Abstract
Brucellosis is endemic in most parts of Egypt, where it is caused mainly by Brucella melitensis biovar 3, and affects cattle and small ruminants in spite of ongoing efforts devoted to its control. Knowledge of the predominant Brucella species/strains circulating in a region is a prerequisite of a brucellosis control strategy. For this reason a study aiming at the evaluation of the phenotypic and genetic heterogeneity of a panel of 17 Brucella spp. isolates recovered from domestic ruminants (cattle, buffalo, sheep, and goat) from four governorates during a period of five years (2002–2007) was carried out using microbiological tests and molecular biology techniques (PCR, MLVA-15, and sequencing). Thirteen strains were identified as B. melitensis biovar 3 while all phenotypic and genetic techniques classified the remaining isolates as B. abortus (n = 2) and B. suis biovar 1 (n = 2). MLVA-15 yielded a high discriminatory power (h = 0.801), indicating a high genetic diversity among the B. melitensis strains circulating among domestic ruminants in Egypt. This is the first report of the isolation of B. suis from cattle in Egypt which, coupled with the finding of B. abortus, suggests a potential role of livestock as reservoirs of several zoonotic Brucella species in the region.
1. Introduction
Since the first description of B. melitensis in Malta in 1897 [1], small ruminant brucellosis (SRB) has become a widespread problem in most Mediterranean countries as well as in other parts of the world (Middle East, Central Asia, and Latin America) [2].
In spite of the lack of precise information on the prevalence of ruminant brucellosis in Egypt, the disease is considered endemic in animals and humans in most parts of the country [3] leading to an estimated yearly economic loss of approximately 60 million Egyptian pounds [4]. Several studies have attempted to determine the incidence of brucellosis in ruminants and humans in some regions of the country leading to a high variability of estimates depending mainly on the analyzed host species, geographic localization, and the serological technique used [5–8]. Predominance of smallholdings that favor close contacts between humans and animals, presence of mixed populations of animals, and consumption of unpasteurized milk and dairy products are among the main major risk factors for Brucella infection present in Egypt [3, 7, 9]. Implementation of control measures of bovine brucellosis (test and slaughter, S19 vaccination) since the 1980's in the country led to a reduction on B. abortus incidence in cattle [3]. However, efforts directed to control small ruminant brucellosis have been less intensive, contributing to an increase of B. melitensis infection (considered the predominant Brucella species in Egypt nowadays) not only in sheep and goats, but also in cattle, buffaloes, and camels [3]. The identification and molecular characterization of prevailing Brucella species are a cornerstone to understand the epidemiology of the disease in a region and implement adequate strategies to control this important zoonosis [10]. For this reason, a study to evaluate the heterogeneity of Brucella spp. isolates recovered from domestic ruminants in different governorates of Egypt was conducted.
2. Materials and Methods
In 2002–2007 a total of 17 Brucella isolates were cultured according to OIE guidelines from samples (milk, aborted fetus, lymph node, and spleen) of domestic ruminants (buffalo, sheep, goat, and cattle) collected by convenience sampling in Assiut, Menofia, Beni-Suef, and Sharkia Governorates (n = 12) and unknown locations (n = 5) (Table 1, Figure 1). All animals were reactors to Buffered Acidified Plate Antigen Test (BAPAT), Rose Bengal Test (RBT), Tube Agglutination Test (TAT), Rivanol Test, and Complement Fixation Test (CFT). All Brucella-like isolates obtained in the following 14 days were classified using phenotypical methods (morphology, CO2 requirements, H2S production, urease, catalase and oxidase activity, nitrate reduction, lactose fermentation, citrate utilization, grow in presence of thionine and fuchsin dyes (at different concentrations: 1 : 50,000 and 1 : 100,000), lysis by Tbilisi phage and agglutination with A and M anti-sera) in the Animal Health Research Institute (Giza, Egypt). Brucella DNA from all isolates was sent to the VISAVET Health Surveillance Centre for genetic identification and characterization. Brucella spp. identification was confirmed using a Brucella-specific PCR [11] and isolates were further characterized using the Bruce-ladder kit (Ingenasa, Tres Cantos, Spain). Isolates identified as Brucella suis were also analyzed using the Ingene Bruce-ladder Suis (Ingenasa) for serovar determination [12]. Finally, the whole panel was subjected to Multilocus Variable Number Tandem-Repeat analysis (MLVA-15) as described before [13]. The number of repetitions found in each locus was determined by band size assessment (according to Le Flèche et al. [13] instructions) and sequencing. Allelic diversity for each locus was determined according to Selander et al. [14] (adapted from Nei [15]). The genetic diversity was also calculated for B. melitensis isolates. Results were compared with those available in the database of Brucella from other African and Middle East countries (http://mlva.u-psud.fr/mlvav4/genotyping/view.php). All MLVA profiles not previously described have been submitted to the MLVA database (http://mlva.u-psud.fr/mlvav4/genotyping/). A cluster analysis was performed using Neighbor Joining Analysis calculating the proportion of loci at which dissimilar alleles occur using MLST Data Analysis-Tree drawing (http://pubmlst.org/cgi-bin/mlstanalyse/mlstanalyse.pl?site=pubmlst&page=treedraw&referer=pubmlst.org).
Table 1.
Brucella spp. isolates recovered from livestock in Egypt (2002–2007) and examined by MLVA-15 technique included in the present study (n = 17). Panel 1 includes eight minisatellite markers (Bruce 06–Bruce 55) and Panel 2 is composed of seven microsatellite markers (Bruce 18–Bruce 30). The number of tandem repeats observed in each marker is indicated in the corresponding cells.
| Id. strain | Sample | Host | Origin | Year | Specie (biovar) | MLVA-15 Panel 1 | MLVA-15 Panel 2 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bruce 06 | Bruce 08 | Bruce 11 | Bruce 12 | Bruce 42 | Bruce 43 | Bruce 45 | Bruce 55 | Bruce 18 | Bruce 21 | Bruce 04 | Bruce 07 | Bruce 09 | Bruce 16 | Bruce 30 | ||||||
| 5 | Lymph node | Cow | ND* | 2002–2007 | B. melitensis bv. 3 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 8 | 8 | 7 | 5 | 9 | 5 | 3 |
| 6 | Fetus | Cow | ND | 2002–2007 | B. melitensis bv. 3 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 8 | 8 | 7 | 5 | 6 | 4 | 3 |
| 7 | Lymph node | Sheep | Sharkia | 2007 | B. melitensis bv. 3 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 7 | 8 | 5 | 7 | 5 | 8 | 3 |
| 8 | Spleen | Cow | ND | 2002–2007 | B. melitensis bv. 3 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 7 | 8 | 6 | 5 | 8 | 4 | 3 |
| 9 | Lymph node | Cow | ND | 2006-2007 | B. melitensis bv. 3 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 8 | 8 | 7 | 5 | 9 | 5 | 3 |
| 11 | Lymph node | Cow | Beni-Suef | 2006-2007 | B. melitensis bv. 3 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 8 | 8 | 7 | 5 | 6 | 4 | 3 |
| 12 | Lymph node | Cow | Beni-Suef | 2006-2007 | B. melitensis bv. 3 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 8 | 8 | 7 | 5 | 9 | 5 | 3 |
| 15 | Lymph node | Cow | Beni-Suef | 2006-2007 | B. melitensis bv. 3 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 8 | 8 | 7 | 5 | 7 | 4 | 3 |
| 16 | Lymph node | Sheep | Beni-Suef | 2006-2007 | B. melitensis bv. 3 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 8 | 8 | 6 | 5 | 9 | 5 | 3 |
| 17 | Lymph node | Goat | Beni-Suef | 2006-2007 | B. melitensis bv. 3 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 8 | 8 | 7 | 5 | 10 | 5 | 3 |
| 18 | Milk | Buffalo | Beni-Suef | 2006-2007 | B. melitensis bv. 3 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 8 | 8 | 7 | 5 | 6 | 4 | 3 |
| 19 | Fetus | Cow | Beni-Suef | 2006-2007 | B. melitensis bv. 3 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 8 | 8 | 7 | 5 | 9 | 5 | 3 |
| 20 | Milk | Buffalo | Assiut | 2007 | B. melitensis bv. 3 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 8 | 8 | 6 | 5 | 6 | 6 | 3 |
|
| ||||||||||||||||||||
| 10 | Milk | Cow | Menofia | 2007 | B. suis bv. 1 | 2 | 3 | 6 | 10 | 4 | 1 | 5 | 2 | 4 | 9 | 6 | 6 | 5 | 5 | 3 |
| 14 | Lymph node | Cow | Beni-Suef | 2006-2007 | B. suis bv. 1 | 2 | 3 | 6 | 10 | 4 | 1 | 5 | 2 | 4 | 9 | 5 | 5 | 8 | 6 | 3 |
|
| ||||||||||||||||||||
| 13 | Spleen | Cow | Beni-Suef | 2006-2007 | B. abortus | 4 | 5 | 4 | 12 | 2 | 3 | 3 | 3 | 6 | 8 | 3 | 7 | 3 | 3 | 5 |
| 4 | Milk | Cow | ND | 2002–2007 | B. abortus | 4 | 5 | 4 | 12 | 2 | 3 | 3 | 3 | 6 | 8 | 3 | 9 | 3 | 3 | 5 |
*ND: non determined.
Figure 1.
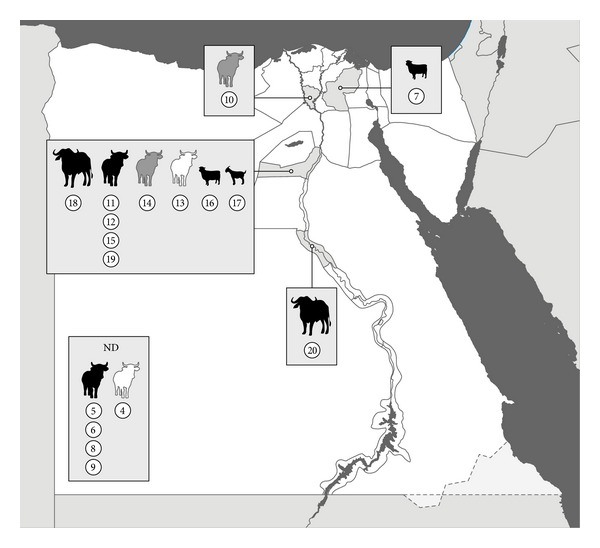
Geographical origin of the 17 Brucella spp. isolates (labeled from 4 to 20) recovered from livestock (buffalo, cattle, sheep, and goats) in Egypt during 2002–2007 (color of the animal indicates the Brucella species: Brucella melitensis, black; B. suis, grey animals, B. abortus, white).
3. Results and Discussion
Ruminant brucellosis is an endemic food-borne disease in most parts of Egypt and other developing countries of Africa. Recent studies [10, 16, 17] have highlighted the need of identifying the animal species infected with members of the genus Brucella to define their potential role in the transmission of this zoonotic pathogen and to determine the prevailing Brucella strains present in a region in order to adopt the most suitable control strategies.
Most (13/17) of the isolates recovered from ruminants in several governorates of Egypt were identified as B. melitensis biovar 3 in agreement with previous reports that described this Brucella species as the most prevalent in Egypt [3, 18] (Table 1). However, the unexpected phenotypic results (H2S production, urease, grow in presence of thionine and fuchsin dyes (at different concentrations: 1 : 50,000 and 1 : 100,000), lysis by Tbilisi phage, and agglutination with A and M anti-sera) of a subset of isolates (n = 4) suggested their identification as non-B. melitensis (Table 2). Molecular identification using the Bruce-ladder kit identified in fact isolates 10–14 and 4–13 as B. suis and B. abortus, respectively. The Ingene Bruce-ladder Suis kit further identified the B. suis isolates as biovar 1.
Table 2.
Differential phenotypic characteristics of Brucella suis (strains 10 and 14) and B. abortus (strains 4 and 13) isolated from cattle in Egypt, 2003–2007. Three reference strains (B. melitensis strain Ether, B. suis strain 1330, and B. abortus strain 544) are included for comparison.
| B. melitensis strain Ether | B. suis strain 1330 | B. abortus strain 544 | Strain 10/14 (B. suis) | Strain 4/13 (B. abortus) | |
|---|---|---|---|---|---|
| H2S production | − | +++ | + | +++ | +++ |
| Urease activity | + in 18–24 h | ++ in <15 min | + in 2 h | ++ in 3–5 min | + in 2 h |
| Growth in presence of dye thionin 1 : 50000 | + | + | − | + | − |
| Growth in presence of dye thionin 1 : 100000 | + | + | − | + | − |
| Growth in presence of dye fuchsin 1 : 50000 | + | − | + | − | + |
| Growth in presence of dye fuchsin 1 : 100000 | + | − | + | − | + |
| Agglutination with A anti-sera | + | + | + | + | + |
| Agglutination with M anti-sera | + | − | − | − | − |
B. suis isolates were cultured from milk (strain 10) and lymph node (strain 14) from two cows from Menofia and Beni-Suef Governorates, respectively. MLVA-15 analyses (Table 1) revealed that both isolates had typical but different B. suis biovar 1 patterns [13]. Zoonotic B. suis has been isolated in cattle elsewhere and it is becoming an emerging problem in several countries as Brazil and Colombia [19]. In this host species B. suis infection appears to be asymptomatic although biovar 1 shedding in milk has been described before [20] in agreement with our results. Although B. suis biovar 1 presence in swine has been reported previously in Egypt, its current distribution is unknown [21], and it had not been reported in cattle before. Brucellosis infection in swine has been described in the country usually by means of serological techniques [3, 22] that cannot distinguish between infections by the different Brucella spp. In Egypt, swine (with an approximate population of 30,000 animals [3]) may live in small groups in contact with other animals and humans [23]. As mentioned for B. melitensis, B. suis could be easily transmitted from swine to other animals and humans in these small holdings. To our knowledge, this is the first detection of the zoonotic biovar 1 of Brucella suis in cattle in Egypt. No information was available regarding potential contact between swine and the cattle from which B. suis was recovered in our study. However, taking into account the presence of a zoonotic B. suis biovar 1 in the region and the high number of reactors reported previously in swine populations located in different areas of Egypt (up to 12.61% using RBT [23]), more efforts are needed in order to determine the importance of this animal species as a source of infection and to avoid spillover to other domestic animals and human.
The presence of B. abortus in cattle in Egypt was also demonstrated here in agreement with previous occasional reports [3]. One of the two different MLVA-15 patterns (Table 1) found in the two B. abortus strains matched existing profiles in the MLVA Bank-Microbes genotyping [13] including B. abortus RB51 strains [24, 25] isolated in USA, Italy, and Portugal. However, results of the Bruce-ladder kit ruled out a possible isolation of this vaccine strain. The other MLVA profile was not present in the MLVA database.
Nevertheless, the most prevalent Brucella species found in the study was B. melitensis as previously described. All isolates belonged to West Mediterranean Group (MLVA8 genotype 51). The genetic heterogeneity existing among the 13 isolates analyzed was high (Table 1), with a total of 8 different genotypes (h = 0.801) (Figure 1), none of which had been included in the MLVA Bank-Microbes genotyping (Brucella Aggregated database, http://mlva.u-psud.fr/mlvav4/genotyping/). A high discriminatory power of MLVA-15 had been previously reported in B. melitensis isolates from Lebanon, Spain, and China [26–28]. In our study, the highest genetic heterogeneity was found in markers bruce 09 and bruce 16 (h = 0.712 and h = 0.596, resp.) while only one allele was found in MLVA markers bruce 21 and bruce 30 from panel 2 and in all markers from panel 1. However, the Neighbor Joining Analysis clustered the Egyptian profiles obtained in this study with B. melitensis isolates in a large cluster with profiles from sheep and human isolates recovered in Algeria and Tunisia (Figure 2). Some authors have previously reported the limited value of panel 1 and panel 2A [27] to differentiate B. melitensis isolates recovered from the same geographical origin/outbreak [27, 28]. Our results also confirm the need of using markers of panel 2B to achieve a sufficient discriminatory capacity when isolates are geographically related.
Figure 2.
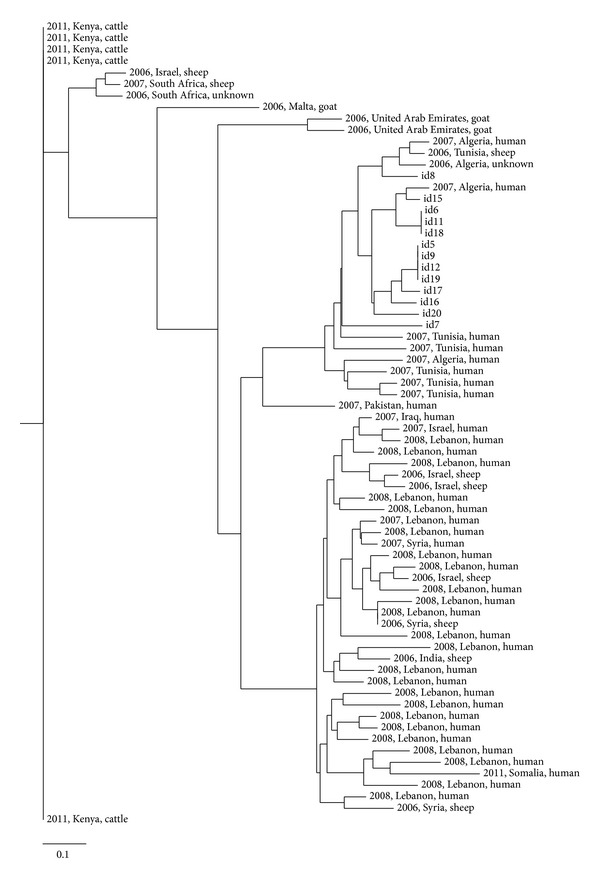
Neighbor Joining Analysis for the MLVA-15 profiles of 13 B. melitensis isolates recovered from domestic ruminants from Egypt in 2002–2007 compared with 57 isolates from Africa and Middle East recovered in 2006–2012 (source: http://mlva.u-psud.fr/mlvav4/genotyping/view.php).
4. Conclusions
The high genetic heterogeneity found in this study and particularly the identification of zoonotic strains of B. suis and B. abortus isolated from samples from domestic ruminants suggest a complex underlying epidemiological situation in Egypt. In addition, our results demonstrate the usefulness of a complete phenotypic and genetic characterization of isolates to avoid misclassification of bacterial species belonging to the Brucella genus. Our study, although performed on a limited sample size, gives an insight in the current disease-causing Brucella species present in domestic ruminants in Egypt. Further studies aiming at the assessment of the prevalence of B. suis in domestic ruminants and swine in Egypt using adequate identification techniques would be needed in order to determine the importance of the infection due to this zoonotic pathogen in livestock in the region.
Acknowledgments
This work is a contribution to the project AGL-2009-12674 (Ministerio de Ciencia e Innovación). Marta Perez-Sancho was a recipient of a Predoctoral Grant from Comunidad de Madrid (IV Plan regional de investigación científica e innovación tecnológica, 20052008) y Fondo Social Europeo.
Conflict of Interests
None of the authors have any conflict of interests relating to this study.
Authors' Contribution
Ahmed M. S. Menshawy and Marta Perez-Sancho contributed equally to this paper.
References
- 1.Bruce D. Note on the discovery of a microorganism in Malta fever. Practitioner. 1887;39:161–170. [Google Scholar]
- 2.FAO. Brucella Melitensis in Eurasia and the Middle East. 2009;10 [Google Scholar]
- 3.Refai M. Incidence and control of brucellosis in the Near East region. Veterinary Microbiology. 2002;90(1–4):81–110. doi: 10.1016/s0378-1135(02)00248-1. [DOI] [PubMed] [Google Scholar]
- 4.AOAD. Arab organization for agriculture development report. pp. 414–474, 1995.
- 5.El Sherbini A, Kabbash I, Schelling E, et al. Seroprevalences and local variation of human and livestock brucellosis in two villages in Gharbia Governorate, Egypt. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2007;101(9):923–928. doi: 10.1016/j.trstmh.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 6.Hegazy YM, Molina-Flores B, Shafik H, Ridler AL, Guitian FJ. Ruminant brucellosis in Upper Egypt (2005–2008) Preventive Veterinary Medicine. 2011;101(3-4):173–181. doi: 10.1016/j.prevetmed.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Holt HR, Eltholth MM, Hegazy YM, El-Tras WF, Tayel AA, Guitian J. Brucella spp. infection in large ruminants in an endemic area of Egypt: cross-sectional study investigating seroprevalence, risk factors and livestock owner’s knowledge, attitudes and practices (KAPs) BMC Public Health. 2011;11, article 341 doi: 10.1186/1471-2458-11-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samaha H, Mohamed TR, Khoudair RM, Ashour HM. Serodiagnosis of brucellosis in cattle and humans in Egypt. Immunobiology. 2009;214(3):223–226. doi: 10.1016/j.imbio.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Hegazy YM, Moawad A, Osman S, Ridler A, Guitian J. Ruminant brucellosis in the Kafr El Sheikh governorate of the Nile Delta, Egypt: prevalence of a neglected zoonosis. PLoS Neglected Tropical Diseases. 2011;5(1, article e944) doi: 10.1371/journal.pntd.0000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godfroid J, Al Dahouk S, Pappas G, et al. A “One Health” surveillance and control of brucellosis in developing countries: moving away from improvisation. Comparative Immunology Microbiology and Infectious Diseases. 2013;36(3):241–248. doi: 10.1016/j.cimid.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Hinić V, Brodard I, Thomann A, et al. Novel identification and differentiation of Brucella melitensis, B. abortus, B. suis, B. ovis, B. canis, and B. neotomae suitable for both conventional and real-time PCR systems. Journal of Microbiological Methods. 2008;75(2):375–378. doi: 10.1016/j.mimet.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 12.López-Goñi I, García-Yoldi D, Marín CM, et al. New Bruce-ladder multiplex PCR assay for the biovar typing of Brucella suis and the discrimination of Brucella suis and Brucella canis . Veterinary Microbiology. 2011;154(1-2):152–155. doi: 10.1016/j.vetmic.2011.06.035. [DOI] [PubMed] [Google Scholar]
- 13.Le Flèche P, Jacques I, Grayon M, et al. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiology. 2006;6, article 9 doi: 10.1186/1471-2180-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selander RK, Caugant DA, Ochman H. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Applied and Environmental Microbiology. 1986;51(5):873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89(3):583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasymbekov J, Imanseitov J, Ballif M, et al. Molecular epidemiology and antibiotic susceptibility of livestock Brucella melitensis isolates from Naryn Oblast, Kyrgyzstan. PLoS Neglected Tropical Diseases. 2013;7(2) doi: 10.1371/journal.pntd.0002047.e2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanogo M, Abatih E, Thys E, Fretin D, Berkvens D, Saegerman C. Importance of identification and typing of Brucellae from West African cattle: a review. Veterinary Microbiology. 2013;164(3-4):202–211. doi: 10.1016/j.vetmic.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Samaha H, Al-Rowaily M, Khoudair RM, Ashour HM. Multicenter study of brucellosis in Egypt. Emerging Infectious Diseases. 2008;14(12):1916–1918. doi: 10.3201/eid1412.071452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corbel MJ. Brucellosis: an overview. Emerging Infectious Diseases. 1997;3(2):213–221. doi: 10.3201/eid0302.970219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ewalt DR, Payeur JB, Rhyan JC, Geer PL. Brucella suis biovar 1 in naturally infected cattle: a bacteriological, serological, and histological study. Journal of Veterinary Diagnostic Investigation. 1997;9(4):417–420. doi: 10.1177/104063879700900414. [DOI] [PubMed] [Google Scholar]
- 21.EFSA. Scientific opinion of the panel on animal health and welfare (AHAW) on a request from the commission on porcine brucellosis (Brucella suis) The EFSA Journal. 2009;1144:1–112. [Google Scholar]
- 22.Zaki R. Brucella infection among ewes, camels and pigs in Egypt. Journal of Comparative Pathology and Therapeutics. 1948;58(2):145–151. doi: 10.1016/s0368-1742(48)80013-5. [DOI] [PubMed] [Google Scholar]
- 23.Barakat AM, El Fadaly HA, Shaapan RM, Khalil FAM. Occupational health hazard of egyptian employees in contact with wastage nourished swine. Journal of American Science. 2011;7(1):808–813. [Google Scholar]
- 24.Ferreira AC, Chambel L, Tenreiro T, et al. MLVA16 typing of Portuguese human and animal Brucella melitensis and Brucella abortus isolates. PLoS ONE. 2012;7(8) doi: 10.1371/journal.pone.0042514.e42514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garofolo G, Ancora M, Di GE. MLVA-16 loci panel on Brucella spp. using multiplex PCR and multicolor capillary electrophoresis. Journal of Microbiological Methods. 2013;92(2):103–107. doi: 10.1016/j.mimet.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 26.Jiang H, Fan M, Chen J, et al. MLVA genotyping of Chinese human Brucella melitensis biovar 1, 2 and 3 isolates. BMC Microbiology. 2011;11, article 256 doi: 10.1186/1471-2180-11-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kattar MM, Jaafar RF, Araj GF, et al. Evaluation of a multilocus variable-number tandem-repeat analysis scheme for typing human Brucella isolates in a region of brucellosis endemicity. Journal of Clinical Microbiology. 2008;46(12):3935–3940. doi: 10.1128/JCM.00464-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valdezate S, Navarro A, Villalón P, Carrasco G, Saéz-Nieto JA. Epidemiological and phylogenetic analysis of Spanish human Brucella melitensis strains by multiple-locus variable-number tandem-repeat typing, hypervariable octameric oligonucleotide fingerprinting, and rpoB typing. Journal of Clinical Microbiology. 2010;48(8):2734–2740. doi: 10.1128/JCM.00533-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


