Abstract
The initial encounter of an unrepaired DNA lesion is likely to be with a replicative DNA polymerase, and the outcome of this event determines whether an error-prone or error-free damage avoidance pathway is taken. To understand the atomic details of this critical encounter, we have determined the crystal structures of the pol α family RB69 DNA polymerase with DNA containing the two most prevalent, spontaneously generated premutagenic lesions, an abasic site and 2′-deoxy-7,8-dihydro-8-oxoguanosine (8-oxodG). Identification of the interactions between these damaged nucleotides and the active site provides insight into the capacity of the polymerase to incorporate a base opposite the lesion. A novel open, catalytically inactive conformation of the DNA polymerase has been identified in the complex with a primed abasic site template. This structure provides the first molecular characterization of the DNA synthesis barrier caused by an abasic site and suggests a general mechanism for polymerase fidelity. In contrast, the structure of the ternary 8-oxodG:dCTP complex is almost identical to the replicating complex containing unmodified DNA, explaining the relative ease and fidelity by which this lesion is bypassed.
Keywords: abasic site, DNA polymerase, lesion, oxidative DNA damage, 8-oxoguanine, RB69, translesion synthesis
Introduction
The accurate replication of DNA is crucial for the maintenance of a functional genome and is accomplished by high-fidelity DNA polymerases. The basic architecture of the polymerase domains, the fingers, palm, and thumb, can be characterized as a ‘right hand' that is capable of holding DNA in its grasp (Joyce and Steitz, 1995; Ollis et al, 1985). DNA polymerases can be subdivided into several families. Family B DNA polymerases include the major replicative DNA polymerases α and δ and the replicative DNA polymerases from bacteriophages T4 and RB69. RB69 DNA polymerase (RB69 pol) contains two additional domains, an N-terminal domain and a 3′–5′ exonuclease domain. These domains, together with the polymerase active site domains, extend from a central opening. Three conformations of RB69 pol have been observed in crystal structures: the apo state without DNA (Wang et al, 1997), the editing mode forming a binary complex with DNA bound to the exonuclease site (Shamoo and Steitz, 1999) and the replicating mode, a ternary complex with DNA and dNTP bound in the polymerase site (Franklin et al, 2001). Differences between these structures indicate the conformational changes that occur during DNA polymerization. The fingers domain undergoes the largest conformational changes during DNA polymerization. In the apo state and editing mode, the enzyme is in the ‘open' conformation, characterized by a rotation of the fingers domain 60° away from the palm domain. In the ‘closed' conformation, Arg 482 and Lys 560 of the fingers domain are involved in hydrogen bonding to the phosphate groups of the incoming dNTP.
Replicative polymerases demonstrate high fidelity and processivity when utilizing normal DNA; however, they can be challenged with modified or damaged DNA, which has escaped the DNA repair machinery. The polymerase may then disengage from the primer/template to allow another DNA polymerase to bypass the DNA lesion (translesion synthesis), or another damage avoidance mechanism to occur, or the replicative polymerase may perform translesion synthesis itself. These events are important in understanding the mechanism by which DNA damage can result in miscoding, leading to mutagenesis and carcinogenesis. Numerous studies have shown that the efficiency and fidelity of translesion synthesis is both polymerase and lesion specific (Shibutani et al, 1991; Moriya et al, 1999; Lehmann, 2002). Understanding the molecular and structural events that take place when a DNA polymerase encounters damage is therefore of major importance.
2′-deoxy-7,8-dihydro-8-oxoguanosine (8-oxodG) is an endogenous premutagenic lesion generated by reactive oxygen species as a result of aerobic respiration. 8-OxodG is one of the most prevalent lesions found in DNA and, if not repaired, can direct incorporation of dATP, resulting in G → T transversions. Structures of the 8-oxodG:dC pair in duplex DNA show little disruption of the overall DNA structure (Oda et al, 1991; Lipscomb et al, 1995), while the glycosidic torsion angle of the modified dG in the miscoding base pair 8-oxodG:dA is syn, allowing the stabilization by two hydrogen bonds (Kouchakdjian et al, 1991; McAuley-Hecht et al, 1994). DNA polymerases from different families are diverse in their fidelity of 8-oxodG translesion synthesis (Shibutani et al, 1991). Therefore, it is likely that the structure of the premutagenic lesion within the polymerase active site determines the miscoding properties of the lesion. Recently, the structure of the repair polymerase, DNA polymerase β (pol β), with an 8-oxodG:dCTP base pair was solved and shows that the 5′-phosphate backbone flips 180° in order to accommodate the oxygen at C8 (Krahn et al, 2003).
About 10 000 abasic sites are created per day in each mammalian cell (Lindahl, 1979). They arise through spontaneous depurination or by the action of DNA glycosylases during base excision repair. The natural abasic site is unstable and we have used tetrahydrofuran (3-hydroxy-2-(hydroxymethyl)tetrahydrofuran), an isosteric and isoelectronic model of the closed form of an abasic site. In vivo and in vitro studies have shown that dA is preferentially incorporated opposite tetrahydrofuran and that this lesion represents a block to DNA synthesis (Takeshita and Eisenberg, 1994; Shibutani et al, 1997).
Here we present the structure of a replicative family B polymerase with two of the most common forms of DNA damage, 8-oxodG and a model abasic site (AP). Based on these two structures and the ability of RB69 pol to accommodate both modified DNA sequences in its active site, we can make predictions as to which other lesions can be tolerated in the active site of a high-fidelity DNA polymerase and which are sterically too demanding to produce the necessary stability required for polymerization. This snapshot of translesion synthesis increases our understanding of miscoding, a first step in mutagenesis.
Results
Structure determination
To prevent degradation of the oligonucleotides during crystallization, an exonuclease-deficient polymerase mutant of RB69 pol was used as described (Wang et al, 1995; Franklin et al, 2001). The dideoxy-terminated primer cannot be extended, and allows capture of the polymerase during nucleotide incorporation opposite or past 8-oxoguanine (8-oxodG) and tetrahydrofuran, the abasic site model (Figure 1B). Two different template strands were designed for both lesions: one, where the lesion is positioned for nascent base pair formation (Figure 1A, (1)), allowing analysis of nucleotide insertion opposite the lesion site, and a second sequence, where the lesion base pair is poised for extension (Figure 1A, (2)). Additionally, an unmodified template strand (Figure 1A, (3)) was used in crystallization trials to study the structures of mismatched nascent base pairs. Except for the site of modification, the DNA sequences were identical to those in the described ternary complex with unmodified DNA and a nascent Watson–Crick dA:dTTP base pair (Franklin et al, 2001) to ensure that the structural changes observed are exclusively due to the specific adduct. Two complexes featuring 8-oxodG (8-oxodG:dCTP complex) and an AP lesion (AP:dG complex) in the polymerase active site were analyzed (Table I). Four additional primer/template/dNTP combinations led to crystals with the DNA bound to the exonuclease active site (see below).
Figure 1.
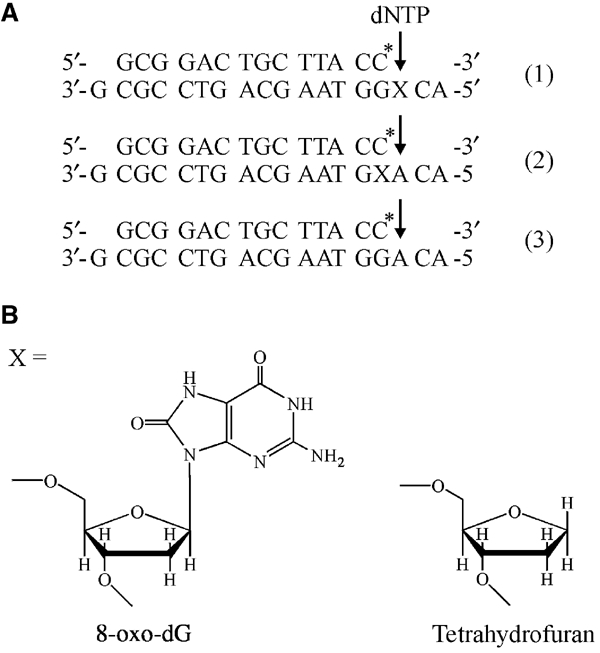
Oligonucleotide sequences and DNA adducts. (A) The 14 nt primers are identical in all trials and terminated by ddC (indicated by C*). Template strands are 18 nt long with a 3′-dG overhang. X denotes the position of the lesion for primer/template combinations (1) and (2), and the arrow the position of dNTP incorporation. (B) Structures of the lesions at position X in the templates 8-oxodG and tetrahydrofuran (abasic site model).
Table 1.
Data collection and refinement statistics
| Data collection statistics | 8-oxodG:dCTP complex | AP:dG complex | 8-oxodG editing complex |
|---|---|---|---|
| Space group | P212121 | P21 | P1 |
| Unit cell dimensions: | |||
| a, b, c (Å) | 80.8, 118.6, 127.2 | 131.9, 122.2, 165.4 | 73.1, 124.7, 126.5 |
| α, β, γ (deg) | 90, 90, 90 | 90, 96.8, 90 | 118.8, 89.7, 106.8 |
| Unique reflections | 30441 | 137338 | 148642 |
| Resolution (highest shell) (Å) | 46.1–2.8 (2.9–2.8) | 47.1–2.7 (2.8–2.7) | 50.0–2.35 (2.43–2.35) |
| Rsym (highest shell) | 0.116 (0.616) | 0.113 (0.434) | 0.045 (0.373) |
| Completeness (highest shell) (%) | 99.9 (100.0) | 94.7 (73.3) | 98.0 (97.1) |
| Average redundancy | 4.5 | 4.8 | 6.3 |
| 〈I〉/〈σ(I)〉 (highest shell) | 11.0 (1.8) | 19.1 (1.8) | 10.1 (2.1) |
| Refinement statistics | |||
| Number of reflections used in refinement | 28854 | 130465 | |
| Final R (Rfree) | 20.4 (27.1) | 21.5 (28.8) | |
| Deviations from ideal values in | |||
| Bond distances (Å) | 0.016 | 0.013 | |
| Bond angles (deg) | 1.87 | 1.54 | |
| Torsion angles (deg) | 7.5 | 6.5 | |
| Chiral centers (Å3) | 0.09 | 0.11 | |
| Planar groups (Å) | 0.006 | 0.005 | |
| Ramachandran statistics (%) | 88.0/10.9/1.0/0.1 | 86.7/12.1/0.8/0.4 | |
| Model contents (number of atoms) | |||
| Protein | 7356 | 29460 | |
| DNA | 652 | 2692 | |
| Waters | 84 | 535 | |
| Bound metals | 3 | 8 | |
|
Other ligands |
28 |
61 |
|
| Rsym=∑hkl∑i ∣Ii−〈I〉∣/∑i 〈I〉, where Ii is the ith measurement and 〈I〉 is the weighted mean of all measurements of I. 〈I〉/〈sigI〉 indicates the average of the intensity divided by its standard deviation. Numbers in parentheses refer to the respective highest resolution data shell in each dataset. Rcryst=∑∣∣Fo∣−∣Fc∣∣/∑∣Fo∣ where Fo and Fc are the observed and calculated structure factor amplitudes. Rfree same as Rcryst for 5% of the data randomly omitted from refinement. Ramachandran statistics indicate the fraction of residues in the most favored, additionally allowed and generously allowed and disallowed regions of the Ramachandran diagram as defined by PROCHECK (Laskowski et al, 1993). | |||
Overall structure of the 8-oxodG:dCTP complex and comparison with the replicative complex
Steady-state kinetics show that, in contrast to other pol α-like DNA polymerases (Shibutani et al, 1991), RB69 pol incorporates dCTP opposite 8-oxodG 20-fold more efficiently than dATP (Table II; see also Figure 6D). The ternary 8-oxodG:dCTP complex formed with DNA sequence 1 (Figure 1A, X=8-oxodG) and dCTP crystallized in the same space group, with identical cell parameters as the replicative complex (Franklin et al, 2001). Refinement of 902 protein residues, the DNA, dCTP, and three calcium ions converged against an R factor of 20.4% (Rfree of 27.1%) for all data from 46.1 to 2.8 Å (Table I). The overall structure of the complex is shown in Figure 2A. The DNA is bound to the polymerase active site similar to that observed for the complex with unmodified DNA. A superposition of both structures, based on the Cα atoms of residues 1–901 and all atoms of the DNA (excluding the 8-oxoguanine base) and the nucleotide triphosphates (excluding C5M of dTTP), reveals a root mean square (rms) displacement of 0.5 Å, showing that the two structures are nearly identical. The largest difference can be found between the riboses of the cytidine next to the 5′-end of the primer, with a maximum displacement of 3.3 Å for the C5′ atoms. The maximum deviation with respect to the protein residues can be found in a loop region (residues 255–259) of the exonuclease domain, with a peak value of 2.4 Å for the Tyr 257 Cα atoms.
Table 2.
Steady-state incorporation kinetics opposite 8-oxodG
| Template | dNTP | Km (μM) | kcat (min−1) | kcat/Km (μM/min) | dCTP:dATP |
|---|---|---|---|---|---|
| 8-oxodG | dCTP | 20 | 31 | 1.6 | 20 |
| dATP | 270 | 22 | 0.08 | ||
| dG | dCTP | 0.64 | 46 | 72 | 5 × 105 |
| dATP | 220 | 0.03 | 0.00014 |
Figure 6.

Modeling of base pairs in the polymerase active site. (A) Model of an 8-oxodG:dATP base pair in the polymerase active site of RB69 pol. The syn conformation of 8-oxodG (green) leads to steric interference between O8 and the Cα atom of Gly 568. (B) Model of an 8-oxodG:dA base pair poised for extension. Modeling the 8-oxodG base in the syn conformation generates only a contact (yellow arrow) to a water molecule. (C) Model of an 8-oxodG:dC base pair poised for extension. The anti conformation of 8-oxodG leads to a steric clash with the phosphate group. (D) Nucleotide selection during insertion opposite 8-oxodG. RB69 pol (0–19 nM) was incubated with P:T (Figure 1A, sequence 1) in the presence of 5 μM dCTP (closed circles) or dATP (open circles), as described in Materials and methods. The % of extended primer was plotted versus the RB69 pol concentration. (E) Extension of an 8-oxodG:dC (closed circles) or an 8-oxodG:dA (open circles) base pair as described above, but using 5 μM dTTP instead of dCTP.
Figure 2.
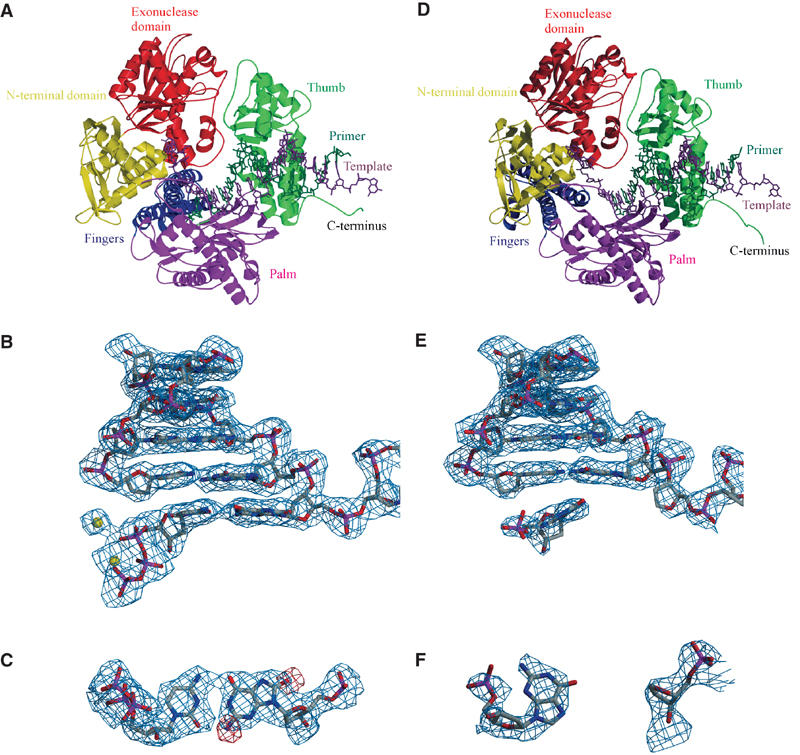
Structure of RB69 pol/DNA complexes. (A) Overall structure of the replicating 8-oxodG:dCTP complex formed between RB69 pol, DNA sequence 1 (Figure 1A, X=8-oxodG), and dCTP. RB69 pol is colour coded according to its five domains (N-terminal domain: residues 1–108 and 340–382 in yellow; 3′–5′ exonuclease domain: residues 109–339 in red; palm domain: residues 383–468 and 573–729 in magenta; fingers domain: residues 469–572 in blue; and thumb domain: residues 730–903 in green) as well as the primer (dark green) and template (dark magenta) DNA strands and the dCTP (dark green). (B) Final SIGMAA-weighted 2Fo−Fc electron density map contoured at 1σ, covering the 8-oxodG:dCTP base pair as well as the four base pairs preceding the nascent base pair. Ca2+ ions are depicted as yellow spheres. (C) Top view of the nascent 8-oxodG:dCTP base pair with the SIGMAA-weighted 2Fo−Fc electron density contoured at 1σ obtained after molecular replacement with the dA:dTTP-containing structure. Difference electron density at 3σ is shown in red and marks the positions of the O8 and N2 groups of 8-oxodG. (D) Overall structure of the AP:dG complex (molecule C) formed by RB69 pol, DNA sequence 1 (Figure 1A, X=AP, including an additional G at the template 3′-end) and dGMP, colour coded as in (A). (E) Electron density map of the DNA (analogous to (B)). (F) Top view of the AP:dGMP ‘pair' featuring the final electron density map contoured at 1σ. Figures 2, 3, 4, 5, 6 and 7 were generated with Molscript (Kraulis, 1991) and Raster3D (Merrit and Murphy, 1994).
The DNA is well defined in the complex (Figure 2B) and in the initial difference electron density map additional residual electron density at 3.0σ was observed, indicating the position of N2 and O8 in 8-oxodG (Figure 2C). Both bases of the 8-oxodG:dCTP base pair retain the anti conformation around the N-glycosidic bond and form Watson–Crick hydrogen bonds. Distances are at 2.7 Å each for N4_dC…O6_8-oxodG and N2_8-oxodG…O2_dC, and 2.8 Å for N1_8-oxodG…N3_dC, in the normal range for dG:dC base pairs in B-form DNA (2.86–2.98 Å for N1…N3 and N4…O6, 2.80–2.85 Å for N2…O2; Chiu and Dickerson, 2000).
The size and shape of the polymerase active site binding pocket plays a crucial role in replication fidelity and thus in lesion tolerance. A rigid active site is formed by a number of highly conserved residues located at the base of the two anti-parallel α-helices of the fingers domain and the β-sheet of the palm domain. Several of these residues (Leu 415, Tyr 416, and Tyr 567 on the dNTP side; Tyr 567 and Gly 568 on the template side) are solely involved in hydrophobic interactions with the DNA and define a tight-fitting binding pocket at the minor groove side of the nascent base pair (Figure 3A). Asp 411 and Asp 623, which coordinate the active site Mg2+ ions, complete the pocket at the minor groove side of the sugar and phosphate backbone of the incoming nucleotide. Other conserved residues in the active site, Asn 564 and Ser 565, as well as the type-conserved Leu 561 form a rigid rear wall to ensure a coplanar base arrangement of the nascent base pair (Figure 3C). A hydrogen bond between the amino group of Asn 564 and the α-phosphate of the dNTP in the ‘closed' state (3.2 Å) may assist in positioning the rear wall of the binding pocket. In the ‘closed' conformation, Arg 482 and Lys 560 of the fingers are involved in hydrogen bonding to the phosphate groups of the incoming dNTP and in this way define the wall of the binding pocket that restricts the width of the active site on the primer side. The amino group of Lys 486 in the ‘closed' conformation is not within hydrogen bonding distance (4.1 Å) to the γ-phosphate, but H-bonding to the incoming dNTP during the transition from the ‘open' to the ‘closed' conformation seems feasible. The size of the binding pocket is sufficient for the consensus base pair shape of a Watson–Crick base pair (Kool, 2002), but shows little tolerance for groups protruding into the minor groove, while the major groove side is relatively unrestricted.
Figure 3.
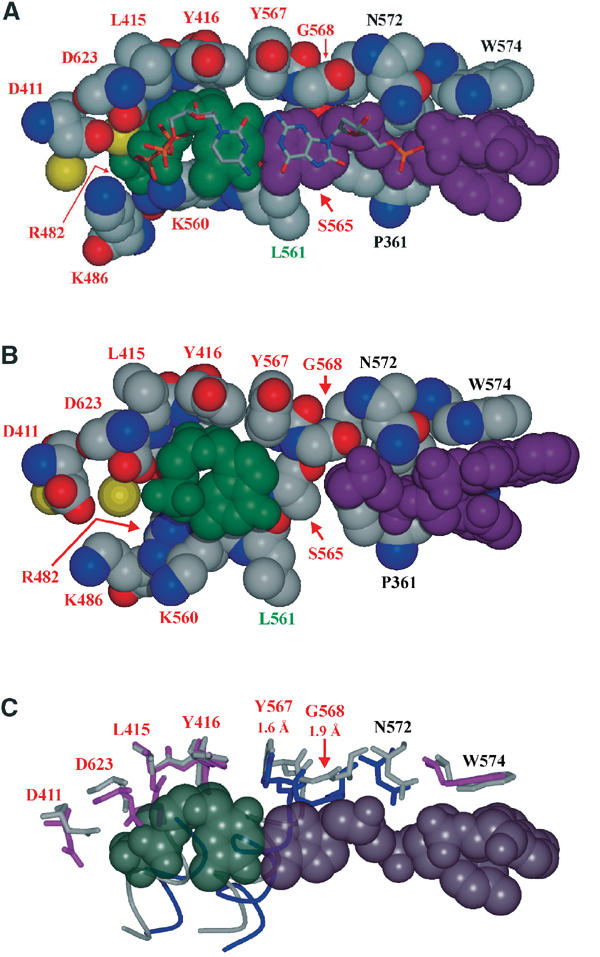
The active site. (A) Superposition of the 8-oxodG:dCTP base pair (stick mode) with the dA:dTTP base pair (CPK mode) of the replicating complex with unmodified DNA. Also shown are the amino acids of the active site (red: highly conserved, green: type conserved, black: not conserved). (B) Polymerase active site with the nascent AP:dGMP ‘pair'. (C) Superposition of active site amino acids of the 8-oxodG:dCTP complex (colour coded according to the different domains as in Figure 2A) and the AP:dG complex (in light gray). Numbers denote displacements of Cα main chain atoms (for a complete list, see supplementary data, Supplementary Table S1). The dA:dTTP base pair has been included as in part A.
Notably, the positions of the amino acids in the 8-oxodG:dCTP complex forming the polymerase active site are identical within the error limits to those in the replicating complex with unmodified DNA. A superposition of the nascent base pairs (Figure 3A) reveals that the 8-oxodG lesion can be accommodated without distortions, presumably because the 8-oxo group of the base projects into the unrestricted major groove of the active site pocket. Another reason might be a sharp kink in the single-stranded template DNA 5′-overhang, which in the structure with the unmodified DNA template prevents a possible C8-H…O5′ base–backbone interaction that stabilizes the anti conformation of purine bases in nucleic acids (Shefter and Trueblood, 1965; Rubin et al, 1972; Sussman et al, 1972). In the 8-oxodG:dCTP complex, however, this kink provides the additional space needed to accommodate the 8-oxo group without steric clashes with the backbone.
Pol β also shows a similar though less pronounced bend in the 5′-phosphodiester bond of the template strand. Nevertheless, the 8-oxo group still adopts the nonmutagenic anti conformation due to a second structural change, the flipping of the 5′-phosphate by 180°. This allows pol β to incorporate preferentially dCTP opposite an 8-oxodG lesion as well (Shibutani et al, 1991).
Overall structure of the AP:dG complex and comparison with other states
Replicative DNA polymerases challenged with an abasic site tend to follow the ‘A-rule', the preferential incorporation of adenine opposite noninstructive lesions. RB69 pol additionally incorporates dGTP opposite an AP site, albeit less efficiently (data not shown). Crystallization of RB69 pol with DNA containing an AP lesion in the template position (Figure 1A, sequence 1, X=AP) and dATP or dGTP was tried; however, only setups with dGTP were successful (see Discussion below on extended template strand). The resulting AP:dG complex crystallized in space group P21, a unit cell that has not been observed previously for RB69 pol, with four molecules (A, B, C, and D) in the asymmetric unit. Refinement of the full-length protein, the DNA, part of the dGTP molecules, and two calcium ions per complex resulted in R=21.5% (Rfree of 28.8%) for all data from 47.1 to 2.7 Å (Table I). As the four complexes only show minor structural differences, the following discussion is based on molecule C. The primer/template DNA is bound to the polymerase active site as seen in the replicating and the 8-oxodG:dCTP complexes (Figure 2D), but the electron density for the incoming nucleotide, dGTP, is poorly defined in all four molecules. In molecule C, sufficient electron density was observed to include a dGMP (Figure 2F), whereas the electron density in molecules B and D only allowed refinement of the guanosine and in molecule A no density was observed for an incoming nucleotide. Possible explanations for this observation include (a) hydrolysis of dGTP to dGMP, (b) decreased binding affinity and thus higher flexibility of dGTP in the active site compared to the replicating complex, (c) partial occupancy of the nucleotide triphosphate in the active site, or (d) a combination of all three.
Superpositions of molecule C with the polymerase in replicating mode (Franklin et al, 2001), editing mode (Shamoo and Steitz, 1999) and the apo enzyme (Wang et al, 1997) (see supplementary data, Supplementary Figure S1) show that the structure of the AP:dG complex can be described as a hybrid structure between the enzyme in replicating and editing or apo mode, revealing a novel structural conformation of RB69 pol. Apart from a slight movement of the exonuclease domain, all domains except the fingers domain adopt the same conformation relative to each other and to the bound DNA as in the replicative complex (Figure 4A). The fingers domain, however, is in the conformation found in the apo enzyme and the editing complex, leaving the polymerase in the open, catalytically inactive state (Figure 4B). The closed conformation seen in the replicative and the 8-oxodG:dCTP complexes is essential for the catalytic formation of the phosphodiester bond, because in this state the three highly conserved residues Arg 482, Lys 486, and Lys 560 at the base of the fingers domain are able to complement the tight-binding pocket on the dNTP side (see Figure 3). In this way, they ensure correct positioning of the incoming nucleotide relative to the other residues and metal ions. A consequence of the open conformation in the AP:dG structure is a significant movement of the active site amino-acid residues belonging to the fingers domain ranging from 1.2 to 7.1 Å in Cα positions compared to the closed replicative complex (Figure 3C), thereby increasing the distance to the DNA. The only residue that moves towards the DNA is the universally conserved Gly 568 (1.9 Å) (Hopfner et al, 1999; Zhao et al, 1999; Rodriguez et al, 2000; Hashimoto et al, 2001), which now fills part of the space occupied by the adenine base of the template strand in the replicative complex (compare Figures 3A and B). In the AP:dG complex, only one Ca2+ ion at the position of the Mg2+ ion that is hypothesized to promote deprotonation of the 3′-OH of the primer strand (Steitz, 1993) was found. The second metal ion, which is required for the stabilization of the pentacovalent transition state of the α-phosphate and compensation of the negative charges, is missing as in the editing complex (Shamoo and Steitz, 1999).
Figure 4.

Comparison of the AP:dG and replicative complexes. (A) Ribbon presentation of the AP:dG complex, colour coded with respect to rms deviations from the replicating complex. The superposition is based on the Cα atoms of the palm domain (residues 383–468, 573–729), as they define a relatively rigid unit of the protein. Rms deviations for the Cα atoms of the other four domains were calculated using RMSPDB (Kleywegt et al, 2001). Gray colour denotes virtually no changes, while red colour shows maximum domain shifts (see depicted colour code). (B) Detail of (A), showing the fingers, palm, and N-terminal domains of the AP:dG complex and additionally the fingers domain of the replicative complex (dark gray) to illustrate the domain shift. The DNA is shown in green/magenta for the AP:dG complex and yellow for the replicating complex.
Subunit interactions of the 8-oxodG:dCTP and AP:dG complexes
Inter-complex contacts in the structure of the 8-oxodG:dCTP complex are limited to the binding of the template 3′-dG overhang of one molecule to a pocket formed by amino acids of the N-terminal domain of a neighboring molecule. Although the precise function of this binding pocket is still unclear, guanine binding to this site has been described for the replicating complex, the editing complex, and the apo enzyme. In the AP:dG complex, a similar interaction can be observed (Figure 5A). Surprisingly, analysis of the electron density map revealed the presence of an additional unpaired guanine base at the 3′-end of the template strand (Figure 5B), indicating that RB69 pol is able to add untemplated nucleotides to the 3′-end of an oligonucleotide. To determine whether RB69 pol exhibits terminal deoxynucleotidyl transferase activity under crystallization conditions, reactions were set up as described in Materials and methods. In the presence of dGTP, nearly 100% of the template strand was extended by one base (Figure 5C). Extension in the presence of dCTP, dATP or dTTP, was much less efficient. One-base extension occurred equally well when the template contained 8-oxodG, and a two-base extension product can be observed as well. The terminal transferase activity in the presence of dGTP may explain why better diffracting crystals of the complex with the AP lesion were obtained with dGTP and not with the more easily inserted dATP.
Figure 5.

Intermolecular interactions in the AP:dG complex. (A) Contacts between neighboring molecules in the AP:dG complex. The 3′-dG of each template strand is bound to a specific binding pocket in the N-terminal domain of an adjacent polymerase molecule. (B) Final SIGMAA-weighted 2Fo−Fc electron density map of the binding pocket at 1σ for the amino acids, part of the template (magenta) as well as the primer strand (green) revealing the presence of an additional unpaired dG at the 3′-end of the template. (C) Terminal transferase activity of RB69 pol in the presence of dideoxy-terminated primed templates (P:T) containing an abasic site (AP) or 8-oxodG. Primer and template bands are indicated with arrows. P:T was incubated with RB69 pol in the absence (lane 1) or presence of dGTP (lanes 2 and 6), dATP (lane 3), dCTP (lane 4), or dTTP (lane 5).
To rule out the possibility that the observed open conformation of the polymerase in the AP:dG complex might be due to packing effects, a superposition of the replicative complex with each of the four molecules of the AP-containing structure was performed. For molecules A, C, and D, the closed formation could also be accommodated in the crystal packing. In the case of molecule B, the ‘closed' conformation would introduce steric clashes, which could nevertheless be easily avoided through minor movements inside the crystal lattice.
Overall structure of the 8-oxodG editing complex
Attempts to crystallize mismatched nascent base pairs showing incorporation of either dCTP or dGTP opposite dA (Figure 1A, sequence 3), a complex featuring incorporation of dATP opposite 8-oxodG (sequence 1, X=8-oxodG) and a complex showing the extension event past an 8-oxodG:dC base pair (sequence 2, X=8-oxodG), led to another new crystal form (space group P1, Table I). Of the three molecules per unit cell, one shows the enzyme in the apo state while the other two present another example for RB69 pol in the editing mode. As in the editing complex with unmodified DNA (Shamoo and Steitz, 1999), the 3′-end of the template, not the 3′-end of the primer, is bound to the exonuclease active site. Accordingly, the 8-oxodG lesion near the 5′-end of the template is exposed to the surrounding solvent, but cannot be localized due to disorder.
Due to the fact that we were not able to capture these potentially interesting complexes with the 3′-end of the primer bound to the polymerase active site and because of the similarity of the 8-oxodG editing complex to the editing complex with unmodified DNA published earlier, no further refinement was performed. Nevertheless, considering the sequence similarity of the DNA ends in both editing complexes, it is unlikely that the presence or nature of the 8-oxodG lesion triggers this switching from the replicating to the editing mode and other reasons must be considered. Furthermore, the results are valuable in suggesting which primer/template combinations are likely to result in the editing complex and which ‘base pairs' can be tolerated in the polymerase active site.
Discussion
Formation of replicating versus editing complexes
A number of attempts have been made to obtain structures of RB69 pol in complex with different DNA sequences to gain further insights into the reaction catalyzed at the replication fork. While the presence of unmodified DNA yielded a model of the binary editing complex, incorporation of a dideoxy-terminated primer and addition of a correct incoming nucleotide triphosphate led to the structure in the replicating mode. These results suggested that the binary editing complex exhibits a greater stability than a binary replicating complex. The opposite seems to be true if a ‘correct' incoming dNTP is provided, suggesting an enhanced stability of the closed ternary replicative complex compared to the binary editing complex. Use of a dideoxy-terminated primer to ‘lock' the polymerase into a ternary complex contributes to this stability and can be demonstrated by gel shift assays, which showed a significant drop in the dissociation constant when a dideoxy-terminated primer was used (data not shown).
The importance of the ‘correct' dNTP becomes apparent in the study with RB69 pol and mismatched nascent base pairs described above that led to the formation of editing complexes (analogous to the 8-oxodG editing complex). Thus, the fit of the nascent base pair into the rigid binding pocket of the polymerase active site seems to be essential to stabilize the DNA in the polymerase active site. Accordingly, an incorrect nucleotide prevents the formation of the catalytically active, closed conformation due to steric exclusion. The resulting open conformation leads to a diminished binding affinity of the DNA in the polymerase active site and thus the editing complex is observed.
Two other DNA/dNTP combinations that led to the formation of an editing complex support this hypothesis: 8-oxodG:dATP (Figure 1A; sequence 1, X=8-oxodG; the 8-oxodG editing complex) and a primed template containing an 8-oxodG:dC base pair at the penultimate position, with a nascent dA:dTTP base pair (sequence 2, X=8-oxodG). Modeling of an 8-oxodG:dATP base pair in the active site of RB69 pol (Figure 6A) indicates that the fixed width of the active site forces one of the two purine bases into the syn conformation. There are fewer steric clashes with 8-oxodG in syn, but the proximity of the O8 group to the backbone atoms of Gly 568 is unfavorable and may lower the stability of the replicative complex such that formation of the editing complex is preferred.
Models of an 8-oxodG:dA (Figure 6B) or 8-oxodG:dC (Figure 6C) base pair in the penultimate position representing the extension from the lesion base pair were also generated. The 8-oxodG:dC model clearly shows a steric clash between the O8 and the phosphate group of 8-oxodG. Due to the proximity to the side chain of Asn 572 on the opposite side of the DNA backbone and other nearby residues, the only way to avoid this clash would be a slight shift of the newly replicated duplex DNA towards the major groove, thereby loosening the grip of the polymerase on the DNA. In contrast, the 8-oxodG:dA model shows that unfavorable steric clashes with the backbone phosphate can again be avoided through the syn conformation of 8-oxodG. A water molecule that forms hydrogen bonds to OH of Tyr 567 (2.7 Å) and N3 of guanine (2.7 Å) in the structure with the unmodified template would be in close proximity to the O8 atom. This water molecule is only weakly present in the 8-oxodG:dCTP complex, and is not observed in any of the four molecules of the AP:dG complex. Although it has been proposed to be of particular importance with respect to the binding affinity of DNA to the polymerase active site (Franklin et al, 2001), the O8 atom of 8-oxodG in the syn conformation could substitute for this water and would form a hydrogen bond to OH of Tyr 567 without the loss of protein–DNA interactions. These structural constraints are reflected in the efficiency of DNA synthesis on these primed templates. As mentioned earlier, incorporation of dATP opposite 8-oxodG by RB69 pol is much less efficient (20-fold) than incorporation of dCTP (Table II and Figure 6D); in contrast, extension of the 8-oxodG:dA base pair is more efficient (Figure 6E). Favored extension of an 8-oxodG:dA base pair is common among DNA polymerases (Shibutani et al, 1991; Miller and Grollman, 1997; Haracska et al, 2002), except for DNA polymerase η (Zhang et al, 2000).
Gly 568 influences DNA binding
According to our hypothesis regarding the relative energetics of replicating and editing complexes, an RB69 pol–DNA complex would only be stable in the polymerase site in its ternary form, requiring the presence of a complementary dNTP that fits into the rigid active site. However, the formation of the AP:dG complex seems to contradict this hypothesis as the poor electron density for the dGTP molecule, the absence of the second catalytic metal ion, and the open conformation of the polymerase rather indicate that AP:dG is a binary complex with DNA bound to the polymerase active site. We propose that this apparent contradiction is a consequence of the nature of the AP lesion. A comparison of the AP:dG and 8-oxodG:dCTP complexes reveals that Gly 568 is displaced by 1.9 Å (Figure 3C) and protrudes into the binding pocket at the AP site (Figure 3B). It is the only active site residue that competes for space with a DNA residue and appears to be ‘pushed back' by the template 8-oxoguanine or adenine base in the ternary complexes in the closed conformation (Figure 3A). We therefore define Gly 568 in the closed complex to be in the strained state (Figure 7, IA/IIA), while Gly 568 protruding into the binding pocket in the open complex adopts the relaxed state (Figure 7, IB/IIB). Thus, for RB69 pol the open conformation and relaxed state seem to be associated. As a consequence, there is a competition between Gly 568 and the template base in the active site, which weakens or even prevents DNA binding (Figure 7, IB, C/IIB, C). The preferred binding of binary complexes with unmodified DNA to the exonuclease active site corroborates this view of diminished binding affinity. The catalytically active, closed complex can only be formed in the presence of a correct dNTP. Here the fingers domain closes down and pushes the nascent base pair against the active site residues. In this way, Gly 568 is moved into its strained state and the phosphodiester bond can be formed (Figure 7, ID). From the experiments presented here, one can conclude that the closed complex exhibiting the strained state (Figure 7, ID) seems to be energetically favored over the open conformation in the relaxed state (Figure 7, IIB), provided that the nascent base pair fits into the tight binding pocket (Figure 7, ID). Therefore, if no binding of a complementary dNTP occurs, for example due to a lesion, the template DNA is released. In the special case of an abasic site as in the AP:dG complex, the absence of a template base allows Gly 568 to adopt the relaxed state even in the closed conformation (Figure 7, IID), facilitating the formation of a stable binary complex at the polymerase active site. Involvement of Tyr 567 in maintaining replication fidelity has been suggested by the increased error frequency of the Y567A mutant (Yang et al, 1999). The lower fidelity of Y567A may be a result of the increased size of the binding pocket at the minor groove side of the nascent base pair. Additionally, it is also feasible that the smaller Ala residue might ease the strain imposed on Gly 568 in the closed conformation. This would enhance the binding affinity of DNA to the active site, increasing the persistence of the complex and the probability for incorporation of an incorrect nucleotide. Further support for the importance of Gly 568 comes from the described 8-oxodG:dA base pair model. The observed steric clash between O8 of 8-oxodG and Gly 568 in the already strained state should weaken DNA binding even further and thus make nucleotide insertion more difficult, in accordance with biochemical data (Figure 6D).
Figure 7.
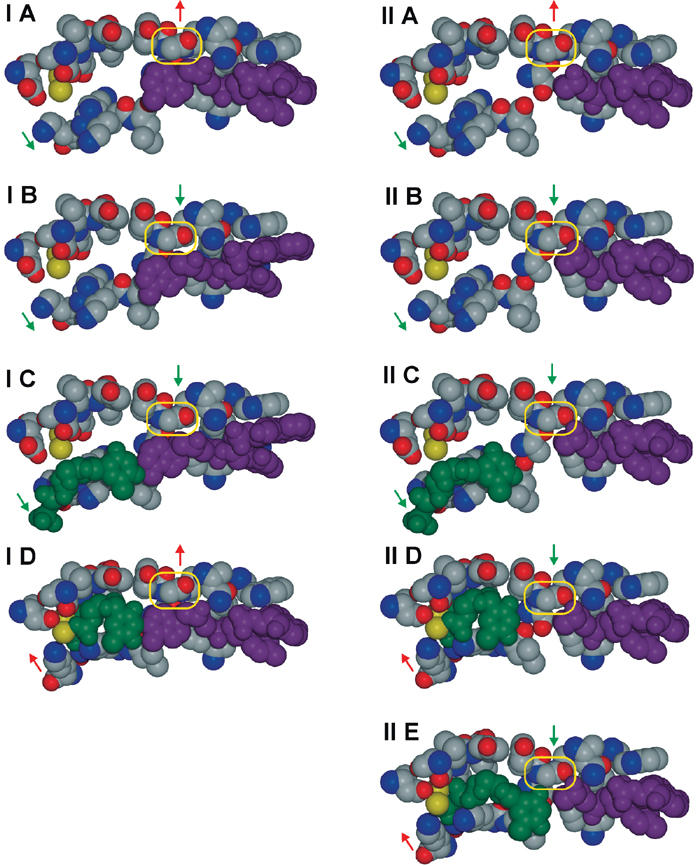
Influence of Gly 568 on DNA binding to the polymerase active site. Column (I) shows the event of nucleotide insertion opposite an unmodified template strand with adenine in the active site, while column (II) depicts the case of a template containing an abasic site as in the AP:dG complex. Vertical arrows specify the strained (red) or the relaxed state (green), respectively. Diagonal arrows indicate whether the polymerase is in the closed (red) or open conformation (green). The template strand is depicted in magenta and the incoming nucleotide in green. The yellow box indicates the position of Gly 568. (IA) and (IIA) show the polymerase in the strained state and the open conformation. Transition into the relaxed state presumably causes the adenine base of the unmodified template to be pushed back (IB), while the AP-containing template is unaffected (IIB). (IC) and (IIC) depict an incoming dNTP bound to the base of the fingers domains. Transition to the closed and strained conformation ensures the correct positioning of all residues to enable the catalytic phosphodiester bond formation (ID). In the case of AP, a closed and relaxed conformation is feasible (IID). The missing complementary base causes the dNTP to be held in place less tightly and phosphodiester bond formation is less efficient (IIE).
Figure 7, part IIE, illustrates why besides dA, RB69 pol also inserts dG opposite an AP site. For phosphodiester bond formation, the correct position of the incoming dNTP is crucial, which is accomplished by the conserved amino acid residues and by its complementary base. Absence of the latter boundary, as in the case of an abasic site, makes the smaller pyrimidine bases less likely to be in the right position than the sterically more demanding purine bases A and G. This, together with the superior stacking ability of adenine and its relatively weak solvation compared to G, C, and T, is also the rationale behind the ‘A-rule' (Kool, 2002).
Based on the insights gained from these studies, we are now able to predict which new lesion/dNTP combinations will be tolerated in the active site of a family B DNA polymerase and which may be sterically too demanding to enable replication. First, lesions in the template strand must allow the tight steric fit of the binding pocket on the minor groove side; hence, any sterically demanding groups may only protrude into the major groove. Second, unless dealing with abasic sites, suitable incoming nucleotides or nucleotide derivatives must be provided to allow the formation of a closed ternary complex. Even if a nascent ‘base pair' passes the requirements of the active site binding pocket, further extension may be inhibited because of an additional size requirement that has to be fulfilled at the template side upon DNA translocation. This restraint is enforced by Asn 572 in connection with the tight arrangement of the surrounding amino acids, which prohibit movement of this residue. The presence of an abasic site, however, leads to a catalytically inactive state that is distinct from the apo or exo states of the polymerase and explains the replication blockage at the molecular level.
Materials and methods
Oligonucleotide synthesis and purification
Modified template oligodeoxynucleotides were synthesized and purified as described (Bodepudi et al, 1992; Shibutani et al, 1993; Takeshita et al, 1987). A ddC was added to the 3′-end of the primer. The homogeneity of the oligonucleotides was confirmed by 32P-labeling and electrophoresis on a 20% denaturing polyacrylamide gel. Oligonucleotides were desalted and annealed prior to use in crystallization and were quantified by A260 using molar extinction coefficients.
Protein expression and purification
The plasmid containing exonuclease-deficient RB69 pol mutant (D222A/D327A) was a generous gift from Dr WH Konigsberg. The coding sequence was cloned into the expression vector pQE30 (Qiagen, USA, BamHI and PstI) and transformed into strain M15[pREP4]. Cultures were grown in LB medium with 50 μg/ml carbenicillin, at 37°C until OD600=0.7 was achieved, transferred to 15°C for 1 h, induced with 0.5 mM IPTG, and grown ∼20 h at 15°C. Cells were harvested, resuspended in buffer (20 mM NaH2PO4/Na2HPO4 (pH 6.0), 1 mM EDTA), and frozen at −80°C. Cells were lysed by incubation with 0.1 mg/ml hen egg white lysozyme (Sigma, USA) for 1 h, followed by three cycles in a French pressure cell. The cleared cell lysate was passed over a Ni-NTA agarose (Qiagen, USA) column equilibrated with buffer A (20 mM NaH2PO4/Na2HPO4 (pH 6.0), 500 mM NaCl, 10% glycerol) and the protein was eluted with increasing imidazole concentrations. After buffer exchange (buffer B, 10 mM Tris–HCl (pH 7.5), 10 mM KCl, 5% glycerol, 3 mM DTT), the protein was applied to a Source Q column (Amersham Biosciences, USA). The protein eluted at 300–350 mM KCl, was dialyzed against buffer C (buffer B with 2.5% glycerol) and concentrated to 12.5 mg/ml.
Complex formation, crystallization, and data collection
RB69 pol and DNA were mixed in an equimolar ratio, incubated at room temperature for 10 min and a 10-fold excess of the corresponding dNTP was added. After an additional 10 min incubation at room temperature, hanging drop vapor diffusion crystallization experiments were set up, resulting in a final enzyme concentration of 8.6 mg/ml. Drops were set up by mixing equal volumes of complex and precipitant (8-oxodG:dCTP complex: 18% PEG 350MME, 220 mM CaCl2, 50 mM Tris–HCl 7.5; AP:dG complex: 10% PEG 350MME, 120 mM CaCl2, 50 mM Tris–HCl 6.5) and equilibrated against reservoir solution. Crystals reached their maximum size (∼200 × 70 × 70 μm3) in 3–6 days (8-oxodG:dCTP) or 2–3 weeks (AP:dG complex) at 22°C. Crystals were cryoprotected by increasing the PEG 350MME concentration to 30% and then flash-cooled in liquid nitrogen.
Data were collected at beam lines X26C (AP:dG) and X25 (8-oxodG:dCTP) at the National Synchrotron Light Source at Brook-haven National Laboratory on an ADSC Quantum 4 detector and a custom-made CCD detector, respectively, at a wavelength of 1.1 Å and a temperature of 100 K. Diffraction data were indexed, integrated, and scaled using the HKL software (Otwinowski and Minor, 1997). Data statistics are given in Table I.
Structure solution and refinement
Crystals of the 8-oxodG:dCTP complex (space group P212121 with a=80.8 Å, b=118.6 Å, and c=127.2 Å) contain one ternary complex in the asymmetric unit. Molecular replacement was performed with the structure of the protein in replicating mode (PDB code 1IG9). After rigid body refinement with Refmac5 (Collaborative Computational Project, 1994; Murshudov et al, 1997), electron density maps showed an identical binding mode for the DNA and the latter was added accordingly. The library for the 8-oxodG base was generated from the crystal structure of 9-ethyl-8-hydroxyguanine monohydrate (CSD entry FURGAA03; Marsh, 1999). Water molecules were either added automatically using ARP/wARP (Lamzin and Wilson, 1993) or fitted into the electron density using the program O (Jones et al, 1991). Three amino acids of the N-terminal His-tag were included as well.
Crystals of the AP:dG complex belong to space group P21 with a=131.9 Å, b=122.2 Å, c=165.4 Å, and β=96.8°. Molecular replacement using MOLREP (Vagin and Teplyakov, 1997) with the apo structure as a search model (PDB code 1IH7) identified four copies of RB69 pol in the asymmetric unit. Electron density maps after rigid body refinement with Refmac5 revealed DNA binding to the polymerase active site in all four molecules and resulted in an initial R factor of 50.1% and a correlation coefficient of 34.0% in the resolution range of 50.0–4.0 Å. Loose noncrystallographic symmetry (NCS) restraints were maintained throughout the entire refinement process. The tightness of restraints was chosen to minimize the free R value. The average rms deviation of equivalent main chain and side chain atoms of NCS-related domains is 0.8 and 1.3 Å, respectively.
Terminal transferase activity studies
Terminal deoxynucleotidyl transferase activity was tested in 10 μl reactions at pH 6.5 with enzyme, DNA, and dNTP concentrations representing those in the crystallization drop, but without addition of salts or precipitants. After 1 week incubation at 22°C, the mixtures were 5′-radiolabeled with [γ-32P]-ATP (Amersham) using T4 polynucleotide kinase (New England Biolabs) according to the manufacturer's protocol, and resolved with 20% denaturing (7 M urea) PAGE. Labeled DNA was visualized using a Molecular Dynamics Storm 840 Phosphorimager.
DNA polymerase assays and kinetics
Oligonucleotide primers were 5′-radiolabeled as described above. Primer/template substrates (sequences as in Figure 1A) were prepared by annealing 5′-radiolabeled primer to the complementary oligonucleotide at a 1:1.2 molar ratio. The standard reaction (10 μl) contained 50 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 5′-32P-primer/template at 100 nM, and 0–2000 μM of the next correct dNTP. RB69 pol (0.16–0.64 nM; 190 nM was used for incorporation of dATP opposite dG) was diluted in a solution containing 50 mM Tris–HCl, pH 7.5, 0.5 mg/ml BSA, and 10% glycerol. Reactions were performed at 25°C for 1 min and quenched by addition of 10 μl of 95% formamide dye mixture (95% formamide, 0.001% xylene cyanol, 0.001% bromphenol blue). Samples were heated at 100°C for 3 min, and aliquots (2 μl) were subjected to 20% denaturing PAGE. The amounts of primer and product were quantified using a Molecular Dynamics Storm 840 PhosphorImager. Values for the Michaelis constant (Km) and vmax were obtained by least-squares nonlinear regression to a rectangular hyperbola. Less than 20% of the primer is extended under the steady-state conditions used in our kinetic studies, ensuring single hit kinetics (Boosalis et al, 1987; Goodman et al, 1993).
Accession numbers
Coordinates and structure factors for the 8-oxodG:dCTP and AP:dG complexes have been deposited in the Protein Data Bank under accession codes 1Q9Y and 1Q9X, respectively.
Supplementary Material
Supplementary Figure S1
Supplementary Table S1
Acknowledgments
This research was supported by DOE grant (DE-FG02-01ER63073) and Pew Scholars Program in the Biomedical Sciences to CK, NIH grants to CK (GM581980) and APG (ES-04068), as well as a Deutsche Forschungsgemeinschaft postdoctoral fellowship to EF. Beamline X26C is supported in part by the State University of New York and its Research Foundation. We thank Ms Cecilia Torres for providing modified oligonucleotides and Hermann Schindelin for critical reading of the manuscript.
References
- Bodepudi V, Shibutani S, Johnson F (1992) Synthesis of 2′-deoxy-7,8-dihydro-8-oxoguanosine and 2′-deoxy-7,8-dihydro-8-oxoadenosine and their incorporation into oligomeric DNA. Chem Res Toxicol 5: 608–617 [DOI] [PubMed] [Google Scholar]
- Boosalis MS, Petruska J, Goodman MF (1987) DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J Biol Chem 262: 14689–14696 [PubMed] [Google Scholar]
- Chiu TK, Dickerson RE (2000) 1 Ang crystal structures of B-DNA reveal sequence-specific binding and groove-specific bending of DNA by magnesium and calcium. J Mol Biol 301: 915–945 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project N (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Franklin MC, Wang J, Steitz TA (2001) Structure of the replicating complex of a pol alpha family DNA polymerase. Cell 105: 657–667 [DOI] [PubMed] [Google Scholar]
- Goodman MF, Creighton S, Bloom LB, Petruska J (1993) Biochemical basis of DNA replication fidelity. Crit Rev Biochem Mol Biol 28: 83–126 [DOI] [PubMed] [Google Scholar]
- Haracska L, Prakash S, Prakash L (2002) Yeast Rev 1 protein is a G template-specific DNA polymerase. J Biol Chem 277: 15546–15551 [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Nishioka M, Fujiwara S, Takagi M, Imanaka T, Inoue T, Kai Y (2001) Crystal structure of DNA polymerase from hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. J Mol Biol 306: 469–477 [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Eichinger A, Engh RA, Laue F, Ankenbauer W, Huber R, Angerer B (1999) Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc Natl Acad Sci USA 96: 3600–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Joyce CM, Steitz TA (1995) Polymerase structures and function: variations on a theme? J Bacteriol 177: 6321–6329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ, Zou JY, Kjeldgaard M, Jones TA (2001) Around O. In International Tables for Crystallography, Volume F. Crystallography of Biological Macromolecules, Rossmann MG, Arnold E (eds), Vol. Chapter 17.1, pp 353–356, 366–367 The Netherlands, Dordrecht: Kluwer Academic Publishers [Google Scholar]
- Kool ET (2002) Active site tightness and substrate fit in DNA replication. Annu Rev Biochem 71: 191–219 [DOI] [PubMed] [Google Scholar]
- Kouchakdjian M, Bodepudi V, Shibutani S, Eisenberg M, Johnson F, Grollman AP, Patel DJ (1991) NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-Oxo-7H-dG(syn).dA(anti) alignment at lesion site. Biochemistry 30: 1403–1412 [DOI] [PubMed] [Google Scholar]
- Krahn JM, Beard WA, Miller H, Grollman AP, Wilson SH (2003) Structure of DNA polymerase beta with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure 121–127 [DOI] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT—a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Lamzin VS, Wilson KS (1993) Automated refinement of protein models. Acta Crystallogr D 49: 129–149 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, McArthur MW, Moss DS, Thornton JM (1993) PROCHECK—a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Lehmann AR (2002) Replication of damaged DNA in mammalian cells: new solutions to an old problem. Mutat Res 509: 23–34 [DOI] [PubMed] [Google Scholar]
- Lindahl T (1979) DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog Nucleic Acid Res Mol Biol 22: 135–192 [DOI] [PubMed] [Google Scholar]
- Lipscomb LA, Peek ME, Morningstar ML, Verghis SM, Miller EM, Rich A, Essigmann JM, Williams LD (1995) X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc Natl Acad Sci USA 92: 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh RE (1999) P1 or P-1? Or something else? Acta Crystallogr B 55: 931–936 [DOI] [PubMed] [Google Scholar]
- McAuley-Hecht KE, Leonard GA, Gibson NJ, Thomson JB, Watson WP, Hunter WN, Brown T (1994) Crystal structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base pairs. Biochemistry 33: 10266–10270 [DOI] [PubMed] [Google Scholar]
- Merrit EA, Murphy MEP (1994) Raster3D Version 2.0—a program for photorealistic molecular graphics. Acta Crystallogr D 50: 869–873 [DOI] [PubMed] [Google Scholar]
- Miller H, Grollman AP (1997) Kinetics of DNA polymerase I (Klenow fragment exo-) activity on damaged DNA templates: effect of proximal and distal template damage on DNA synthesis. Biochemistry 36: 15336–15342 [DOI] [PubMed] [Google Scholar]
- Moriya M, Pandya GA, Johnson F, Grollman AP (1999) Cellular response to exocyclic DNA adducts. IARC Sci Publ 150: 263–270 [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Oda Y, Uesugi S, Ikehara M, Nishimura S, Kawase Y, Ishikawa H, Inoue H, Ohtsuka E (1991) NMR studies of a DNA containing 8-hydroxydeoxyguanosine. Nucleic Acids Res 19: 1407–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA (1985) Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature 313: 762–766 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. In Methods Enzymology, Carter CWJ, Sweet RM (eds),Vol. 276, pp 307–326. New York: Academic Press [DOI] [PubMed] [Google Scholar]
- Rodriguez AC, Park HW, Mao C, Beese LS (2000) Crystal structure of a pol alpha family DNA polymerase from the hyperthermophilic archaeon Thermococcus sp. 9 degrees N-7. J Mol Biol 299: 447–462 [DOI] [PubMed] [Google Scholar]
- Rubin J, Brennan T, Sundaralingam M (1972) Crystal and molecular structure of a naturally occurring dinucleoside monophosphate. Uridylyl-(3′–5′)-adenosine hemihydrate. Conformational ‘rigidity' of the nucleotide unit and models for polynucleotide chain folding. Biochemistry 11: 3112–3128 [DOI] [PubMed] [Google Scholar]
- Shamoo Y, Steitz TA (1999) Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell 99: 155–166 [DOI] [PubMed] [Google Scholar]
- Shefter E, Trueblood KN (1965) Crystal and molecular structure of D(+)-barium uridine-5′-phosphate. Acta Crystallogr 1067–1077 [DOI] [PubMed] [Google Scholar]
- Shibutani S, Bodepudi V, Johnson F, Grollman AP (1993) Translesional synthesis on DNA templates containing 8-oxo-7,8-dihydrodeoxyadenosine (8-oxodA). Biochemistry 32: 4615–4621 [DOI] [PubMed] [Google Scholar]
- Shibutani S, Takeshita M, Grollman AP (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349: 431–434 [DOI] [PubMed] [Google Scholar]
- Shibutani S, Takeshita M, Grollman AP (1997) Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the ‘A rule'. J Biol Chem 23: 13916–13922 [DOI] [PubMed] [Google Scholar]
- Steitz TA (1993) DNA- and RNA-dependent DNA polymerases. Curr Opin Struct Biol 3: 31–38 [Google Scholar]
- Sussman JL, Seeman NC, Kim S-H, Berman HM (1972) Crystal structure of a naturally occurring dinucleoside phosphate: uridylyl 3′,5′-adenosine phosphate model for RNA chain folding. J Mol Biol 66: 403–421 [DOI] [PubMed] [Google Scholar]
- Takeshita M, Chang CN, Johnson F, Will S, Grollman AP (1987) Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J Biol Chem 262: 10171–10179 [PubMed] [Google Scholar]
- Takeshita M, Eisenberg W (1994) Mechanism of mutation on DNA templates containing synthetic abasic sites: study with a double strand vector. Nucleic Acids Res 22: 1897–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Crystallogr 30: 1022–1025 [Google Scholar]
- Wang CC, Yeh LS, Karam JD (1995) Modular organisation of T4 DNA polymerase. J Biol Chem 270: 26558–26564 [DOI] [PubMed] [Google Scholar]
- Wang J, Sattar AK, Wang CC, Karam JD, Konigsberg WH, Steitz TA (1997) Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell 89: 1087–1099 [DOI] [PubMed] [Google Scholar]
- Yang G, Lin T-C, Karam J, Konigsberg WH (1999) Steady-state kinetic characterization of RB69 DNA polymerase mutants that affect dNTP incorporation. Biochemistry 38: 8094–8101 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Wu X, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z (2000) Error-prone lesion bypass by human DNA polymerase eta. Nucleic Acids Res 28: 4717–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Jeruzalmi D, Moarefi I, Leighton L, Lasken R, Kuriyan J (1999) Crystal structure of an archaebacterial DNA polymerase. Struct Fold Des 7: 1189–1199 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Table S1


