facilitating the interaction of P/CAF with the estrogen receptor (ER) and possibly providing a mechanism for CycD1-stimulated ER transcriptional activity and oncogenic potential in breast cancer (8, 44). The zinc finger transcription factor INSM1 has also been shown to bind to CycD1, leading to cell cycle arrest and inhibition of proliferation, thereby suggesting a possible mechanism for pancreatic endocrine cell differentiation where INSM1 plays an important role (45). CycD3 was shown to be a coactivator of the nuclear receptor PPARγ in adipogenesis (12). CycD1 has been reported to inhibit PPARγ-mediated adipogenesis via HDAC recruitment (6). CycD1/CDK4 was also shown to phosphorylate and inhibit the DNA-binding ability of BRCA1 in breast cancer cells (46). Interestingly, a genome-wide location approach revealed association of CycD1 with regulatory genomic regions enriched for binding sites of nuclear effectors of growth factor signaling (5).
Our study provides evidence for a role for CycD2 as a transcription modulator and identifies GATA4 as an important interacting partner for CycD2 in the heart. The finding that a cell-specific transcription factor mediates CycD2 recruitment to DNA-regulatory sequences may provide a paradigm for cell-specific effects of Cyclin Ds.
Materials and Methods
Plasmids.
Plasmids used were either previously described (18, 47, 48) or obtained by PCR-mediated amplification and cloned in the indicated vector using standard procedures. Constructs were confirmed by sequencing.
Cell Cultures and Transfections.
All cell lines were maintained and transfected as described previously (18, 47). CDK inhibitor was obtained from Calbiochem (catalog no. 219476), and RO506220 was obtained from Roche (49).
ChIP.
ChIP assays were performed as described previously (18). Primers sequences are available upon request.
Transgenic Mice.
All animal experiments were carried out in accordance with institutional guidelines for animal care. Experiments were approved by the institutional Animal Ethics Committee of the University of Ottawa, which conforms to that of the NIH (assurance no. A5043-01). At end points, mice were handled as detailed previously (50). Transgenic mice used were fully characterized in previous reports (18).
Protein and RNA Analysis.
Coimmunoprecipitation assays, Western blots, and electrophoretic mobility-shift assays were carried out using nuclear extracts from 293T cells overexpressing the appropriate protein as described previously (51, 52). The immunofluorescence assay was performed as described previously (53). The primary antibodies used were as follows: anti-Flag M2 (Cell Signaling; 2368) at a 1/700 dilution and anti-CycD2 (Abcam; ab308) at a 1/500 dilution. The secondary antibodies used were as follows: anti-mouse Alexa Fluor 488 (Molecular Probes; A-11029) and anti-rabbit Alexa Fluor 546 (Molecular Probes; A-11035), both at a 1/500 dilution. Hoechst staining (Molecular Probes; H1398) at a 1/5,000 dilution was used to mark nuclei. Images were taken on Zeiss AxioObserver.D1 microscope.
In Vitro Pull-Down and Kinase Assays.
Production of the recombinant GST-GATA4 proteins and pull-down assays were carried out as described previously (31, 54). Kinase assay was performed as described previously (54). The kinases used were as follows: MAPK2 (14-173; Cedarlane); ELK-1 (9184; Cell Signaling); and CDK4/CycD1 (7530; Cell Signaling).
Xenopus Embryos and Explants.
The work with Xenopus was approved by Cardiff University’s Ethical Review Committee and was carried out under a license from the United Kingdom Home Office. Xenopus embryos were obtained and cultured as described previously (55). Details are provided in SI Materials and Methods.
Statistics.
The data are presented as means ± SEM; P < 0.05 by Student t test is considered statistically significant.
Supplementary Material
Acknowledgments
We thank Janie Beauregard, Chantal Lefebvre, and Sarah Black for technical support, Hélène Touchette for editorial assistance, Jamie Whitcomb for reagents, and Dr. Hiba Komati for discussions and suggestions. This work was supported by grants from the Canadian Institutes of Health Research, the Heart and Stroke Foundation of Canada, and the British Heart Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312993111/-/DCSupplemental.
References
- 1.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18(22):2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 2.Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7(3):331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 3.Hirai H, Sherr CJ. Interaction of D-type cyclins with a novel myb-like transcription factor, DMP1. Mol Cell Biol. 1996;16(11):6457–6467. doi: 10.1128/mcb.16.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuura I, et al. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430(6996):226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 5.Bienvenu F, et al. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature. 2010;463(7279):374–378. doi: 10.1038/nature08684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu M, et al. Cyclin D1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipogenesis through histone deacetylase recruitment. J Biol Chem. 2005;280(17):16934–16941. doi: 10.1074/jbc.M500403200. [DOI] [PubMed] [Google Scholar]
- 7.Fu M, et al. Cyclin D1 represses p300 transactivation through a cyclin-dependent kinase-independent mechanism. J Biol Chem. 2005;280(33):29728–29742. doi: 10.1074/jbc.M503188200. [DOI] [PubMed] [Google Scholar]
- 8.McMahon C, Suthiphongchai T, DiRenzo J, Ewen ME. P/CAF associates with cyclin D1 and potentiates its activation of the estrogen receptor. Proc Natl Acad Sci USA. 1999;96(10):5382–5387. doi: 10.1073/pnas.96.10.5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schiewer MJ, et al. Cyclin D1 repressor domain mediates proliferation and survival in prostate cancer. Oncogene. 2009;28(7):1016–1027. doi: 10.1038/onc.2008.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W, et al. Cyclin D3 interacts with human activating transcription factor 5 and potentiates its transcription activity. Biochem Biophys Res Commun. 2004;321(4):954–960. doi: 10.1016/j.bbrc.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, et al. Cyclin D1 repression of peroxisome proliferator-activated receptor gamma expression and transactivation. Mol Cell Biol. 2003;23(17):6159–6173. doi: 10.1128/MCB.23.17.6159-6173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarruf DA, et al. Cyclin D3 promotes adipogenesis through activation of peroxisome proliferator-activated receptor gamma. Mol Cell Biol. 2005;25(22):9985–9995. doi: 10.1128/MCB.25.22.9985-9995.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comstock CE, et al. Cyclin D1 is a selective modifier of androgen-dependent signaling and androgen receptor function. J Biol Chem. 2011;286(10):8117–8127. doi: 10.1074/jbc.M110.170720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Augello MA, et al. Convergence of oncogenic and hormone receptor pathways promotes metastatic phenotypes. J Clin Invest. 2013;123(1):493–508. doi: 10.1172/JCI64750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukaszewicz AI, Anderson DJ. Cyclin D1 promotes neurogenesis in the developing spinal cord in a cell cycle-independent manner. Proc Natl Acad Sci USA. 2011;108(28):11632–11637. doi: 10.1073/pnas.1106230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawai CM, et al. Therapeutic targeting of the cyclin D3:CDK4/6 complex in T cell leukemia. Cancer Cell. 2012;22(4):452–465. doi: 10.1016/j.ccr.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozar K, et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118(4):477–491. doi: 10.1016/j.cell.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 18.Yamak A, et al. Cyclin D2 rescues size and function of GATA4 haplo-insufficient hearts. Am J Physiol Heart Circ Physiol. 2012;303(8):H1057–H1066. doi: 10.1152/ajpheart.00250.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rojas A, et al. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol Cell Biol. 2008;28(17):5420–5431. doi: 10.1128/MCB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasumarthi KB, Nakajima H, Nakajima HO, Soonpaa MH, Field LJ. Targeted expression of cyclin D2 results in cardiomyocyte DNA synthesis and infarct regression in transgenic mice. Circ Res. 2005;96(1):110–118. doi: 10.1161/01.RES.0000152326.91223.4F. [DOI] [PubMed] [Google Scholar]
- 21.Zhong W, et al. Hypertrophic growth in cardiac myocytes is mediated by Myc through a Cyclin D2-dependent pathway. EMBO J. 2006;25(16):3869–3879. doi: 10.1038/sj.emboj.7601252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angelis E, et al. A cyclin D2-Rb pathway regulates cardiac myocyte size and RNA polymerase III after biomechanical stress in adult myocardium. Circ Res. 2008;102(10):1222–1229. doi: 10.1161/CIRCRESAHA.107.163550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyons I, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9(13):1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 24.Wianny F, et al. G1-phase regulators, cyclin D1, cyclin D2, and cyclin D3: Up-regulation at gastrulation and dynamic expression during neurulation. Dev Dyn. 1998;212(1):49–62. doi: 10.1002/(SICI)1097-0177(199805)212:1<49::AID-AJA5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 25.Vernon AE, Philpott A. The developmental expression of cell cycle regulators in Xenopus laevis. Gene Expr Patterns. 2003;3(2):179–192. doi: 10.1016/s1567-133x(03)00006-1. [DOI] [PubMed] [Google Scholar]
- 26.Nakajima K, et al. Coordinated regulation of differentiation and proliferation of embryonic cardiomyocytes by a jumonji (Jarid2)-cyclin D1 pathway. Development. 2011;138(9):1771–1782. doi: 10.1242/dev.059295. [DOI] [PubMed] [Google Scholar]
- 27.Peng T, Wang L, Zhou SF, Li X. Mutations of the GATA4 and NKX2.5 genes in Chinese pediatric patients with non-familial congenital heart disease. Genetica. 2010;138(11-12):1231–1240. doi: 10.1007/s10709-010-9522-4. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W, et al. GATA4 mutations in 486 Chinese patients with congenital heart disease. Eur J Med Genet. 2008;51(6):527–535. doi: 10.1016/j.ejmg.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Butler TL, et al. GATA4 mutations in 357 unrelated patients with congenital heart malformation. Genet Test Mol Biomarkers. 2010;14(6):797–802. doi: 10.1089/gtmb.2010.0028. [DOI] [PubMed] [Google Scholar]
- 30.Yang YQ, et al. GATA4 loss-of-function mutations in familial atrial fibrillation. Clin Chim Acta. 2011;412(19-20):1825–1830. doi: 10.1016/j.cca.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 31.Charron F, et al. Tissue-specific GATA factors are transcriptional effectors of the small GTPase RhoA. Genes Dev. 2001;15(20):2702–2719. doi: 10.1101/gad.915701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coqueret O. Linking cyclins to transcriptional control. Gene. 2002;299(1-2):35–55. doi: 10.1016/s0378-1119(02)01055-7. [DOI] [PubMed] [Google Scholar]
- 33.Gallagher JM, Komati H, Roy E, Nemer M, Latinkić BV. Dissociation of cardiogenic and postnatal myocardial activities of GATA4. Mol Cell Biol. 2012;32(12):2214–2223. doi: 10.1128/MCB.00218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Latinkić BV, Kotecha S, Mohun TJ. Induction of cardiomyocytes by GATA4 in Xenopus ectodermal explants. Development. 2003;130(16):3865–3876. doi: 10.1242/dev.00599. [DOI] [PubMed] [Google Scholar]
- 35.Grépin C, Nemer G, Nemer M. Enhanced cardiogenesis in embryonic stem cells overexpressing the GATA-4 transcription factor. Development. 1997;124(12):2387–2395. doi: 10.1242/dev.124.12.2387. [DOI] [PubMed] [Google Scholar]
- 36.Koshiba-Takeuchi K, et al. Reptilian heart development and the molecular basis of cardiac chamber evolution. Nature. 2009;461(7260):95–98. doi: 10.1038/nature08324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5(3):320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 38.Ratineau C, Petry MW, Mutoh H, Leiter AB. Cyclin D1 represses the basic helix-loop-helix transcription factor, BETA2/NeuroD. J Biol Chem. 2002;277(11):8847–8853. doi: 10.1074/jbc.M110747200. [DOI] [PubMed] [Google Scholar]
- 39.Mullany LK, et al. Distinct proliferative and transcriptional effects of the D-type cyclins in vivo. Cell Cycle. 2008;7(14):2215–2224. doi: 10.4161/cc.7.14.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamamori-Adachi M, Goto I, Yamada K, Kitajima S. Differential regulation of cyclin D1 and D2 in protecting against cardiomyocyte proliferation. Cell Cycle. 2008;7(23):3768–3774. doi: 10.4161/cc.7.23.7239. [DOI] [PubMed] [Google Scholar]
- 41.Kowalczyk A, et al. The critical role of cyclin D2 in adult neurogenesis. J Cell Biol. 2004;167(2):209–213. doi: 10.1083/jcb.200404181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen S, Shimoda M, Chen J, Matsumoto S, Grayburn PA. Transient overexpression of cyclin D2/CDK4/GLP1 genes induces proliferation and differentiation of adult pancreatic progenitors and mediates islet regeneration. Cell Cycle. 2012;11(4):695–705. doi: 10.4161/cc.11.4.19120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pestell RG. New roles of cyclin D1. Am J Pathol. 2013;183(1):3–9. doi: 10.1016/j.ajpath.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neuman E, et al. Cyclin D1 stimulation of estrogen receptor transcriptional activity independent of cdk4. Mol Cell Biol. 1997;17(9):5338–5347. doi: 10.1128/mcb.17.9.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang T, Liu WD, Saunee NA, Breslin MB, Lan MS. Zinc finger transcription factor INSM1 interrupts cyclin D1 and CDK4 binding and induces cell cycle arrest. J Biol Chem. 2009;284(9):5574–5581. doi: 10.1074/jbc.M808843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kehn K, et al. Functional consequences of cyclin D1/BRCA1 interaction in breast cancer cells. Oncogene. 2007;26(35):5060–5069. doi: 10.1038/sj.onc.1210319. [DOI] [PubMed] [Google Scholar]
- 47.Aries A, Paradis P, Lefebvre C, Schwartz RJ, Nemer M. Essential role of GATA-4 in cell survival and drug-induced cardiotoxicity. Proc Natl Acad Sci USA. 2004;101(18):6975–6980. doi: 10.1073/pnas.0401833101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997;16(18):5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burgess A, et al. Inhibition of S/G2 phase CDK4 reduces mitotic fidelity. J Biol Chem. 2006;281(15):9987–9995. doi: 10.1074/jbc.M512714200. [DOI] [PubMed] [Google Scholar]
- 50.Paradis P, Dali-Youcef N, Paradis FW, Thibault G, Nemer M. Overexpression of angiotensin II type I receptor in cardiomyocytes induces cardiac hypertrophy and remodeling. Proc Natl Acad Sci USA. 2000;97(2):931–936. doi: 10.1073/pnas.97.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, et al. Convergence of protein kinase C and JAK-STAT signaling on transcription factor GATA-4. Mol Cell Biol. 2005;25(22):9829–9844. doi: 10.1128/MCB.25.22.9829-9844.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Debrus S, et al. The zinc finger-only protein Zfp260 is a novel cardiac regulator and a nuclear effector of α1-adrenergic signaling. Mol Cell Biol. 2005;25(19):8669–8682. doi: 10.1128/MCB.25.19.8669-8682.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Georges R, Nemer G, Morin M, Lefebvre C, Nemer M. Distinct expression and function of alternatively spliced Tbx5 isoforms in cell growth and differentiation. Mol Cell Biol. 2008;28(12):4052–4067. doi: 10.1128/MCB.02100-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Komati H, Maharsy W, Beauregard J, Hayek S, Nemer M. ZFP260 is an inducer of cardiac hypertrophy and a nuclear mediator of endothelin-1 signaling. J Biol Chem. 2011;286(2):1508–1516. doi: 10.1074/jbc.M110.162966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sive HL, Grainger RM, Harland RM, editors. Early Development of Xenopus laevis: A Laboratory Manual. Woodbury, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.