Abstract
Kallipyga massiliensis strain ph2T is the type strain of Kallipyga massiliensis gen. nov., sp. nov., the type species of the new genus Kallipyga within the family Clostridiales Incertae Sedis XI. This strain, whose genome is described here, was isolated from the fecal flora of a 26-year-old woman suffering from morbid obesity. K. massiliensis is an obligate anaerobic coccus. Here we describe the features of this organism, together with the complete genome sequence and annotation. The 1,770,679 bp long genome (1 chromosome but no plasmid) contains 1,575 protein-coding and 50 RNA genes, including 4 rRNA genes.
Keywords: Kallipyga massiliensis, genome, culturomics, taxonogenomics
Introduction
Kallipyga massiliensis strain ph2T (CSUR=P241, DSM=26229) is the type strain of K. massiliensis gen. nov., sp. nov. This bacterium was isolated from the stool sample of an obese French patient as part of a study aiming at individually cultivating all species occurring within human feces [1-3]. It is a Gram-positive, anaerobic, indole-negative coccus. Defining the taxonomic status of bacterial isolates remains a challenging task. The taxonomic molecular tools currently available, including 16S rRNA sequence similarity, G + C content and DNA–DNA hybridization (DDH) [4,5], although considered as gold standards, have limitations [6,7]. The 16S rRNA sequence similarity and G+C content thresholds do not apply uniformly to all species or genera, and the DDH method lacks intra- and inter-laboratory reproducibility [5]. The advent of high-throughput genome sequencing and proteomic analysis [8] has granted unprecedented access to exhaustive genetic and protein information for bacterial isolates. We recently proposed a polyphasic approach to describe new bacterial species in which genome sequences and MALDI-TOF spectra are used along with phenotypic characteristics [9-30].
The family Clostridiales Incertae Sedis XI (Garrity and Holt 2001) was created in 2001 [31] and currently includes the 11 following genera: Anaerococcus (Ezaki et al. 2001) [32], Dethiosulfatibacter (Takii et al. 2007) [33], Finegoldia (Murdoch and Shah 2000) [34], Gallicola (Ezaki et al. 2001) [32], Helcococcus (Collins et al. 1993) [35], Parvimonas (Tindall and Euzéby 2006) [36], Peptoniphilus (Ezaki et al. 2001) [32], Sedimentibacter (Breitenstein et al. 2002) [37], Soehngenia (Parshina et al. 2003) [38], Sporanaerobacter (Hernandez-Eugenio et al. 2002) [39] and Tissierella (Collins and Shah 1986) [40]. Currently, 31 species with validly published names are reported in this family [41]. The species listed in the Clostridiales Incertae Sedis XI are mostly comprised of Gram-positive, obligate anaerobic cocci. Members belonging to this family were identified as pathogens in both humans and animals. In humans, they were often isolated from patients with septic arthritis, necrotizing pneumonia, prosthetic joint infection and other clinical conditions associated with vaginal discharges and ovarian, peritoneal and sacral abscesses [42-46].
Here we present a summary classification and a set of features for K. massiliensis gen. nov., sp. nov., strain ph2T (CSUR=P241, DSM=26229) together with the description of the complete genomic sequencing and annotation. These characteristics support the circumscription of the genus Kallipyga and its type species, K. massiliensis within the Clostridiales Incertae Sedis XI family.
Classification and features
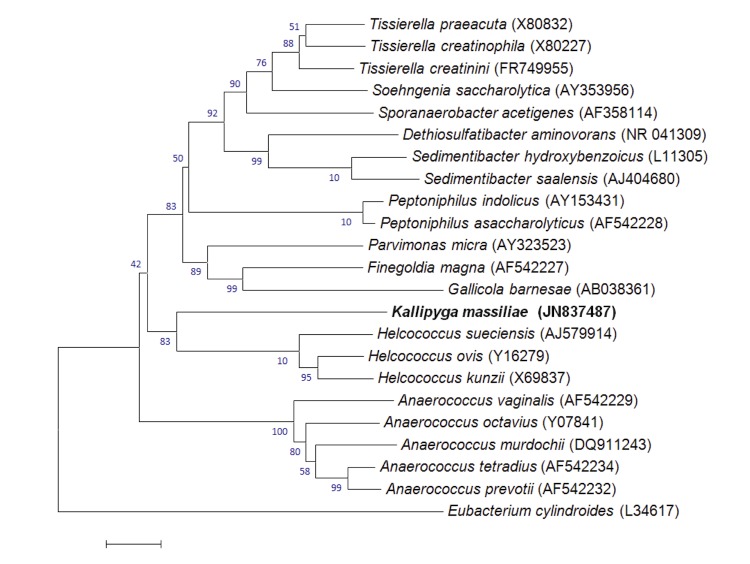
A stool sample was collected from a 26-year-old woman living in Marseille (France). She suffered from morbid obesity and had a body mass index of 48.2 (118.8 kg, 1.57 meter). At the time of stool sample collection she did not take any medication and was not on a diet. The patient gave an informed and signed consent, and the agreement of the ethics committee of the Institut Fédératif de Recherche (IFR48, Faculty of Medicine, Marseille, France) was obtained under reference 09-022. Another four new bacterial species, Alistipes obesi, Peptoniphilus grossensis, P. obesi and Enorma massiliensis [25-27,33], were also isolated from this specimen using various culture conditions. The fecal specimen was preserved at -80°C after collection. Strain ph2 T (Table 1) was isolated in 2011 by anaerobic culture on 5% sheep blood-enriched agar in anaerobic atmosphere at 37°C, following 26 days in a blood culture bottle with rumen and sheep blood. The 16S rRNA nucleotide sequence (GenBank accession number JN837487) of Kallipyga massiliensis strain ph2T was 86.09% similar to Helcococcus sueciensis, the phylogenetically closest species (Figure 1). This value was lower than the 95.0% 16S rRNA gene sequence threshold recommended by Stackebrandt and Ebers (2006) to delineate a new genus without carrying out DNA-DNA hybridization [5].
Table 1. Classification and general features of Kallipyga massiliensis strain ph2T according to the MIGS recommendations [45].
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Domain Bacteria | TAS [47] | ||
| Phylum Firmicutes | TAS [48-50] | ||
| Class Clostridia | TAS [51,52] | ||
| Current classification | Order Clostridiales | TAS [53,54] | |
| Family Clostridiales Incertae Sedis XI | TAS [55] | ||
| Genus Kallipyga | IDA | ||
| Species Kallipyga massiliensis | IDA | ||
| Type strain ph2T | IDA | ||
| Gram stain | Positive | IDA | |
| Cell shape | Cocci | IDA | |
| Motility | Non-motile | IDA | |
| Sporulation | Non-sporulating | IDA | |
| Temperature range | Mesophile | IDA | |
| Optimum temperature | 37°C | IDA | |
| MIGS-6.3 | Salinity | unknown | IDA |
| MIGS-22 | Oxygen requirement | Anaerobic | IDA |
| Carbon source | Unknown | NAS | |
| Energy source | Unknown | NAS | |
| MIGS-6 | Habitat | Human gut | IDA |
| MIGS-15 | Biotic relationship | Free living | IDA |
| MIGS-14 | Pathogenicity Biosafety level Isolation |
Unknown 2 Human feces |
NAS |
| MIGS-4 | Geographic location | France | IDA |
| MIGS-5 | Sample collection time | January 2011 | IDA |
| MIGS-4.1 | Latitude | 43.296482 | IDA |
| MIGS-4.1 | Longitude | 5.36978 | IDA |
| MIGS-4.3 | Depth | Surface | IDA |
| MIGS-4.4 | Altitude | 0 m above sea level | IDA |
Evidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [56]. If the evidence is IDA, then the property was directly observed for a live isolate by one of the authors or an expert mentioned in the acknowledgements.
Figure 1.
Phylogenetic tree highlighting the position of Kallipyga massiliensis strain ph2T relative to other type strains within the Clostridiales Incertae Sedis XI family. Genbank accession numbers are indicated in parentheses. Sequences were aligned using CLUSTALW, and phylogenetic inferences obtained using the maximum-likelihood method in MEGA software. Numbers at the nodes are percentages of bootstrap values obtained by repeating the analysis 500 times to generate a majority consensus tree. Eubacterium cylindroides was used as outgroup. The scale bar represents a 2% nucleotide sequence divergence.
By comparison to the Genbank database [57], strain ph2T also exhibited a nucleotide sequence similarity greater than 98.7% with 16 sequences from uncultured bacteria from the human skin microbiome [58]. These bacteria are most likely classified within the same species as strain ph2T.
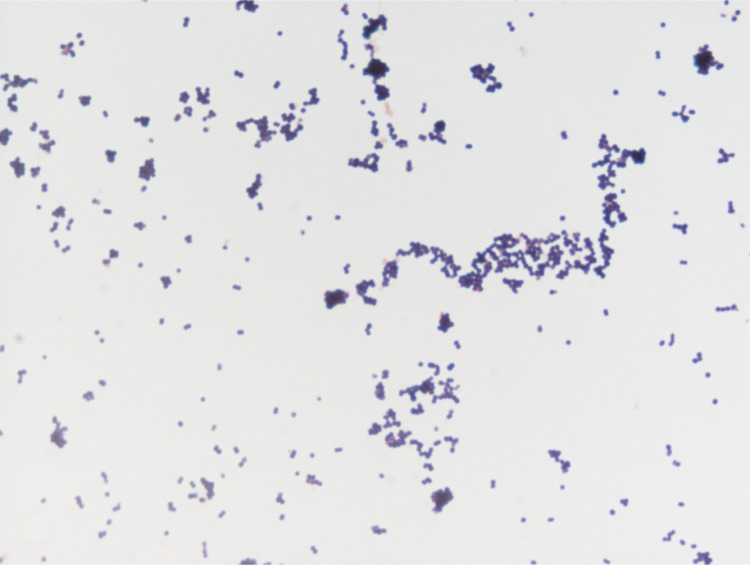
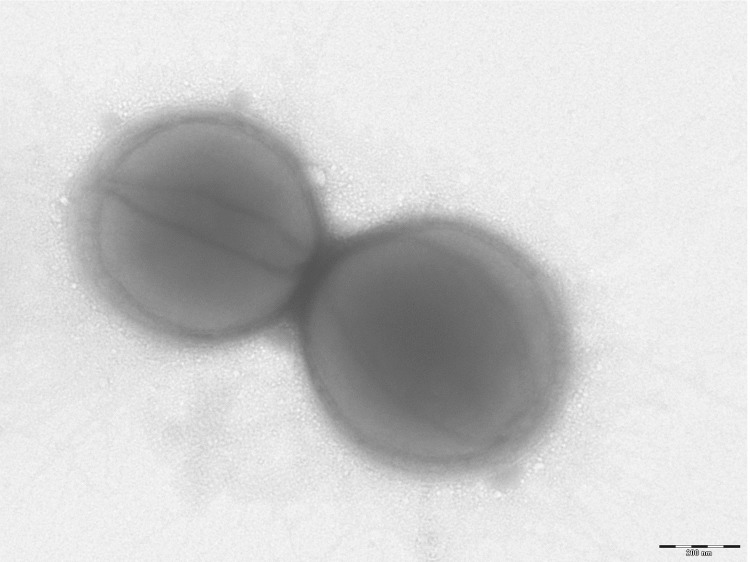
Different growth temperatures (25, 30, 37, 45°C) were tested; no growth occurred at 25°C and 30°C, growth occurred between 37°C and 45°C, and optimal growth was observed at 37°C. Colonies were bright grey with a diameter of 1.0 mm on 5% blood-enriched Columbia agar. Growth of the strain was tested under anaerobic and microaerophilic conditions using GENbag anaer and GENbag microaer systems, respectively (BioMérieux), and in the presence of air, with or without 5% CO2. Optimal growth was obtained anaerobically. No growth was observed under aerobic and microaerophilic conditions. Gram staining showed Gram-positive cocci (Figure 2). A motility test was negative. Cells grown on agar are Gram-positive, have a diameter in electron microscopy ranging from 0.57µm to 0.78µm (mean, 0.67 µm, Figure 3) and are mostly grouped in pairs, short chains or small clumps.
Figure 2.
Gram staining of K massiliensis strain ph2T
Figure 3.
Transmission electron microscopy of K. massiliensis strain ph2T using a Morgani 268D (Philips) at an operating voltage of 60kV. The scale bar represents 200 nm.
Strain ph2T exhibited neither catalase or oxidase activities. Using API 32A (BioMerieux), nitrate reduction, indole formation and urease production were negative. A positive reaction was obtained for α-galactosidase, arginine dihydrolase and arginine arylamidase, α-glucosidase and β-glucosidase. Strain ph2T did not ferment mannose or raffinose. Negative reactions were observed for β-galactosidase, β-galactosidase-6-phosphate, α-arabinosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, glutamic acid decarboxylase, proline arylamidase, leucyl glycine arylamidase, phenylalanine arylamidase, pyroglutamic acid arylamidase, tyrosine arylamidase, alanine arylamidase, glycine arylamidase, histidine arylamidase, glutamyl glutamic acid arylamidase, and serine arylamidase. Using an API Zym (BioMerieux), positive reactions were observed for esterase lipase, leucine arylamidase, α-glucosidase, β-glucosidase and acid phosphatase. Negative reactions were obtained for esterase, lipase, valine and cysteine arylamidase, trypsine, α-chymotrypsine, naphthol-AS-BI-phosphohydrolase, α-galactosidase, β-galactosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase and α-fucosidase. Using an API 50CH (BioMerieux), K. massiliensis weakly fermented D-ribose, D-glucose, D-fructose and aesculin. By comparison with its closest phylogenetic neighbors, K. massiliensis differed from Finegoldia magna in α-galactosidase and α-glucosidase production, D-ribose and D-fructose fermentation. It also differed from Helcococcus kunzii in oxygen requirement, α-galactosidase and leucine arylamidase production, D-ribose, D-glucose and esculin utilization. It differed from Parvimonas micra in alkaline phosphatase, glutamyl glutamic acid arylamidase, β-glucosidase, phenylalanine arylamidase and histidine arylamidase production and D-glucose fermentation. It differed from Peptoniphilus indolicus in α-galactosidase, indole, α-glucosidase, β-glucosidase, dihydrolase phenylalanine, phenylalanine arylamidase and histidine arylamidase production, and D-glucose fermentation (Table 2).
Table 2. Differential characteristics of Kallipyga massiliensis gen. nov., sp. nov., strain ph2T, Finegoldia magna strain ATCC 29328, Helcococcus kunzii strain ATCC 51366, Parvimonas micra strain ATCC 33270 and Peptoniphilus indolicus strain ATCC 29427T.
| Properties | K. massiliensis | F. magna | H. kunzii | P. micra | P. indolicus |
|---|---|---|---|---|---|
| Cell diameter (µm) | 0.67 | na | na | 0.3-0.7 | na |
| Oxygen requirement | anaerobic | anaerobic | facultative anaerobic | anaerobic | anaerobic |
| Colony color | bright gray | var | na | na | |
| Gram stain | + | + | + | + | + |
| Salt requirement | - | na | +/- | na | - |
| Motility | - | - | - | na | - |
| Endospore formation | - | - | na | na | - |
| Production of | |||||
| Alkaline Phosphatase | - | +/- | - | + | - |
| Catalase | - | +/- | - | na | na |
| Oxidase | - | na | na | na | na |
| Nitrate reductase | - | - | - | na | na |
| Urease | - | - | na | na | - |
| α-galactosidase | + | - | - | na | - |
| β-galactosidase | - | - | - | na | - |
| Indole | - | - | na | - | + |
| Arginine arylamidase | + | + | na | + | + |
| Glutamyl glutamic acid arylamidase |
- | na | na | + | na |
| Arginine dihydrolase | + | +/- | na | na | - |
| α-glucosidase | + | - | na | na | - |
| β-glucosidase | + | na | na | - | - |
| β-glucuronidase | - | - | - | na | na |
| Phenylalanine arylamidase | - | - | na | + | + |
| Esterase lipase | + | na | na | na | na |
| Leucine arylamidase | + | + | - | na | + |
| Cystine arylamidase | - | na | na | na | na |
| Histidine arylamidase | - | -/w | na | + | + |
| Fermentation of | |||||
| D-mannose | - | - | na | na | - |
| D-ribose | w | - | - | na | na |
| D-glucose | w | -/w | - | - | - |
| D-fructose | w | + | na | na | na |
| Esculin | w | na | + | na | na |
| Isolated from | human gut | human | human | human | mastitis of cattle |
na = data not available; var = variable; w = weak
K. massiliensis is susceptible to amoxicillin, amoxicillin-clavulanic acid, gentamicin 500, penicillin, imipenem, vancomycin, rifampicin and nitrofurantoin, but resistant to ciprofloxacin, metronidazole, gentamicin 10, trimethoprim/sulfamethoxazole, ceftriaxon, erythromycin and doxycycline.
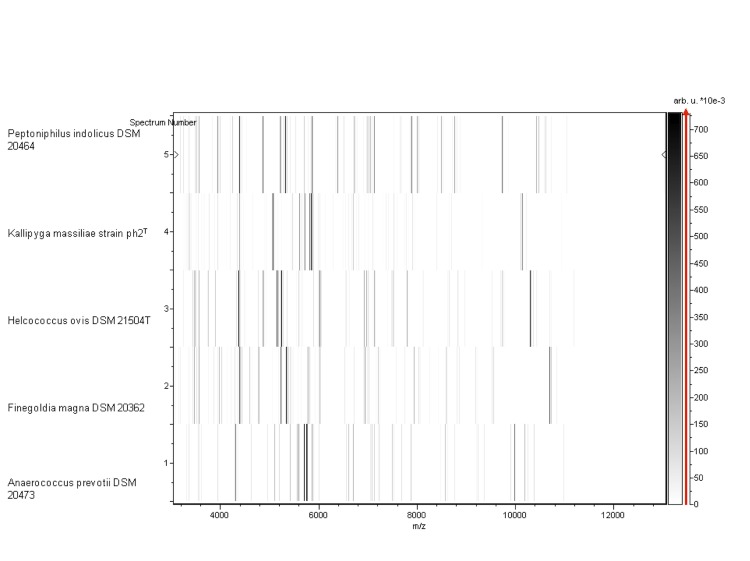
Matrix-assisted laser-desorption/ionization time-of-flight (MALDI-TOF) MS protein analysis was carried out as previously described [59] using a Microflex spectrometer (Bruker Daltonics, Germany). Twelve distinct deposits were done for strain ph2T from 12 isolated colonies. The twelve ph2T spectra were imported into our database and compared to spectra from 3,769 bacteria using the MALDI BioTyper software (version 2.0, Bruker). A score enabled the presumptive identification and discrimination of the tested species from those in a database: a score > 2 with a validly published species enabled the identification at the species level; a score > 1.7 but < 2 enabled the identification at the genus level; and a score < 1.7 did not enable any identification. For strain ph2T, no significant score was obtained, suggesting that our isolate was not a member of any known species or genus (Figures 4 and 5). A broader study incorporating MALDI-TOF and 16S rDNA and genomic DNA identity data may be conducted to define taxonomic criteria at the family level.
Figure 4.
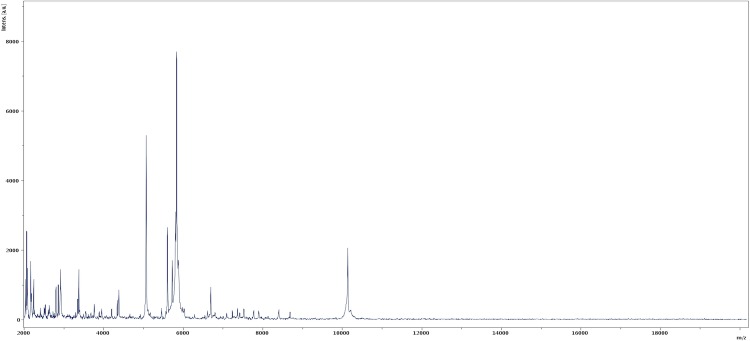
Reference mass spectrum from K. massiliensis strain ph2T. Spectra from 12 individual colonies were compared and a reference spectrum was generated.
Figure 5.
Gel view comparing Kallipyga massiliensis sp. nov strain ph2T to other phylogenetically close species. The gel view displays the raw spectra of loaded spectrum files arranged in a pseudo-gel like look. The x-axis records the m/z value. The left y-axis displays the running spectrum number originating from subsequent spectra loading. The peak intensity is expressed by a Gray scale scheme code. The color bar and the right y-axis indicate the relation between the color a peak is displayed with and the peak intensity in arbitrary units. Displayed species are indicated on the left.
Genome sequencing information
Genome project history
The organism was selected for sequencing on the basis of its phylogenetic position and 16S rRNA similarity to members of the family Clostridiales Incertae Sedis XI and is part of a study of the human digestive flora aiming at isolating all bacterial species within human feces [1-3]. It was the thirty-sixth genome from the family Clostridiales Incertae Sedis XI to be sequenced and the first genome of K. massiliensis gen. nov., sp. nov. The GenBank accession number is CAHC00000000 and consists of 22 contigs. Table 3 shows the project information and its association with MIGS version 2.0 compliance [60].
Table 3. Project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High-quality draft |
| MIGS-28 | Libraries used | 454 GS paired-end 3- kb libraries |
| MIGS-29 | Sequencing platform | 454 GS FLX Titanium |
| MIGS-31.2 | Sequencing coverage | 51.23 × |
| MIGS-30 | Assemblers | Newbler |
| MIGS-32 | Gene calling method | Prodigal |
| Genbank Date of Release | May 30, 2012 | |
| NCBI project ID | CAHC00000000 | |
| MIGS-13 | Project relevance | Study of the human gut microbiome |
Growth conditions and DNA isolation
Kallipyga massiliensis gen. nov., sp. nov., strain ph2T (CSUR= P241, DSM=26229) was grown anaerobically on 5% sheep blood-enriched Columbia agar at 37°C. Three petri dishes were spread and the bacteria cultivated were resuspended in 3 × 100µl of G2 buffer (EZ1 DNA Tissue kit, Qiagen). A first mechanical lysis was performed by glass powder on the Fastprep-24 device (MP Biomedicals, USA) using 2 × 20 seconds cycles. DNA was then treated with 2.5µg/µL lysozyme for 30 minutes at 37°C and extracted using the BioRobot EZ1 Advanced XL (Qiagen). The DNA was then concentrated and purified on a QIAamp kit (Qiagen). The yield and concentration was measured by the Quant-it Picogreen kit (Invitrogen) on the Genios Tecan fluorometer at 78.2ng/µl.
Genome sequencing and assembly
DNA (5 µg) was mechanically fragmented on a Hydroshear device (Digilab, Holliston, MA, USA) with an enrichment size at 3-4kb. The DNA fragmentation was visualized through the Agilent 2100 BioAnalyzer on a DNA labchip 7500 with an optimal size of 3.179kb. A 3kb paired-end library was constructed according to the 454 GS FLX Titanium paired-end protocol (Roche). Circularization and nebulization were performed and generated a pattern with an optimal at 600 bp. After PCR amplification through 17 cycles followed by double size selection, the single stranded paired-end library was quantified on the Quant-it Ribogreen kit (Invitrogen) on the Genios Tecan fluorometer at 58 pg/µL. The library concentration equivalence was calculated as 1.77E+08 molecules/µL. The library was stored at -20°C until further use.
The paired-end library was clonally amplified with 0.5 cpb and 1 cbp in 2 SV-emPCR reactions with the GS Titanium SV emPCR Kit (Lib-L) v2 (Roche). The yields of the emPCR were essentially the same at 12.3 and 12%, in the range of 5 to 20% recommended by the Roche procedure.
Approximately 790,000 beads were loaded on 1/4 region of a GS Titanium PicoTiterPlate PTP Kit 70x75 and sequenced with the GS FLX Titanium Sequencing Kit XLR70 (Roche). The run was performed overnight and then analyzed on the cluster through the gsRunBrowser and gsAssembler (Roche). A total, of 261,794 passed filter wells were obtained and generated 90.68 Mb with a length average of 346 bp. The global passed filter sequences were assembled using Newbler with 90% identity and 40 bp as overlap. The final assembly identified 3 scaffolds and 22 large contigs (> 1,500 bp) generating a genome size of 1.77 Mb which corresponds to a coverage of 51.23× genome equivalent.
Genome annotation
Open Reading Frames (ORFs) were predicted using Prodigal [61] with default parameters. However, the predicted ORFs were excluded if they spanned a sequencing gap region. The predicted bacterial protein sequences were searched against the GenBank [57] and Clusters of Orthologous Groups (COG) databases using BLASTP. The tRNAs and rRNAs were predicted using the tRNAScan-SE [62] and RNAmmer [63] tools, respectively. Lipoprotein signal peptides and numbers of transmembrane helices were predicted using SignalP [64] and TMHMM [65], respectively. ORFans were identified if their BLASTP E-value was lower than 1e-03 for alignment length greater than 80 amino acids. If alignment lengths were smaller than 80 amino acids, we used an E-value of 1e-05. Such parameter thresholds have already been used in previous works to define ORFans. Artemis [66] and DNA Plotter [67] were used for data management and visualization of genomic features, respectively. Mauve alignment tool (version 2.3.1) was used for multiple genomic sequence alignment [68]. To estimate the mean level of nucleotide sequence similarity at the genome level between K. massiliensis and four other members of the family Clostridiales Incertae Sedis XI (Table 6), orthologous proteins were detected using the Proteinortho software [69] and genomes compared two by two. For each pair of genomes, we determined the mean percentage of nucleotide sequence identity among orthologous ORFs using BLASTn.
Genome properties
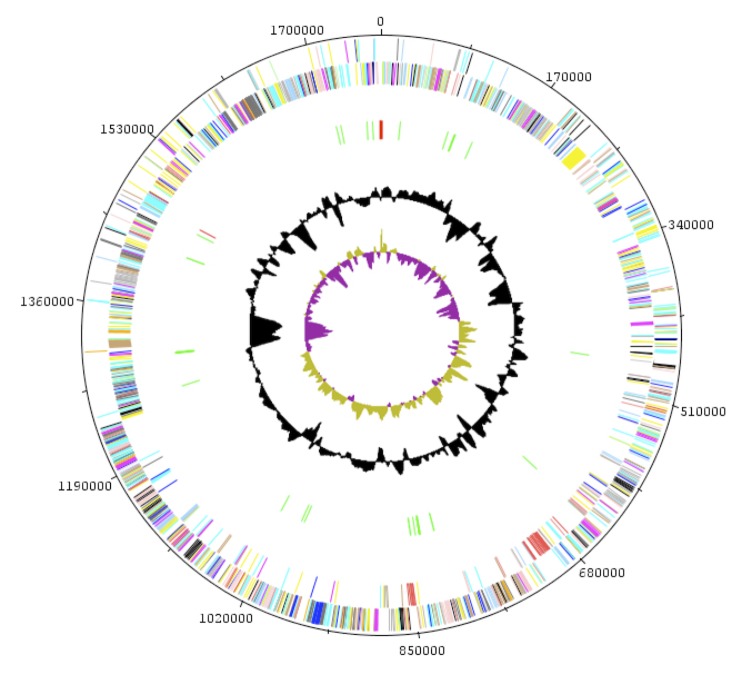
The genome is 1,770,679 bp long (one chromosome, no plasmid) with a G+C content of 51.40% (Figure 6 and Table 4). Of the 1,625 predicted chromosomal genes, 1,575 were protein-coding genes and 50 were RNAs. A total of 1,238 genes (76.18%) were assigned a putative function. Forty-two genes were identified as ORFans (2.66%) and the remaining genes were annotated as hypothetical proteins. The properties and statistics of the genome are summarized in Tables 4 and 5. The distribution of genes into COGs functional categories is presented in Table 5.
Figure 6.
Graphical circular map of the chromosome. From the outside in, the outer two circles show the genes on the forward and reverse directions (colored by COG categories). The third circle marks the rRNA operon in red and tRNA genes in green. The fourth circle shows the G+C% content plot. The inner-most circle shows GC skew, with purple and olive indicating negative and positive values, respectively.
Table 4. Nucleotide content and gene count levels of the chromosome.
| Attribute | Value | % of totala |
|---|---|---|
| Genome size (bp) | 1,770,679 | |
| DNA coding region (bp) | 1,590,528 | 89.82 |
| DNA G+C content (bp) | 910,129 | 51.40 |
| Total genes | 1,625 | 100 |
| RNA genes | 50 | 3.07 |
| Protein-coding genes | 1,575 | 96.92 |
| Genes with function prediction | 1,238 | 76.18 |
| Genes assigned to COGs | 1,165 | 71.69 |
| Genes with peptide signals | 90 | 5.53 |
| Genes with transmembrane helices | 405 | 24.92 |
a The total is based on either the size of the genome in base pairs or the total number of protein-coding genes in the annotated genome
Table 5. Number of genes associated with the 25 general COG functional categories.
| Code | Value | % agea | Description |
|---|---|---|---|
| J | 131 | 4.86 | Translation |
| A | 0 | 0.032 | RNA processing and modification |
| K | 75 | 5.31 | Transcription |
| L | 107 | 5.74 | Replication, recombination and repair |
| B | 0 | 0 | Chromatin structure and dynamics |
| D | 15 | 0.78 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 51 | 1.53 | Defense mechanisms |
| T | 22 | 1.69 | Signal transduction mechanisms |
| M | 72 | 3.42 | Cell wall/membrane biogenesis |
| N | 1 | 0 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 0 | 0 | Extracellular structures |
| U | 13 | 0.84 | Intracellular trafficking and secretion |
| O | 45 | 2.47 | Posttranslational modification, protein turnover, chaperones |
| C | 66 | 4.53 | Energy production and conversion |
| G | 88 | 2.87 | Carbohydrate transport and metabolism |
| E | 67 | 6.16 | Amino acid transport and metabolism |
| F | 47 | 2.05 | Nucleotide transport and metabolism |
| H | 34 | 2.34 | Coenzyme transport and metabolism |
| I | 28 | 4.01 | Lipid transport and metabolism |
| P | 59 | 4.14 | Inorganic ion transport and metabolism |
| Q | 4 | 0.81 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 127 | 8.15 | General function prediction only |
| S | 113 | 5.93 | Function unknown |
| - | 410 | 26.03 | Not in COGs |
a The total is based on the total number of protein-coding genes in the annotated genome
Genomic comparison of K. massiliensis and other members of the family Clostridiales Incertae Sedis XI.
Currently, 35 genomes are available for members of the family Clostridiales Incertae Sedis XI. Here, we compared the genome sequence of K. massiliensis strain ph2T with those of Finegoldia magna strain ATCC 29328, Helcococcus kunzii strain ATCC 51366, Peptoniphilus indolicus strain ATCC 29427 and Parvimonas micra strain ATCC 33270. The draft genome of K. massiliensis (1.77Mb) is smaller than all other genomes except P. micra (1.70Mb) and exhibits a higher G+C content (51.40) (Table 6A). The gene content of K. massiliensis is also lower than the other four genomes used for comparison (Table 6B). In addition, K. massiliensis shared 653, 549, 592 and 548 orthologous genes with F.magna, H. kunzii,P. indolicus and P.micra respectively. The average nucleotide sequence identity ranged from 58.61 to 69.17% among Clostridiales Incertae Sedis XI family species, and from 58.61 to 59.97% between K. massiliensis and other species, thus confirming its new genus status (Table 6B).
Table 6A. Genomic comparison of K. massiliensis gen. nov., sp. nov., strain ph2T with four other members of the family Clostridiales Incertae Sedis XI†.
| Species | Strain | Genome accession number | Genome size (Mb) | G+C content |
|---|---|---|---|---|
| K. massiliensis | ph2T | CAHC00000000 | 1,770,679 | 51.40 |
| F. magna | ATCC 29328 | NC_010376 | 1,797,577 | 32.1 |
| H. kunzii | ATCC 51366 | AGEI01000000 | 2,083,191 | 29.40 |
| P. indolicus | ATCC 29427 | AGBB01000000 | 2,101,630 | 31.70 |
| P. micra | ATCC 33270 | ABEE02000000 | 1,703,772 | 28.70 |
†Species and strain names, GenBank genome accession numbers, sizes and G+C contents.
Table 6B. Genomic comparison of K. massiliensis gen. nov., sp. nov., strain ph2T with four other members of the family Clostridiales Incertae Sedis XI†.
| K. massiliensis | F. magna | H. kunzii | P. indolicus | P. micra | |
|---|---|---|---|---|---|
| K. massiliensis | 1,568 | 635 | 549 | 592 | 548 |
| F. magna | 59.22 | 1,656 | 629 | 687 | 665 |
| H. kunzii | 59.06 | 68.20 | 1,878 | 561 | 560 |
| P. indolicus | 59.97 | 67.98 | 67.57 | 2,205 | 615 |
| P. micra | 58.61 | 69.17 | 68.52 | 68.64 | 1,597 |
† Numbers of orthologous proteins shared between genomes (upper right), average percentage of nucleotide similarity of orthologous proteins shared between genomes (lower left). Bold numbers indicate numbers of proteins per genome.
Conclusion
On the basis of phenotypic, phylogenetic and genomic analyses, we formally propose the creation of Kallipya massiliensis gen. nov., sp. nov., that contains the strain ph2T. This bacterium has been found in France.
Description of Kallipyga gen. nov.
Kallipyga (cal.li.pi’ga N.L. fem. N. Kallipyga of the Greek epithet kallipygos, said of a statue of Aphrodite having beautifully proportioned buttocks).
Gram-positive cocci. Strictly anaerobic. Mesophilic. Non-Motile. Does not exhibit catalase, oxidase and indole production nor nitrate reduction. Positive for α-galactosidase, arginine dihydrolase, arginine arylamidase, and α- and β-glucosidase. Habitat: human digestive tract. Type species: Kallipyga massiliensis.
Description of Kallipyga massiliensis gen. nov., sp. nov.
Kallipyga massiliensis (mas.il’ien’sis. L. gen. fem. n. massiliensis, of Massilia, the Latin name of Marseille where was cultivated strain ph2T). It has been isolated from the feces of an obese French patient.
Gram-positive cocci. Strictly anaerobic. Mesophilic. Optimal growth at 37°C. Non-motile and non-sporulating. Colonies are bright grey with 1.0 mm in diameter on blood-enriched Columbia agar. Cells are cocci with a diameter ranging from 0.57 µm to 0.78 µm with a mean diameter of 0.67. Catalase and oxidase activities are negative. Nitrate reduction and indole production are absent. Negative reactions were observed for β-galactosidase, β-galactosidase-6-phosphate, α-arabinosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, glutamic acid decarboxylase, proline arylamidase, leucyl glycine arylamidase, phenylalanine arylamidase, pyroglutamic acid arylamidase, tyrosine arylamidase, alanine arylamidase, glycine arylamidase, histidine arylamidase, glutamyl glutamic acid arylamidase, and serine arylamidase, mannose and raffinose fermentation, esterase, lipase, valine and cystine arylamidase, trypsine, α-chymotrypsine, naphthol-AS-BI-phosphohydrolase, β-galactosidase, β-Glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase and α-fucosidase. Positive reactions were observed for α-galactosidase, arginine dihydrolase and arginine arylamidase, α and β-glucosidase, esterase lipase, leucine arylamidase and acid phosphatase. Cells weakly oxidized D-ribose, D-glucose, D-fructose and aesculin. Cells are susceptible to amoxicillin, amoxicillin-clavulanic acid, gentamicin 500, penicillin, imipenem, vancomycin, rifampin and nitrofurantoine. The 16S rRNA and genome sequences are deposited in GenBank under accession numbers JN837487 and CAHC00000000, respectively. The G+C content of the genome is 51.40%. The type strain ph2T (= CSUR P241 = DSM 26229) was isolated from the fecal flora of an obese patient in France.
Acknowledgements
The authors thank the Xegen Company (www.xegen.fr) for automating the genomic annotation process. This study was funded by the Mediterranee-Infection Foundation.
References
- 1.Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect 2012; 18:1185-1193 [DOI] [PubMed] [Google Scholar]
- 2.Dubourg G, Lagier JC, Armougom F, Robert C, Hamad I, Brouqui P. The gut microbiota of a patient with resistant tuberculosis is more comprehensively studied by culturomics than by metagenomics. Eur J Clin Microbiol Infect Dis 2013; 32:637-645 10.1007/s10096-012-1787-3 [DOI] [PubMed] [Google Scholar]
- 3.Pfleiderer A, Lagier JC, Armougom F, Robert C, Vialettes B, Raoult D. Culturomics identified 11 new bacterial species from a single anorexia nervosa stool sample. Eur J Clin Microbiol Infect Dis. 2013 doi: 10.1007/s10096-013-1900-2. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 4.Tindall BJ, Rossello-Mora R, Busse HJ, Ludwig W, Kampfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 2010; 60:249-266 10.1099/ijs.0.016949-0 [DOI] [PubMed] [Google Scholar]
- 5.Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 2006; 33:152-155 [Google Scholar]
- 6.Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematic. Int J Syst Bacteriol 1987; 37:463-464 10.1099/00207713-37-4-463 [DOI] [Google Scholar]
- 7.Rossello-Mora R. DNA-DNA Reassociation Methods Applied to Microbial Taxonomy and Their Critical Evaluation. In: Stackebrandt E (ed), Molecular Identification, Systematics, and population Structure of Prokaryotes. Springer, Berlin, 2006; p. 23-50. [Google Scholar]
- 8.Welker M, Moore ER. Applications of whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry in systematic microbiology. Syst Appl Microbiol 2011; 34:2-11 10.1016/j.syapm.2010.11.013 [DOI] [PubMed] [Google Scholar]
- 9.Kokcha S, Mishra AK, Lagier JC, Million M, Leroy Q, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Bacillus timonensis sp. nov. Stand Genomic Sci 2012; 6:346-355 10.4056/sigs.2776064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagier JC, El Karkouri K, Nguyen TT, Armougom F, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Anaerococcus senegalensis sp. nov. Stand Genomic Sci 2012; 6:116-125 10.4056/sigs.2415480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra AK, Gimenez G, Lagier JC, Robert C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Alistipes senegalensis sp. nov. Stand Genomic Sci 2012; 6:304-314 10.4056/sigs.2625821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagier JC, Armougom F, Mishra AK, Ngyuen TT, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Alistipes timonensis sp. nov. Stand Genomic Sci 2012; 6:315-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishra AK, Lagier JC, Robert C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Clostridium senegalense sp. nov. Stand Genomic Sci 2012; 6:386-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mishra AK, Lagier JC, Robert C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Peptoniphilus timonensis sp. nov. Stand Genomic Sci 2012; 7:1-11 10.4056/sigs.2956294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra AK, Lagier JC, Rivet R, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Paenibacillus senegalensis sp. nov. Stand Genomic Sci 2012; 7:70-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagier JC, Gimenez G, Robert C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Herbaspirillum massiliense sp. nov. Stand Genomic Sci 2012; 7:200-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roux V, El Karkouri K, Lagier JC, Robert C, Raoult D. Non-contiguous finished genome sequence and description of Kurthia massiliensis sp. nov. Stand Genomic Sci 2012; 7:221-232 10.4056/sigs.3206554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kokcha S, Ramasamy D, Lagier JC, Robert C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Brevibacterium senegalense sp. nov. Stand Genomic Sci 2012; 7:233-245 10.4056/sigs.3256677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramasamy D, Kokcha S, Lagier JC, N’Guyen TT, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Aeromicrobium massilense sp. nov. Stand Genomic Sci 2012; 7:246-257 10.4056/sigs.3306717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagier JC, Ramasamy D, Rivet R, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Cellulomonas massiliensis sp. nov. Stand Genomic Sci 2012; 7:258-270 10.4056/sigs.3316719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagier JC, El Karkouri K, Rivet R, Couderc C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Senegalemassilia anaerobia sp. nov. Stand Genomic Sci 2013; 7:343-356 10.4056/sigs.3246665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishra AK, Hugon P, Lagier JC, Nguyen TT, Robert C, Couderc C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Peptoniphilus obesi sp. nov. Stand Genomic Sci 2013; 7:357-369 10.4056/sigs.32766871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mishra AK, Lagier JC, Nguyen TT, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Peptoniphilus senegalensis sp. nov. Stand Genomic Sci 2013; 7:370-381 10.4056/sigs.3366764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagier JC, El Karkouri K, Mishra AK, Robert C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Enterobacter massiliensis sp. nov. Stand Genomic Sci 2013; 7:399-412 10.4056/sigs.3396830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hugon P, Ramasamy D, Lagier JC, Rivet R, Couderc C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Alistipes obesi sp. nov. Stand Genomic Sci 2013; 7:427-439 10.4056/sigs.3336746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishra AK, Hugon P, Robert C, Couderc C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Peptoniphilus grossensis sp. nov. Stand Genomic Sci 2012; 7:320-330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mishra AK, Hugon P, Lagier JC, Nguyen TT, Couderc C, Raoult D, Fournier PE. Non contiguous-finished genome sequence and description of Enorma massiliensis gen. nov., sp. nov., a new member of the Family Coriobacteriaceae. Stand Genomic Sci 2013; 8:290-305 10.4056/sigs.3426906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramasamy D, Lagier JC, Gorlas A, Raoult D, Fournier PE. Non contiguous-finished genome sequence and description of Bacillus massiliosenegalensis sp. nov. Stand Genomic Sci 2013; 8:264-278 10.4056/sigs.3496989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramasamy D, Lagier JC, Nguyen TT, Raoult D, Fournier PE. Non contiguous-finished genome sequence and description of of Dielma fastidiosa gen. nov., sp. nov., a new member of the Family Erysipelotrichaceae. Stand Genomic Sci 2013; 8:336-351 10.4056/sigs.3567059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mishra AK, Lagier JC, Robert C, Raoult D, Fournier PE. Genome sequence and description of Timonella senegalensis gen. nov., sp. nov., a new member of the suborder Micrococcinae. Stand Genomic Sci 2013; 8:318-335 10.4056/sigs.3476977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garrity GM, Holt JG. Taxonomic outline of the Archae and Bacteria In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, second edition, Volume, Springer-Verlag, New York, 2001; p. 155-166. [Google Scholar]
- 32.Ezaki T, Kawamura Y, Li N, Li ZY, Zhao L, Shu S. Proposal of the genera Anaerococcus gen. nov., Peptoniphilus gen. nov. and Gallicola gen. nov. for members of the genus Peptostreptococcus. Int J Syst Evol Microbiol 2001; 51:1521-1528 [DOI] [PubMed] [Google Scholar]
- 33.Takii S, Hanada S, Tamaki H, Ueno Y, Sekiguchi Y, Ibe A, Matsuura K. Dethiosulfatibacter aminovorans gen. nov., sp. nov., a novel thiosulfate-reducing bacterium isolated from coastal marine sediment via sulfate-reducing enrichment with Casamino acids. Int J Syst Evol Microbiol 2007; 57:2320-2326 10.1099/ijs.0.64882-0 [DOI] [PubMed] [Google Scholar]
- 34.Murdoch DA, Shah HN. Reclassification of Peptostreptococcus magnus (Prevot 1933) Holdeman and Moore 1972 as Finegoldia magna comb. nov. and Peptostreptococcus micros (Prevot 1933) Smith 1957 as Micromonas micros comb. nov. Anaerobe 1999; 5:555-559 10.1006/anae.1999.0197 [DOI] [Google Scholar]
- 35.Collins MD, Facklam RR, Rodrigues UM, Ruoff KL. Phylogenetic analysis of some Aerococcus-like organisms from clinical sources: description of Helcococcus kunzii gen. nov., sp. nov. Int J Syst Bacteriol 1993; 43:425-429 10.1099/00207713-43-3-425 [DOI] [PubMed] [Google Scholar]
- 36.Tindall BJ, Euzeby JP. Proposal of Parvimonas gen. nov. and Quatrionicoccus gen. nov. as replacements for the illegitimate, prokaryotic, generic names Micromonas Murdoch and Shah 2000 and Quadricoccus Maszenan et al. 2002, respectively. Int J Syst Evol Microbiol 2006; 56:2711-2713 10.1099/ijs.0.64338-0 [DOI] [PubMed] [Google Scholar]
- 37.Breitenstein A, Wiegel J, Haertig C, Weiss N, Andreesen JR, Lechner U. Reclassification of Clostridium hydroxybenzoicum as Sedimentibacter hydroxybenzoicus gen. nov., comb. nov., and description of Sedimentibacter saalensis sp. nov. Int J Syst Evol Microbiol 2002; 52:801-807 10.1099/ijs.0.01998-0 [DOI] [PubMed] [Google Scholar]
- 38.Parshina SN, Kleerebezem R, Sanz JL, Lettinga G, Nozhevnikova AN, Kostrikina NA, Lysenko AM, Stams AJ. Soehngenia saccharolytica gen. nov., sp. nov. and Clostridium amygdalinum sp. nov., two novel anaerobic, benzaldehyde-converting bacteria. Int J Syst Evol Microbiol 2003; 53:1791-1799 10.1099/ijs.0.02668-0 [DOI] [PubMed] [Google Scholar]
- 39.Hernandez-Eugenio G, Fardeau ML, Cayol JL, Patel BK, Thomas P, Macarie H, Garcia JL, Ollivier B. Sporanaerobacter acetigenes gen. nov., sp. nov., a novel acetogenic, facultatively sulfur-reducing bacterium. Int J Syst Evol Microbiol 2002; 52:1217-1223 10.1099/ijs.0.01992-0 [DOI] [PubMed] [Google Scholar]
- 40.Collins MD, Shah HN. Reclassification of Bacteroides praeacutus Tissier (Holdeman and Moore) in a new genus, Tissierella, as Tissierella praeacuta comb. nov. Int J Syst Bacteriol 1986; 36:461-463 10.1099/00207713-36-3-461 [DOI] [Google Scholar]
- 41.List of Prokaryotic names with standing nomenclature (LPSN). http://www.bacterio.cict.fr
- 42.Jain S, Bui V, Spencer C, Yee L. Septic arthritis in a native joint due to Anaerococcus prevotii. J Clin Pathol 2007; 61:775-776 10.1136/jcp.2007.053421 [DOI] [PubMed] [Google Scholar]
- 43.Ulger-Toprak N, Liu C, Summanen PH, Finegold SM. Murdochiella asaccharolytica gen. nov., sp. nov., a Gram-stain-positive, anaerobic coccus isolated from human wound specimens. Int J Syst Evol Microbiol 2010; 60:1013-1016 10.1099/ijs.0.015909-0 [DOI] [PubMed] [Google Scholar]
- 44.Labutti K, Pukall R, Steenblock K, Glavina Del Rio T, Tice H, Copeland A, Cheng JF, Lucas S, Chen F, Nolan M, et al. Complete genome sequence of Anaerococcus prevotii type strain (PC1). Stand Genomic Sci 2009; 1:159-165 10.4056/sigs.24194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veloo AC, Erhard M, Welker M, Welling GW, Degener JE. Identification of Gram-positive anaerobic cocci by MALDI-TOF mass spectrometry. Syst Appl Microbiol 2011; 34:58-62 10.1016/j.syapm.2010.11.005 [DOI] [PubMed] [Google Scholar]
- 46.Pépin J, Deslandes S, Giroux G, Sobela F, Khonde N, Diakite S, Demeule S, Labbe AC, Carrier N, Frost E. The complex vaginal flora of West African women with bacterial vaginosis. PLoS ONE 2011; 6:e25082 10.1371/journal.pone.0025082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archae, Bacteria, and Eukarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibbons NE, Murray RGE. Proposals Concerning the Higher Taxa of Bacteria. Int J Syst Bacteriol 1978; 28:1-6 10.1099/00207713-28-1-1 [DOI] [Google Scholar]
- 49.Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 119-169. [Google Scholar]
- 50.Murray RGE. The Higher Taxa, or, a Place for Everything...? In: Holt JG (ed), Bergey's Manual of Systematic Bacteriology, First Edition, Volume 1, The Williams and Wilkins Co., Baltimore, 1984, p. 31-34. [Google Scholar]
- 51.List of new names and new combinations previously effectively, but not validly, published. List no. 132. Int J Syst Evol Microbiol 2010; 60:469-472 10.1099/ijs.0.022855-0 [DOI] [PubMed] [Google Scholar]
- 52.Rainey FA. Class II. Clostridia class nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 3, Springer-Verlag, New York, 2009, p. 736 [Google Scholar]
- 53.Skerman VBD, Sneath PHA. Approved list of bacterial names. Int J Syst Bact 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [Google Scholar]
- 54.Prevot AR. In: Hauduroy P, Ehringer G, Guillot G, et al.(eds.), Dictionnaire des bactéries pathogènes, Second Edition, Masson, Paris, 1953, p. 1-692. [Google Scholar]
- 55.Garrity GM, Holt J. Taxonomic outline of the Archaea and Bacteria In: Garrity GM, Boone DR, Castenholz RW (eds.), Bergey's Manual of Systematic Bacteriology, Second Edition, Springer-Verlag, New York, 2003, p.155-166. [Google Scholar]
- 56.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res 2012; 40:D48-D53 10.1093/nar/gkr1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kong HH, Grice EA, Conlan S, Deming CB, Freeman AF, Beatson M, Nomicos E, Young AC, Bouffard GG, Blakesley RW, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 2012; 22:850-859 10.1101/gr.131029.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seng P, Drancourt M, Gouriet F, La SB, Fournier PE, Rolain JM, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 2009; 49:543-551 10.1086/600885 [DOI] [PubMed] [Google Scholar]
- 60.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prodigal. http://prodigal.ornl.gov/
- 62.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955-964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100-3108 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 2004; 340:783-795 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- 65.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001; 305:567-580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 66.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics 2000; 16:944-945 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- 67.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 2009; 25:119-120 10.1093/bioinformatics/btn578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 2004; 14:1394-1403 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lechner M, Findeib S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: Detection of (Co-)orthologs in large-scale analysis. BMC Bioinformatics 2011; 12:124 10.1186/1471-2105-12-124 [DOI] [PMC free article] [PubMed] [Google Scholar]