Abstract
Artemisinin-based combination therapies (ACTs) are the main option to treat malaria, and their efficacy and susceptibility must be closely monitored to avoid resistance. We assessed the association of Plasmodium falciparum polymorphisms and ex vivo drug susceptibility with clinical effectiveness. Patients enrolled in an effectiveness trial comparing artemether-lumefantrine (n = 96), fixed-dose artesunate-amodiaquine (n = 96), and sulfadoxine-pyrimethamine (n = 48) for the treatment of uncomplicated malaria 2007 in Benin were assessed. pfcrt, pfmdr1, pfmrp1, pfdhfr, and pfdhps polymorphisms were analyzed pretreatment and in recurrent infections. Drug susceptibility was determined in fresh baseline isolates by Plasmodium lactate dehydrogenase enzyme-linked immunosorbent assay (ELISA). A majority had 50% inhibitory concentration (IC50) estimates (the concentration required for 50% growth inhibition) lower than those of the 3D7 reference clone for desethylamodiaquine, lumefantrine, mefloquine, and quinine and was considered to be susceptible, while dihydroartemisinin and pyrimethamine IC50s were higher. No association was found between susceptibility to the ACT compounds and treatment outcome. Selection was observed for the pfmdr1 N86 allele in artemether-lumefantrine recrudescences (recurring infections) (4/7 [57.1%] versus 36/195 [18.5%]), and of the opposite allele, 86Y, in artesunate-amodiaquine reinfections (new infections) (20/22 [90.9%] versus 137/195 [70.3%]) compared to baseline infections. The importance of pfmdr1 N86 in lumefantrine tolerance was emphasized by its association with elevated lumefantrine IC50s. Genetic linkage between N86 and Y184 was observed, which together with the low frequency of 1246Y may explain regional differences in selection of pfmdr1 loci. Selection of opposite alleles in artemether-lumefantrine and artesunate-amodiaquine recurrent infections supports the strategy of multiple first-line treatment. Surveillance based on clinical, ex vivo, molecular, and pharmacological data is warranted.
INTRODUCTION
Half of the world's population lives at risk for malaria, and more than half a million people die from the disease every year (1). An effective treatment is essential for malaria control, and the emergence and spread of chloroquine resistance have contributed significantly to malaria-attributed mortality (2, 3). Spread of antimalarial drug resistance in most areas where Plasmodium falciparum is endemic prompted the World Health Organization to recommend the use of artemisinin-based combination therapies (ACTs), which have now been adopted by most countries where P. falciparum malaria is endemic as a first-line treatment for uncomplicated malaria (1). Artemether-lumefantrine (AL) and artesunate-amodiaquine (ASAQ) are the ACTs most used in Africa. For intermittent preventive treatment (IPT) of pregnant women the antifolate sulfadoxine-pyrimethamine (SP) is the main present option (1). In Southeast Asia, resistance to artemisinin derivatives has been suggested to emerge, associated with delayed parasite clearance (4–6). Emergence and spread of resistance to artemisinins and the partner drugs would raise a serious problem for malaria control and need to be closely monitored. P. falciparum drug resistance is assessed by clinical efficacy/effectiveness trials, by in vitro susceptibility assays, and by the use of molecular markers associated with parasite drug resistance (7). The relevance of in vitro susceptibility testing for artemisinin derivatives is being discussed (8, 9). However, it is important for susceptibility testing of ACT partner drugs and for detecting true drug resistance to avoid confounding host factors. The molecular mechanisms of artemisinin resistance have not been elucidated yet, but a region in chromosome 13 has been identified that may be associated with the delayed parasite clearance phenotype (10).
Decreased sensitivity or parasite tolerance to artemisinin partner drugs has been observed in Africa, where single nucleotide polymorphisms (SNPs) in pfcrt, pfmdr1, and pfmrp1 have been selected in recurrent infections after AL and ASAQ treatments (11–18). The involvement of these genes in partner drug tolerance is supported by the association of pfcrt and pfmdr1 with variation in in vitro susceptibility to lumefantrine (LUM) and monodesethylamodiaquine (DEAQ), the active metabolite of amodiaquine (AQ) (17, 19, 20).
Molecular characterization of the P. falciparum altered response to ACTs as described above has mainly been assessed in East Africa, and only a few studies have been performed in West Africa, showing selection patterns similar to those seen in East Africa (21–23). As East and West Africa may differ in terms of drug use and parasite genetics (24), it is of importance to further describe the basis of drug tolerance and resistance also in West Africa.
We performed a comprehensive study in Benin to characterize the genetic bases of the P. falciparum response to antimalarial drugs, and the ex vivo drug susceptibility, in an ACT effectiveness study performed before ACT implementation. To our knowledge, this is the first study exploring the association of P. falciparum alleles and ex vivo susceptibility with in vivo efficacy of the currently used ACT and SP in Benin.
MATERIALS AND METHODS
Clinical trial and blood sampling.
The effectiveness of SP and unsupervised AL and ASAQ fixed-dose formulation for the treatment of uncomplicated malaria was compared during a randomized effectiveness noninferiority trial including 240 children, described elsewhere (25). Briefly, P. falciparum isolates were collected from children <5 years old between May and November 2007 in Allada and Sekou, southern Benin. Blood sampling for ex vivo drug susceptibility testing was collected in EDTA tubes at inclusion and in case of treatment failure. For molecular analysis, blood was spotted on filter paper at inclusion, on days 3, 7, 14, 21, 28, 35, and 42 during follow-up, and in case of infection recurrence. Recurrent infections were classified as recrudescences (reappearances) or reinfections (new infections) based on msp1 and msp2 analyses, as previously described (25). Concentrations of lumefantrine or DEAQ in blood on day 3 were determined using high-pressure liquid chromatography with electrochemical (DEAQ) or UV (LUM) detection from blood samples collected on Whatman 3MM filter paper (25). Both drug assays had a lower limit of quantification of 0.02 μg/ml.
Ex vivo drug susceptibility assay.
DEAQ was provided by SAPEC (Barbengo, Switzerland), and LUM was provided by Novartis Pharma (Basel, Switzerland). Quinine (QN), mefloquine (MQ), dihydroartemisinin (DHA), and pyrimethamine (PYR) were obtained from Sigma-Aldrich Company (St. Louis, MO). Chloroquine (CQ) was also tested, but the results were inconclusive due to technical difficulties. Antimalarial drug stock solutions were made in the appropriate solvent, while dilutions were made in water, except for lumefantrine, which was diluted in ethanol. The dilutions were distributed and dried in 96-well tissue culture plates in the following range of concentrations: QN in ethanol, 25 to 3,200 nM; MQ in methanol, 3.12 to 400 nM; DHA in water, 0.25 to 64 nM; PYR in ethanol, 50 to 40,000 nM; DEAQ in water, 7.5 to 1,920 nM; and LUM in ethanol, 1.25 to 320 nM. The reference laboratory P. falciparum clones 3D7 (Africa) and W2 (Indochina) were used to control each batch of plates. For each drug tested, three control wells were drug free, and each concentration was studied in duplicate or triplicate. Parasitized blood samples were washed and cultured in drug-coated plates for 42 h before freezing, as described previously (26, 27). Briefly, after washes, blood samples were resuspended in 1.5% hematocrit in RPMI 1640 (Gibco, Invitrogen Life Technologies, Auckland, New Zealand) supplemented with 25 mM HEPES (Invitrogen, Cergy Pontoise, France), 25 mM NaHCO3 (Sigma-Aldrich), and 10% human serum (Abcys Biowest, Paris, France). PYR susceptibility was assessed in RPMI SP 241 medium (Gibco BRL, Paisley, United Kingdom) with a low concentration of folic acid and p-aminobenzoic acid. For initial parasitemias of >1%, a dilution was made by adding uninfected O+ group erythrocytes to obtain 0.5% to 1% parasite density. The plates were incubated for 42 h at 37°C with 5% CO2 and then frozen at −20°C. The in vitro assay output was determined by Plasmodium lactate dehydrogenase (PLDH) production with a commercial enzyme-linked immunosorbent assay (ELISA)-malaria antigen test (DiaMed AG, Cressier sur Morat, Switzerland), as previously described (27). Optical density was measured with a spectrophotometer (E960; Fisher Bioblock Scientific, Illkirch, France). Fifty percent inhibitory concentrations (IC50s) with their 95% confidence intervals (CIs) were calculated with ICEstimator software (http://www.antimalarial-icestimator.net) (28).
DNA extraction.
DNA was extracted from blood spots on filter paper using the QIAamp DNA blood minikit (Qiagen, Hilden, Germany) according to the manufacturer's protocol for dried blood spots.
pfmrp1 SNPs.
pfmrp1 single nucleotide polymorphisms (SNPs) in the I876V and K1466R codons were analyzed by pyrosequencing. Details regarding the pyrosequencing method (29) and the specific PCR amplification and pyrosequencing of pfmrp1 SNPs in the I876V and K1466R codons have been previously described (30). Briefly, extracted DNA was amplified in a nested PCR using GoTaq polymerase (Promega, Madison, WI). Primers used in amplification and pyrosequencing reactions are shown in Table 1. In the nested PCR, one of the primers was biotinylated, allowing the purification of specific single-strand products with streptavidin Sepharose beads (Amersham Bioscience, Little Chalfont, United Kingdom) for the pyrosequencing reaction. The reagents used for pyrosequencing were provided by Biotage (Uppsala, Sweden). Nucleotide dispensation order was GATACTGAT and CGACGATGT for I876V and K1466R, respectively. The definition for a mixed-genotype infection was a pyrosequencing result between 10% and 90% for both genotypes, and for a single genotype (pure) infection, it was above 90%. For pfmrp1 I876V, the results were adjusted against a standard curve derived from different proportions of mixes of the reference laboratory strains 3D7 and W2.
TABLE 1.
Primers used in PCR, sequencing, and pyrosequencing reactions
| Gene | Reaction or locus | Primer name | Primer sequence (5′ to 3′)b |
|---|---|---|---|
| pfdhfr | Outer | AMP1F | TTTATATTTTCTCCTTTTTA |
| AMP2R | CATTTTATTATTCGTTTTCT | ||
| 51 and 108 | AMP3F | TGATGGAACAAGTCTGCGAC | |
| dhfrR4 | ATAACATTTATCCTATTGCTTAAAGGTT | ||
| 59 | F | GAAATGTAATTCCCTAGATATGgAATATT | |
| dhfrR2 | TTTGGAATGCTTTCCCAG | ||
| 164 | dhfr_164_r | CCTTTAAGCAATAGGATAAATGTTATATTG | |
| F/ | AAATTCTTGATAAACAACGGAACCTttTA | ||
| pfcrt | Outer | CRTP1 | CCGTTAATAATAAATACACGCAG |
| CRTP2 | CGGATGTTACAAAACTATAGTTACC | ||
| 76 | CRTD1 | TGTGCTCATGTGTTTAAACTT | |
| CRTD2 | CAAAACTATAGTTACCAATTTTG | ||
| pfmdr1 | Outer | A1 | TGTTGAAAGATGGGTAAAGAGCAGAAAGAG |
| A3 | TACTTTCTTATTACATATGACACCACAAACA | ||
| 86 and 184 | A2 | GTCAAACGTGCATTTTTTATTAATGACCATTTA | |
| A4 | AAAGATGGTAACCTCAGTATCAAAGAAGAG | ||
| Outer | O1 | AGAAGATTATTTCTGTAATTTGATACAAAAAGC | |
| O2 | ATGATTCGATAAATTCATCTATAGCAGCAA | ||
| 1246a | 1246f | ATGATCACATTATATTAAAAAATGATATGACAAAT | |
| pfdhps | Outer | R2 | AACCTAAACGTGCTGTTCAA |
| R/ | AATTGTGTGATTTGTCCACAA | ||
| 437 and 540 | K | TGCTAGTGTTATAGATATAGGatGAGcATC | |
| K/ | CTATAACGAGGTATTgCATTTAATgCAAGAA | ||
| pfmrp1 | 876 outer | A2626G PS First fw | AATATTCCATTCAATGAAAATTAC |
| A2626G PS First rev | CAACGTACTTTTATTCATTGAGA | ||
| 876 nest | A2626G PS Nest fw | Biotin-TATTCCATTCAATGAAAATTACCT | |
| A2626G PS Nest rev | TATGGAAGGATCTAAAGATGTAAA | ||
| 876 seq | A2626G PS Seq rev | GGAAGGATCTAAAGATGTAA | |
| 1466 outer | A4397G PS First fw | AATAAAGAACATTCAGACACAAT | |
| A4397G PS First rev | TGATTTTCCTACTATCCCAATT | ||
| 1466 nest | A4397G PS Nest fw | TGGATACTGTATATCGTTTTCTGC | |
| A4397G PS Nest rev | Biotin-CCCAATTTTTTGATTTTTTAAAGC | ||
| 1466 seq | A4397G PS Seq fw | TGATTATACTCACATAGAAA |
Primer pair O2-1246f was used for the seminested PCR.
Lowercase letters indicate variant nucleotides.
pfcrt and pfmdr1 SNPs.
The pfcrt K76T and pfmdr1 N86Y and Y184F codons were simultaneously amplified in a multiplex PCR. Two separated nested PCRs were performed: one duplex for determining the genotype at both pfcrt 76 and pfmdr1 86 and one simplex for pfmdr1 184. Primers are shown in Table 1. All PCRs were performed with 200 μM deoxynucleoside triphosphates (dNTPs) and 0.5 U of GoTaq polymerase (Promega). The PCR master mix contained various concentrations of MgCl2 and primers according to the reaction: first multiplex PCR, 3.5 mM MgCl2 and 10 nM for each of primers CRTP1 and CRTP2 (31) and A1 and A3 (32); nested PCR for SNPs at codons pfcrt 76 and pfmdr1 86, 3.5 mM MgCl2 and 300 nM for primers CRTD1 and CRTD2 and 100 nM for primers A2 and A4 (31, 32); nested PCR for SNPs at codon pfmdr1 184, 2 mM MgCl2 and 100 nM for primers A2 and A4. The nested amplification product sizes were 145 bp and 560 bp for pfcrt and pfmdr1, respectively.
Genotyping of the pfmdr1 D1246Y codon was derived from conditions previously published (32). The first PCR master mix contained 3.5 mM MgCl2 and 100 nM (each) primers O1 and O2. The seminested PCR mixture contained 3 mM MgCl2 and 250 nM primers O2 and 1246f. The amplification product was 344 bp.
pfdhfr SNPs.
The N51I, C59R, S108N, and I164L codons were analyzed as described previously (33), with minor modifications. The first PCR master mix contained 3 mM MgCl2 and 100 nM (each) primers AMP1F and AMP2R. The nested PCRs were all performed with 3 mM MgCl2 but differed in the concentration of primers (100 nM primers AMP3F and dhfrR4 for SNPs at codons 51 and 108 and 300 nM for primers F [34] and dhfrR2 for SNPs at codon 59 or dhfr_164_r and F/ [34] for SNPs at codon 164). The results were products of 376 bp for the dhfrR4-AMP3F primer pair, 189 bp for dhfrR2-F, and 168 bp for dhfr_164_r-F/.
pfdhps SNPs.
Genotyping of the pfdhps A437G and K540E codons was carried out as described by Duraisingh et al. (34). The mixtures for both the first and the nested PCRs contained 2 mM MgCl2 and 100 nM (each) primers R2 and R/ (first) and K and K/ (nest). The result was a product of 438 bp.
RFLP.
The genotype of each SNP (except pfmrp1) was determined by restriction fragment length polymorphisms (RFLP) of nested PCR products using appropriate restriction enzymes purchased from New England BioLabs (Ipswich, MA). Four microliters of the nested PCR product was incubated with restriction enzymes in a total volume of 20 μl according to the manufacturer's instructions. The products obtained from the duplex PCR with the D1-D2 and A2-A4 primer pairs were digested with ApoI for SNPs at the pfcrt K76 and pfmdr1 N86 codons. For SNPs at the pfmdr1 184F and 1246Y codons, the PCR products obtained with the A2-A4 and O2-1246f primer pairs were digested with DraI and EcoRV, respectively. For the pfdhfr gene, the PCR product obtained with the AMP3F-dhfrR4 primer pair was digested with AluI and Tsp509I for SNPs at the pfdhfr S108 and N51 codons, respectively, the one obtained with dhfrR2-F was digested with XmnI for SNP at the pfdhfr 59R codon, and the one obtained with the dhfr_164_r-F/ primer pair was digested with DraI for SNP at the pfdhfr 164L codon. For the pfdhps gene, the PCR product was digested with AvaII and FokI for SNPs at the pfdhps 437G and 540E codons, respectively. The products of digestion were resolved on 2 to 2.5% agarose gels (Invitrogen, Life Technologies, Carlsbad, CA). DNAs from 6 laboratory P. falciparum strains (3D7, HB3, 7G8, W2, Dd2, and FCR3) were used as controls for the PCR and the digestion.
pfcrt, pfmdr1, and pfmrp1 SNPs were genotyped in the samples of all three treatment arms, while pfdhfr and pfdhps loci were tested only in the SP arm.
Statistical analysis.
In the molecular analysis, patients were excluded from the pretreatment population due to missing blood samples or filter papers, infection with other Plasmodium species, or failure to meet the inclusion criteria. Additional patients were excluded from the molecular selection analysis of recurrent infections because of wrong dosage, intake of other antimalarial drugs during the follow-up, and early treatment failure. Additional exclusion criteria for the molecular prediction analysis included loss to follow-up, consent withdrawal, and adverse events (Fig. 1). The correlation between ex vivo drug susceptibilities was established by a correlation matrix with Bonferroni adjustment to calculated significance levels. The data were log transformed since they were not normally distributed according to the Shapiro-Wilk W test. Multiple linear regression analysis was used to estimate the relations between drugs. Reported P values are two tailed. Fisher's two-tailed test was used to evaluate the difference in genotype frequencies between pretreatment samples and recrudescences and reinfections. Mixed-genotype infections were analyzed together with the nonselected genotypes against the selected genotype, as previously suggested (35). To evaluate an association between the pfmdr1 N86 and Y184 genotypes, observed and expected frequencies were compared with the χ2 test. Mixed-genotype infections were removed from this analysis. The association between alleles and ex vivo susceptibility was determined with the Mann-Whitney test. Mixed-genotype infections were excluded from the Mann-Whitney analysis. The association of pfmdr1 haplotypes and ex vivo susceptibility was assessed with the Kruskal-Wallis test. Significant associations were further tested post hoc by the Mann-Whitney test. The relation between genotype at the baseline and in vivo treatment outcome was assessed by the χ2 test. The association between pfmdr1 86 genotype and drug concentration was assessed with the Mann-Whitney test. Statistical significance for all of described tests was defined as a P value of <0.05. Fisher's exact two-tailed test and the χ2 test were performed with GraphPad QuickCalcs (GraphPad Software Inc., San Diego, CA) and the Mann-Whitney and Kruskal-Wallis tests with VassarStats (http://vassarstats.net/).
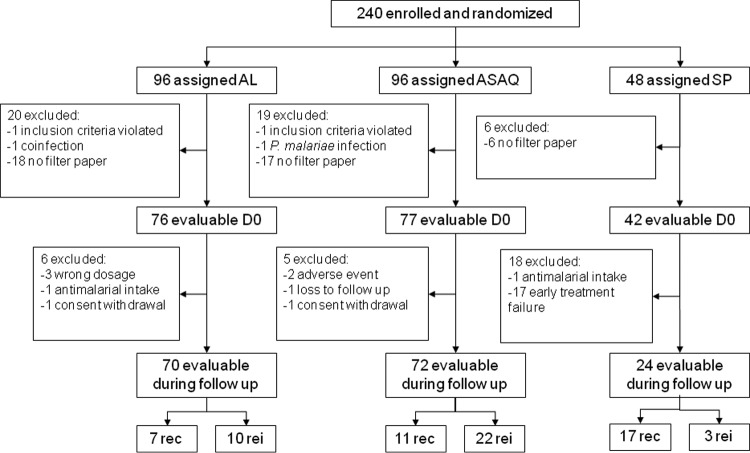
FIG 1.
Flowchart describing excluded and evaluable patients for molecular analysis pretreatment (D0) and during the follow-up for recredescences (rec) and reinfections (rei).
RESULTS
P. falciparum in vitro drug susceptibility at baseline.
Minimal blood volume for isolates to be tested ex vivo was obtained for 118 children before treatment (day 0). Results from the PLDH ELISA ex vivo drug susceptibility assay were available for 58 to 101 isolates depending on the drug tested (Table 2), resulting in a mean of 71.5% successful assays. For these isolates, the geometric mean IC50s of DEAQ, LUM, MQ, and QN were similar to or lower than the corresponding IC50 for 3D7 (considered to be drug susceptible), and a majority of the isolates had IC50 estimates lower than that of 3D7 (Table 2). The geometric mean PYR IC50 of the tested isolates was considerably higher (3,577 nM) than that of 3D7 (876.9 nM), and 52/58 (89.7%) isolates had higher individual IC50s than 3D7. Susceptibility to DHA was also reduced in the isolates (3.1 nM) compared to that of 3D7 (1.9 nM), and 73/95 (76.8%) isolates had higher IC50s than 3D7. Susceptibilities to MQ and LUM were strongly correlated (Table 3), while an inverse correlation was observed between IC50s to MQ and DEAQ. DHA was correlated with QN and PYR. In univariate analysis, there was a positive correlation between DEAQ, QN, and PYR susceptibilities that was confirmed in multivariate analysis (data not shown).
TABLE 2.
Ex vivo drug susceptibilities of field isolates from Benin and reference clones
| Drug | No. of isolates tested | Geometric mean IC50 (nM) | 95% confidence interval (nM) | 3D7 geometric mean IC50 (nM) | W2 geometric mean IC50 (nM) | % of isolates (no./total) with a higher IC50 than 3D7 |
|---|---|---|---|---|---|---|
| DEAQ | 101 | 19.6 | 6.1–62.5 | 20.6 | 79.5 | 34.6 (35/101) |
| LUM | 90 | 9.3 | 1.9–49.6 | 23.2 | 29.9 | 8.9 (8/90) |
| DHA | 95 | 3.1 | 0.7–12.8 | 1.9 | 1.2 | 76.8 (73/95) |
| MQ | 79 | 9.8 | 2.1–45.4 | 49.9 | 45.7 | 3.8 (3/79) |
| QN | 83 | 71.2 | 23.0–220.6 | 129.9 | 528.7 | 15.7 (13/83) |
| PYR | 58 | 3,577 | 194–66,057 | 876.9 | NDa | 89.7 (52/58) |
ND, not done.
TABLE 3.
Correlations between ex vivo drug susceptibilities
| Drug | Correlation |
||||
|---|---|---|---|---|---|
| DEAQ | LUM | DHA | MQ | QN | |
| LUM | 0.121 | ||||
| DHA | 0.176 | 0.153 | |||
| MQ | −0.243a | 0.613a | −0.019 | ||
| QN | 0.289a | 0.096 | 0.393a | −0.076 | |
| PYR | 0.391a | 0.090 | 0.402a | −0.172 | 0.373a |
Statistical significance (P < 0.05) with Bonferroni adjustment.
Relation between in vitro drug susceptibility at baseline and treatment outcome.
No relation was found between day 42 treatment outcome and day 0 IC50 estimates for DHA, LUM, and DEAQ (Mann-Whitney U test, P > 0.05), as shown in Table 4. PYR IC50s were high in isolates from patients with in vivo failure to SP. However, only one isolate from a patient successfully treated with SP could be analyzed.
TABLE 4.
Relation between ex vivo drug susceptibility at baseline and treatment outcome at day 42
| Ex vivo agent tested pretreatment | Treatment | Treatment success |
Recrudescencea |
||
|---|---|---|---|---|---|
| No. of tested ex vivo isolates | IC50 ± SE (nM) | No. of tested ex vivo isolates | IC50 ± SE (nM) | ||
| DEAQ | ASAQ | 27 | 21.3 ± 2.7 | 8 | 31.8 ± 9.3 |
| LUM | AL | 32 | 15.1 ± 2.1 | 2 | 9.6 ± 3.4 |
| DHA | ASAQ or AL | 56 | 4.2 ± 0.4 | 9 | 3.6 ± 0.7 |
| PYR | SP | 1 | 1,001 | 13 | 7,390 ± 2,340 |
Recrudescence defined by msp1 and msp2 analyses.
P. falciparum genetic diversity.
Genotyping was successful in pfcrt and pfmdr1 loci in all of the 195 available baseline samples included in the study and in pfmrp1 loci in 183/195 (94%) samples. pfdhr and pfdhps loci were successfully assessed in samples from all SP patients. At baseline, the frequencies of alleles previously associated with resistance, pfcrt 76T (chloroquine resistance) and the pfdhfr triple mutant 108N 51I 59R and pfdhps 437G (SP resistance), were close to 100% in the parasite population. The pfmrp1 mutation 876V was almost absent. No deviation from the wild-type genotype was identified in pfdhfr codon 164 or pfdhps codon 540 (Table 5). Consequently, these nonpolymorphic loci were not informative in further assessments of P. falciparum polymorphisms with in vivo and ex vivo drug susceptibilities. pfmdr1 loci were found to be polymorphic, with mutations 86Y, 184F, and 1246Y identified in 81.6%, 79.5%, and 11.8% of the patients, respectively. The mutant allele pfmrp1 1466R was found in 13.2% (Table 5). An association between the pfmdr1 86 and 184 genotypes was found (χ2 = 13.82; P = 0.0032) where N86 plus Y184 was observed in a higher frequency (11.9%) than expected (4.5%) based on individual alleles, as well as 86Y plus 184F (observed, 88.9%; expected, 62.2%); N86 plus 184F (observed, 9.5%; expected, 16.3%) and 86Y plus Y184 (observed, 12.5%; expected, 17%) were observed in lower frequencies than expected (Table 6).
TABLE 5.
Number and frequency of pfcrt, pfmdr1, pfmrp1, pfdhfr, and pfdhps genotypes in P. falciparum infections pretreatment and after treatment with ASAQ, AL, and SP
| Genotype | Pretreatment |
Posttreatmenta |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ASAQ |
AL |
SP |
||||||||||||
| Recrudescences |
Reinfections |
Recrudescences |
Reinfections |
Recrudescences |
Reinfections |
|||||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | |
| pfcrt 76 | ||||||||||||||
| K | 4 | 2.1 | 1 | 9.1 | 1 | 4.8 | 0 | 0.0 | 0 | 0.0 | ||||
| K/T | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | ||||
| T | 191 | 97.9 | 10 | 90.9 | 20 | 95.2 | 7 | 100.0 | 10 | 100.0 | ||||
| pfmdr1 86 | ||||||||||||||
| N | 36 | 18.5 | 1 | 9.1 | 1 | 4.5 | 4b | 57.1 | 2 | 20.0 | ||||
| N/Y | 22 | 11.3 | 2 | 18.2 | 1 | 4.5 | 1 | 14.3 | 0 | 0.0 | ||||
| Y | 137 | 70.3 | 8 | 72.7 | 20b | 90.9 | 2 | 28.6 | 8 | 80.0 | ||||
| pfmdr1 184 | ||||||||||||||
| Y | 40 | 20.5 | 2 | 18.2 | 3 | 14.3 | 3 | 42.9 | 2 | 20.0 | ||||
| Y/F | 9 | 4.6 | 0 | 0.0 | 0 | 0.0 | 1 | 14.3 | 1 | 10.0 | ||||
| F | 146 | 74.9 | 9 | 81.8 | 18 | 85.7 | 3 | 42.9 | 7 | 70.0 | ||||
| pfmdr1 1246 | ||||||||||||||
| D | 172 | 88.2 | 11 | 100.0 | 19 | 86.4 | 7 | 100.0 | 10 | 100.0 | ||||
| D/Y | 11 | 5.6 | 0 | 0.0 | 2 | 9.1 | 0 | 0.0 | 0 | 0.0 | ||||
| Y | 12 | 6.2 | 0 | 0.0 | 1 | 4.5 | 0 | 0.0 | 0 | 0.0 | ||||
| pfmrp1 876 | ||||||||||||||
| I | 178 | 97.3 | 11 | 100.0 | 20 | 90.9 | 7 | 100.0 | 9 | 90.0 | 16 | 94.1 | 3 | 100.0 |
| I/V | 2 | 1.1 | 0 | 0.0 | 1 | 4.5 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| V | 3 | 1.6 | 0 | 0.0 | 1 | 4.5 | 0 | 0.0 | 1 | 10.0 | 1 | 5.9 | 0 | 0.0 |
| pfmrp1 1466 | ||||||||||||||
| K | 159 | 86.9 | 10 | 90.9 | 18 | 81.8 | 6 | 85.7 | 8 | 80.0 | 13 | 76.5 | 3 | 100.0 |
| K/R | 10 | 5.5 | 1 | 9.1 | 2 | 9.1 | 1 | 14.3 | 0 | 0.0 | 1 | 5.9 | 0 | 0.0 |
| R | 14 | 7.7 | 0 | 0.0 | 2 | 9.1 | 0 | 0.0 | 2 | 20.0 | 3 | 17.6 | 0 | 0.0 |
| pfdhfr | ||||||||||||||
| Triple mutant 108N 51I 59R | 41 | 100.0 | 17 | 100.0 | 3 | 100.0 | ||||||||
| 164L | 41 | 0.0 | 17 | 0.0 | 3 | 0.0 | ||||||||
| pfdhps | ||||||||||||||
| 437G | 39 | 95.1 | 17 | 100.0 | 3 | 100.0 | ||||||||
| 540E | 41 | 0.0 | 17 | 0.0 | 3 | 0.0 | ||||||||
Recrudescences (reappearances) and reinfections (new infections) were defined by msp1 and msp2 analyses.
P < 0.05.
TABLE 6.
Frequency of pfmdr1 86 plus 184 haplotypes and the association with ex vivo drug susceptibility (IC50 in nM) at baselinea
| pfmdr1 86 plus 184 haplotypeb | Frequency |
DEAQ |
LUM |
DHA |
MQ |
QN |
PYR |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | Mean ± SEM | No. | Mean ± SEM | No. | Mean ± SEM | No. | Mean ± SEM | No. | Mean ± SEM | No. | Mean ± SEM | |
| N + F | 16 | 9.5 | 13 | 20.3 ± 3,6 | 13 | 21.1 ± 5.1* A | 12 | 4.21 ± 1.1 | 12 | 29.6 ± 6.1*** C | 10 | 86.8 ± 14.2 | 5 | 12,447.6 ± 4,457.4 |
| N + Y | 20 | 11.9 | 11 | 23.8 ± 4.4 | 12 | 14.6 ± 2.4* B | 12 | 4.0 ± 0.8 | 12 | 17.0 ± 3.7* D | 11 | 109.9 ± 16.5* E | 6 | 9,676.9 ± 3,483.2 |
| Y + F | 111 | 88.9 | 55 | 23.2 ± 2.2 | 48 | 9.4 ± 1.2* AB | 50 | 4.0 ± 0.4 | 42 | 8.7 ± 0.9*** C/* D | 43 | 75.6 ± 6.4* EF | 32 | 7,001.3 ± 1,408.8 |
| Y + Y | 21 | 12.5 | 7 | 25.1 ± 5.9 | 6 | 10.7 ± 1.9 | 8 | 3.3 ± 0.6 | 4 | 9.7 ± 1.3 | 6 | 111.6 ± 10.6* F | 5 | 15,243 ± 5,894.3 |
*, P < 0.05; **, P < 0.01; ***, P < 0.001. Difference in the ex vivo drug sensitivity between the haplotypes was assessed by Kruskal-Wallis followed by the Mann-Whitney U test to distinguish between which two haplotypes there was a difference. The same letter is placed by haplotype pairs between which there was a significant difference.
Samples containing mixed alleles in either or both loci were excluded from the analysis.
Selection of polymorphisms in recurrent infections.
To determine if any of the studied SNPs was under selection, the allele prevalence was compared between baseline infections and recrudescences and reinfections. The frequency of the selected genotype was compared with the frequency of the deselected genotype together with the mixed genotype. The numbers of recrudescences and reinfections distinguished by msp1 and msp2 analyses were 11 and 22 in the ASAQ arm, 7 and 10 in the AL arm, and 17 and 3 in the SP arm, respectively. The molecular analyses of recurrent infections were successful for all samples except for one reinfection in the ASAQ arm. As shown in Table 5, we observed a statistically significant selection of the pfmdr1 N86 allele in AL recrudescences (57.1% versus 18.5%; P = 0.030) and of the opposite allele, 86Y, in ASAQ reinfections (90.9% versus 70.3%; P = 0.044). It was assessed whether the genotype of baseline isolates could predict treatment outcome in the different treatment groups. Although pfmdr1 N86 and pfmdr1 86Y isolates were shown to be selected in recurrent infections in the AL and ASAQ arms, respectively, the presence of these mutations in baseline isolates did not predict the occurrence of treatment failure.
Blood drug concentrations at day 3 and reinfection.
It was assessed whether the pfmdr1 86 genotype in reinfections was associated with drug concentration at day 3 and the day of reinfection. In the AL arm, the two reinfections with organisms carrying N86 were associated with significantly higher blood LUM concentrations (mean, 3.36 ± 0.68 μg/ml; P = 0.04) and were mainly reinfecting earlier (day 28) than those carrying 86Y (mean, 0.74 ± 0.12 μg/ml) that were reinfecting later (day 35). In the ASAQ arm, there were no differences between reinfections with organisms carrying N86 and 86Y: the two groups had the same mean blood DEAQ concentration and day of reinfection.
Association between polymorphisms and ex vivo drug susceptibility.
Association between the IC50s to DEAQ, LUM, DHA, MQ, QN, and PYR and P. falciparum polymorphisms was assessed in the baseline samples. The loci pfmdr1 86, 184, and 1246 and pfmrp1 1466 were included in the analyses (Table 7), while pfcrt 76, pfmrp1 876, and pfdhfr and pfdhps loci could not be evaluated because of limited genetic variation in the samples with available IC50 estimates.
TABLE 7.
Association between P. falciparum polymorphisms and ex vivo drug susceptibility (IC50 in nM) at baselinea
| Locus | Alleleb | DEAQ |
LUM |
DHA |
MQ |
QN |
PYR |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | IC50, nM (mean ± SEM) | No. | IC50, nM (mean ± SEM) | No. | IC50, nM (mean ± SEM) | No. | IC50, nM (mean ± SEM) | No. | IC50, nM (mean ± SEM) | No. | IC50, nM (mean ± SEM) | ||
| pfmdr1 86 | N | 24 | 21.9 ± 2.8 | 25 | 18.0 ± 2.9** | 24 | 4.1 ± 0.7 | 24 | 23.3 ± 3.7*** | 21 | 98.9 ± 11.0 | 11 | 9894.0 ± 2542.3 |
| Y | 63 | 24.0 ± 2.1 | 54 | 9.5 ± 1.1 | 59 | 3.8 ± 0.3 | 46 | 8.7 ± 0.8 | 49 | 78.2 ± 6.0 | 37 | 8115.1 ± 1490.0 | |
| pfmdr1 184 | Y | 20 | 24.0 ± 3.1 | 20 | 15.2 ± 2.2* | 69 | 4.0 ± 0.3 | 18 | 14.8 ± 2.6 | 19 | 106.2 ± 10.9* | 13 | 10512.7 ± 2940.4 |
| F | 77 | 22.1 ± 1.7 | 67 | 12.0 ± 1.5 | 22 | 3.7 ± 0.5 | 60 | 13.1 ± 1.7 | 61 | 76.4 ± 5.6 | 43 | 6904.9 ± 1174.4 | |
| pfmdr1 1246 | D | 91 | 23.0 ± 1.9 | 83 | 13.0 ± 1.3 | 84 | 4.0 ± 0.3 | 73 | 13.8 ± 1.6 | 75 | 80.8 ± 5.4* | 44 | 6584.9 ± 1167.2 |
| Y | 5 | 27.3 ± 8.4 | 6 | 10.7 ± 1.9 | 6 | 3.1 ± 0.8 | 4 | 9.7 ± 1.3 | 5 | 117.9 ± 10.3 | 4 | 17069.2 ± 7235.2 | |
| pfmrp1 1466 | K | 79 | 23.6 ± 2.1 | 71 | 12.4 ± 1.3 | 71 | 3.9 ± 0.3 | 63 | 13.9 ± 1.8 | 63 | 83.2 ± 6.1 | 47 | 7637.4 ± 1146.2 |
| R | 7 | 21.8 ± 7.8 | 7 | 16.2 ± 5.5 | 8 | 4.5 ± 1.8 | 4 | 11.6 ± 2.8 | 8 | 66.8 ± 9.7 | 2 | 1169.0 ± 1070.4 | |
*, P < 0.05; **, P < 0.01; ***, P < 0.001. Difference in the vivo drug sensitivity between the two alleles was assessed using the Mann-Whitney U test.
Samples containing mixed alleles in the specific locus were excluded from the analysis.
We found an association between both the pfmdr1 allele N86 (P = 0.0039) and pfmdr1 Y184 (P = 0.0466) and decreased ex vivo susceptibility to LUM. pfmdr1 N86 was strongly associated with decreased MQ susceptibility (P < 0.0001). pfmdr1 Y184 (P = 0.0105) and 1246Y (P = 0.0332) were associated with decreased QN susceptibility (Table 7). Evaluation of the pfmdr1 86 plus 184 haplotype showed a decreased susceptibility to LUM in parasites with N86 plus 184F (P = 0.011) and N86 plus Y184 (P = 0.044) compared to 86Y plus 184F (Table 6). A similar association was seen for MQ. While there was no significant difference between N86 plus 184F and N86 plus Y184 parasite susceptibilities to LUM (P = 0.81), N86 plus 184F was associated with a slightly higher IC50 to MQ than was N86 plus Y184 (P = 0.111), although statistical significance was not reached. N86 plus Y184 (P = 0.038) and 86Y plus Y184 (P = 0.017) haplotypes were associated with decreased QN susceptibility in comparison to that of the 86Y plus 184F haplotype.
DISCUSSION
P. falciparum polymorphisms in pfcrt, pfmdr1, pfmrp1, dhfr, and dhps in conjunction with ex vivo drug susceptibility and with AL, ASAQ, and SP treatment effectiveness was assessed in southern Benin. Although AL was adopted in 2004 in Benin to replace CQ as first-line therapy for uncomplicated malaria, there was an insufficient supply and when the clinical trial was done, in 2007, AL still was not widely available (25). SP, previously used as a second-line treatment, was reserved for IPT of pregnant women. Resistance to CQ and SP in vivo was high in Benin previous to this trial (36). The history of drug usage and treatment efficacy is reflected in the parasite genetic diversity in the area. The CQ resistance-associated allele, pfcrt 76T (31), was close to fixation in the baseline parasite population, and pfmdr1 86Y, related to CQ resistance (37), was present at a high prevalence. The triple dhfr haplotype (108N, 51I, and 59R) and dhps 437G, associated with a higher risk of clinical SP failure (38), had increased from a previously high frequency (39) to 100% or almost 100% in this study. The extensive spread of SP resistance was confirmed by ex vivo susceptibility testing, in which the geometric mean IC50 of the tested isolates was considerably higher than that of 3D7. A vast majority of baseline isolates had IC50 estimates of LUM, MQ, and QN lower than those of 3D7, suggesting that these isolates are sensitive to the drugs. However, 36% of the isolates had higher DEAQ IC50 estimates than 3D7, which may represent a decreased sensitivity to DEAQ in these isolates. Decreased sensitivity to DEAQ may depend on the widespread resistance to CQ in the country, since a strong positive correlation has been shown between ex vivo susceptibilities to DEAQ and CQ (40, 41).
A majority of isolates had a DHA IC50 higher than that of 3D7. Current in vitro methodology was developed to assess the growth of long-acting drugs that are mainly effective on schizont stages and may not be adapted for the rapidly acting artemisinins that act on ring stages, as well as mature stages (42). A new method assessing in vitro susceptibility to artemisinins by pulsing ring stage parasites with DHA has demonstrated differences between isolates from regions with different levels of artemisinin resistance in Cambodia. The difference was not observed with the classical [3H]hypoxanthine uptake inhibition assay (43). As methods taking into account artemisinin-related phenomena such as parasite clearance (A. Ndour, presented at the ASTMH 60th annual meeting, Philadelphia, PA, 4 to 8 December 2011) and ring stage survival (43) are being evaluated, DHA susceptibility measured by classical in vitro methods, as in this study, is difficult to interpret. It would be of interest to follow up on changes in DHA susceptibility after the implementation of ACT.
Ex vivo susceptibilities to MQ and LUM were strongly and positively correlated, probably as a consequence of the association of pfmdr1 N86 with decreased susceptibility to both MQ and LUM. These results are consistent with previous studies showing the correlation of MQ and LUM ex vivo susceptibilities (44, 45) and reduced MQ and LUM susceptibility in isolates with pfmdr1 N86 (19, 46). Decreased susceptibility to LUM was also associated with pfmdr1 Y184, inconsistent with the selection of pfmdr1 184F after AL treatment (12, 14, 16, 47). This may be a consequence of the genetic linkage observed between the pfmdr1 alleles N86 and Y184 in this study. N86 was strongly associated with decreased susceptibility to LUM and is probably more important for the phenotype than the 184 genotype. This is supported by the haplotype analyses showing decreased LUM susceptibility of both N86 plus Y184 and N86 plus 184F haplotypes and no difference between the two haplotypes. The association between Y184 and decreased susceptibility to LUM may therefore be due to a higher proportion of N86 in isolates with Y184 than in those carrying 184F. Genetic linkage between N86 and Y184 warrants further investigation, as it may explain the regional differences in the selection of recurrent infections by AL treatment, where pfmdr1 N86 and Y184 were selected in Burkina Faso (23), while N86 and 184F have been selected in East Africa (12, 14, 16, 47).
In the ASAQ arm, reinfecting parasites carrying pfmdr1 86Y were selected. Reinfections represented the majority of the recurrent infections after ASAQ treatment. Subtherapeutic levels of long-half-life drugs can select for tolerant or resistant reinfecting parasites by suppressing the growth of sensitive parasites for weeks or months after treatment (48–50). DEAQ is likely to exert selection pressure on reinfections, as the metabolism of AQ to DEAQ is rapid and DEAQ has a longer half-life. Selection of pfmdr1 86Y by ASAQ has been observed in Mali in West Africa (21) and in East Africa (13–15). In East Africa, the pfmdr1 haplotype (86Y, Y184, and 1246Y) was observed to be selected. In our study, 1246Y was present at a low frequency in the baseline infections (11.8%) compared to populations in East Africa (22 to 83%) (13–15) and Y184 was associated with N86, explaining why this pfmdr1 haplotype was rare in Benin (9%) and was less likely to be selected posttreatment. The difference in frequency of 1246Y and the pfmdr1 haplotype between East and West Africa was further underlined in a study assessing the genetic diversity of resistance-associated polymorphisms in travelers (24), providing a rationale for the differences in selection patterns between regions.
After AL treatment, the opposite allele, pfmdr1 N86, was selected in recrudescent parasites. Selection of this allele after AL treatment has been observed in East (12, 14, 16, 17) and West (22, 23) Africa, mainly in reinfections. In this study, selection of reinfections was not observed. However, the pfmdr1 86 locus was still important for reinfections, since it was demonstrated that the reinfecting organisms carrying pfmdr1 N86 were exposed to significantly higher concentrations of LUM at day 3 and occurred earlier, suggesting that they could sustain higher LUM concentrations than parasites with 86Y. A limitation in this study was that only two reinfecting parasites carried N86. However, the importance of considering the pharmacology when studying parasite genetics in clinical trials is supported by a similar observation by Malmberg and colleagues that reinfecting parasites carrying N86 sustain 12-fold-higher LUM concentrations than 86Y parasites, made by a novel model based on day 7 concentrations (51). In this study, the clinical relevance of the pfmdr1 N86 allele in LUM was supported by its association with decreased ex vivo susceptibility to LUM, as also seen in Kenyan isolates (19).
The samples in this study were collected from an effectiveness trial, which may blur the selection analyses since all patients did not receive a complete treatment. Although adherence, based on drug blister recovery and drug level measurements, was suboptimal in some patients, there was no association with treatment outcome (25), so we consider that the recurrent infections were under drug pressure, resulting in the observed selection.
In transfection studies, pfmdr1 SNPs including 1246Y have been implicated in QN resistance (52). Here we show an association between pfmdr1 Y184 and 1246Y, respectively, and reduced ex vivo susceptibility to QN. Studies on the influence of pfmdr1 on quinine in vitro susceptibility have had inconsistent outcomes (53), maybe as a consequence of differences in the parasite genetic background. Further variation in susceptibility to QN may be explained by the Na+/H+ exchanger (pfnhe-1) that was demonstrated to be involved in QN resistance (54).
More reinfections were observed in the ASAQ arm than in the AL arm. A higher proportion of patients had undetectable levels of DEAQ than LUM on day 7 (tested in a subset of the patients), and it was suggested that the difference in the posttreatment prophylaxis between the two treatments could be due to fast elimination of AQ metabolites (25). An additional factor favoring reinfections following ASAQ treatment could be that the alleles selected by ASAQ (pfcrt 76T and pfmdr1 86Y) were present at high prevalence in the parasite population of the area, while the alleles that allow reinfections to survive AL treatment (pfcrt K76 and pfmdr1 N86) were present at low frequency at the baseline. Furthermore, a slight decrease in ex vivo susceptibility to DEAQ was observed, while the parasite population seems to be sensitive to LUM.
No association was observed between ex vivo susceptibility of pretreatment parasites to DHA, DEAQ, LUM, and PYR, at the baseline and outcome in the respective treatment arms. The number of patients in each treatment arm was low, especially in the SP arm, and drug sensitivity data were available for less than half of the patients, so the study may not have had enough power to detect ex vivo susceptibility differences between treatment successes and failures. This may account for the lack of an association. The relation between AQ in vivo efficacy and in vitro DEAQ susceptibility may be confounded by interindividual variations in pharmacokinetic factors, such as the metabolism of AQ to DEAQ by CYP2C8 (55, 56), and the complex dynamics between AQ, DEAQ, and possibly other metabolites (57). In vitro susceptibility to artemisinins and in vivo efficacy of artesunate have been associated (5), but inconsistently (4).
This study assessed parasite drug sensitivity in Benin before the introduction of ACTs as a first-line antimalarial treatment. It shows ex vivo susceptibility to LUM and a slight decrease in DEAQ susceptibility and a phenotypic and genetic parasite profile associated with chloroquine and/or SP resistance. Selection of specific alleles in recurrent infections after ACT treatment confirms the tolerance of certain parasite strains to the partner drugs, as in East Africa. The results from the molecular and in vitro analysis underline the importance of performing these studies in different regions because of regional parasite genetic differences between West and East Africa. Selection of opposite alleles among recurrent parasites following AL or ASAQ treatment emphasizes the benefits of the strategy of multiple first-line treatments (58) where drug pressure for both ACTs would be reduced because of inversed tolerance and resistance mechanisms. ACT including new partner drugs with potentially different mechanisms of resistance, such as piperaquine or pyronaridine, should be encouraged to increase the options. For a comprehensive picture of drug resistance emergence and spread, it is important to continue clinical, ex vivo, molecular, and pharmacological surveillance after ACT implementation.
ACKNOWLEDGMENT
This study was partly supported by Sanofi-Aventis.
Footnotes
Published ahead of print 7 October 2013
REFERENCES
- 1.WHO 2011. World malaria report. WHO, Geneva, Switzerland: http://www.who.int/malaria/world_malaria_report_2011/en/ [Google Scholar]
- 2.Snow RW, Trape JF, Marsh K. 2001. The past, present and future of childhood malaria mortality in Africa. Trends Parasitol. 17:593–597. 10.1016/S1471-4922(01)02031-1 [DOI] [PubMed] [Google Scholar]
- 3.Trape JF, Pison G, Preziosi MP, Enel C, Desgrees du Lou A, Delaunay V, Samb B, Lagarde E, Molez JF, Simondon F. 1998. Impact of chloroquine resistance on malaria mortality. C. R. Acad. Sci. III 321:689–697. 10.1016/S0764-4469(98)80009-7 [DOI] [PubMed] [Google Scholar]
- 4.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 361:455–467. 10.1056/NEJMoa0808859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noedl H, Se Y, Sriwichai S, Schaecher K, Teja-Isavadharm P, Smith B, Rutvisuttinunt W, Bethell D, Surasri S, Fukuda MM, Socheat D, Chan Thap L. 2010. Artemisinin resistance in Cambodia: a clinical trial designed to address an emerging problem in Southeast Asia. Clin. Infect. Dis. 51:e82–e89. 10.1086/657120 [DOI] [PubMed] [Google Scholar]
- 6.Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, ler Moo C, Al-Saai S, Dondorp AM, Lwin KM, Singhasivanon P, Day NP, White NJ, Anderson TJ, Nosten F. 2012. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet 379:1960–1966. 10.1016/S0140-6736(12)60484-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO 2010. Global report on antimalarial efficacy and drug resistance: 2000–2010 WHO, Geneva, Switzerland: http://www.who.int/malaria/publications/atoz/9789241500470/en/index.html [Google Scholar]
- 8.O'Brien C, Henrich PP, Passi N, Fidock DA. 2011. Recent clinical and molecular insights into emerging artemisinin resistance in Plasmodium falciparum. Curr. Opin. Infect. Dis. 24:570–577. 10.1097/QCO.0b013e32834cd3ed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thanh NV, Toan TQ, Cowman AF, Casey GJ, Phuc BQ, Tien NT, Hung NM, Biggs BA. 2010. Monitoring for Plasmodium falciparum drug resistance to artemisinin and artesunate in Binh Phuoc Province, Vietnam: 1998–2009. Malar. J. 9:181. 10.1186/1475-2875-9-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheeseman IH, Miller BA, Nair S, Nkhoma S, Tan A, Tan JC, Al Saai S, Phyo AP, Moo CL, Lwin KM, McGready R, Ashley E, Imwong M, Stepniewska K, Yi P, Dondorp AM, Mayxay M, Newton PN, White NJ, Nosten F, Ferdig MT, Anderson TJ. 2012. A major genome region underlying artemisinin resistance in malaria. Science 336:79–82. 10.1126/science.1215966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahlström S, Ferreira PE, Veiga MI, Sedighi N, Wiklund L, Martensson A, Farnert A, Sisowath C, Osorio L, Darban H, Andersson B, Kaneko A, Conseil G, Bjorkman A, Gil JP. 2009. Plasmodium falciparum multidrug resistance protein 1 and artemisinin-based combination therapy in Africa. J. Infect. Dis. 200:1456–1464. 10.1086/606009 [DOI] [PubMed] [Google Scholar]
- 12.Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ, Dorsey G. 2006. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob. Agents Chemother. 50:1893–1895. 10.1128/AAC.50.5.1893-1895.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmgren G, Hamrin J, Svard J, Martensson A, Gil JP, Bjorkman A. 2007. Selection of pfmdr1 mutations after amodiaquine monotherapy and amodiaquine plus artemisinin combination therapy in East Africa. Infect. Genet. Evol. 7:562–569. 10.1016/j.meegid.2007.03.005 [DOI] [PubMed] [Google Scholar]
- 14.Humphreys GS, Merinopoulos I, Ahmed J, Whitty CJ, Mutabingwa TK, Sutherland CJ, Hallett RL. 2007. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob. Agents Chemother. 51:991–997. 10.1128/AAC.00875-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nsobya SL, Dokomajilar C, Joloba M, Dorsey G, Rosenthal PJ. 2007. Resistance-mediating Plasmodium falciparum pfcrt and pfmdr1 alleles after treatment with artesunate-amodiaquine in Uganda. Antimicrob. Agents Chemother. 51:3023–3025. 10.1128/AAC.00012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sisowath C, Ferreira PE, Bustamante LY, Dahlstrom S, Martensson A, Bjorkman A, Krishna S, Gil JP. 2007. The role of pfmdr1 in Plasmodium falciparum tolerance to artemether-lumefantrine in Africa. Trop. Med. Int. Health 12:736–742. 10.1111/j.1365-3156.2007.01843.x [DOI] [PubMed] [Google Scholar]
- 17.Sisowath C, Petersen I, Veiga MI, Martensson A, Premji Z, Bjorkman A, Fidock DA, Gil JP. 2009. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J. Infect. Dis. 199:750–757. 10.1086/596738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sisowath C, Stromberg J, Martensson A, Msellem M, Obondo C, Bjorkman A, Gil JP. 2005. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem). J. Infect. Dis. 191:1014–1017. 10.1086/427997 [DOI] [PubMed] [Google Scholar]
- 19.Mwai L, Kiara SM, Abdirahman A, Pole L, Rippert A, Diriye A, Bull P, Marsh K, Borrmann S, Nzila A. 2009. In vitro activities of piperaquine, lumefantrine, and dihydroartemisinin in Kenyan Plasmodium falciparum isolates and polymorphisms in pfcrt and pfmdr1. Antimicrob. Agents Chemother. 53:5069–5073. 10.1128/AAC.00638-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nsobya SL, Kiggundu M, Nanyunja S, Joloba M, Greenhouse B, Rosenthal PJ. 2010. In vitro sensitivities of Plasmodium falciparum to different antimalarial drugs in Uganda. Antimicrob. Agents Chemother. 54:1200–1206. 10.1128/AAC.01412-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Djimdé AA, Fofana B, Sagara I, Sidibe B, Toure S, Dembele D, Dama S, Ouologuem D, Dicko A, Doumbo OK. 2008. Efficacy, safety, and selection of molecular markers of drug resistance by two ACTs in Mali. Am. J. Trop. Med. Hyg. 78:455–461 [PubMed] [Google Scholar]
- 22.Happi CT, Gbotosho GO, Folarin OA, Sowunmi A, Hudson T, O'Neil M, Milhous W, Wirth DF, Oduola AM. 2009. Selection of Plasmodium falciparum multidrug resistance gene 1 alleles in asexual stages and gametocytes by artemether-lumefantrine in Nigerian children with uncomplicated falciparum malaria. Antimicrob. Agents Chemother. 53:888–895. 10.1128/AAC.00968-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Somé AF, Sere YY, Dokomajilar C, Zongo I, Rouamba N, Greenhouse B, Ouedraogo JB, Rosenthal PJ. 2010. Selection of known Plasmodium falciparum resistance-mediating polymorphisms by artemether-lumefantrine and amodiaquine-sulfadoxine-pyrimethamine but not dihydroartemisinin-piperaquine in Burkina Faso. Antimicrob. Agents Chemother. 54:1949–1954. 10.1128/AAC.01413-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dippmann AK, Bienzle U, Harms G, Mockenhaupt FP. 2008. pfmdr1 mutations in imported African Plasmodium falciparum isolates. Trans. R. Soc. Trop. Med. Hyg. 102:1148–1150. 10.1016/j.trstmh.2008.04.001 [DOI] [PubMed] [Google Scholar]
- 25.Faucher JF, Aubouy A, Adeothy A, Cottrell G, Doritchamou J, Gourmel B, Houze P, Kossou H, Amedome H, Massougbodji A, Cot M, Deloron P. 2009. Comparison of sulfadoxine-pyrimethamine, unsupervised artemether-lumefantrine, and unsupervised artesunate-amodiaquine fixed-dose formulation for uncomplicated Plasmodium falciparum malaria in Benin: a randomized effectiveness noninferiority trial. J. Infect. Dis. 200:57–65. 10.1086/599378 [DOI] [PubMed] [Google Scholar]
- 26.Le Bras J, Deloron P. 1983. In vitro study of drug sensitivity of Plasmodium falciparum: evaluation of a new semi-micro test. Am. J. Trop. Med. Hyg. 32:447–451 [DOI] [PubMed] [Google Scholar]
- 27.Kaddouri H, Nakache S, Houze S, Mentre F, Le Bras J. 2006. Assessment of the drug susceptibility of Plasmodium falciparum clinical isolates from Africa by using a Plasmodium lactate dehydrogenase immunodetection assay and an inhibitory maximum effect model for precise measurement of the 50-percent inhibitory concentration. Antimicrob. Agents Chemother. 50:3343–3349. 10.1128/AAC.00367-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Nagard H, Vincent C, Mentre F, Le Bras J. 2011. Online analysis of in vitro resistance to antimalarial drugs through nonlinear regression. Comput. Methods Programs Biomed. 104:10–18. 10.1016/j.cmpb.2010.08.003 [DOI] [PubMed] [Google Scholar]
- 29.Dahlström S, Veiga MI, Ferreira P, Martensson A, Kaneko A, Andersson B, Bjorkman A, Gil JP. 2008. Diversity of the sarco/endoplasmic reticulum Ca(2+)-ATPase orthologue of Plasmodium falciparum (PfATP6). Infect. Genet. Evol. 8:340–345. 10.1016/j.meegid.2008.02.002 [DOI] [PubMed] [Google Scholar]
- 30.Dahlström S, Veiga MI, Martensson A, Bjorkman A, Gil JP. 2009. Polymorphism in PfMRP1 (Plasmodium falciparum multidrug resistance protein 1) amino acid 1466 associated with resistance to sulfadoxine-pyrimethamine treatment. Antimicrob. Agents Chemother. 53:2553–2556. 10.1128/AAC.00091-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, Coulibaly D, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 344:257–263. 10.1056/NEJM200101253440403 [DOI] [PubMed] [Google Scholar]
- 32.Duraisingh MT, Jones P, Sambou I, von Seidlein L, Pinder M, Warhurst DC. 2000. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol. Biochem. Parasitol. 108:13–23. 10.1016/S0166-6851(00)00201-2 [DOI] [PubMed] [Google Scholar]
- 33.Maïga O, Djimde AA, Hubert V, Renard E, Aubouy A, Kironde F, Nsimba B, Koram K, Doumbo OK, Le Bras J, Clain J. 2007. A shared Asian origin of the triple-mutant dhfr allele in Plasmodium falciparum from sites across Africa. J. Infect. Dis. 196:165–172. 10.1086/518512 [DOI] [PubMed] [Google Scholar]
- 34.Duraisingh MT, Curtis J, Warhurst DC. 1998. Plasmodium falciparum: detection of polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes by PCR and restriction digestion. Exp. Parasitol. 89:1–8. 10.1006/expr.1998.4274 [DOI] [PubMed] [Google Scholar]
- 35.Dahlstrom S. 2009. Role of PfATP6 and pfMRP1 in Plasmodium falciparum resistance to antimalarial drugs. Ph.D. thesis. Karolinska Institutet, Stockholm, Sweden [Google Scholar]
- 36.Aubouy A, Fievet N, Bertin G, Sagbo JC, Kossou H, Kinde-Gazard D, Kiniffo R, Massougbodji A, Deloron P. 2007. Dramatically decreased therapeutic efficacy of chloroquine and sulfadoxine-pyrimethamine, but not mefloquine, in southern Benin. Trop. Med. Int. Health 12:886–894. 10.1111/j.1365-3156.2007.01859.x [DOI] [PubMed] [Google Scholar]
- 37.Basco LK, Le Bras J, Rhoades Z, Wilson CM. 1995. Analysis of pfmdr1 and drug susceptibility in fresh isolates of Plasmodium falciparum from subsaharan Africa. Mol. Biochem. Parasitol. 74:157–166. 10.1016/0166-6851(95)02492-1 [DOI] [PubMed] [Google Scholar]
- 38.Gesase S, Gosling RD, Hashim R, Ord R, Naidoo I, Madebe R, Mosha JF, Joho A, Mandia V, Mrema H, Mapunda E, Savael Z, Lemnge M, Mosha FW, Greenwood B, Roper C, Chandramohan D. 2009. High resistance of Plasmodium falciparum to sulphadoxine/pyrimethamine in northern Tanzania and the emergence of dhps resistance mutation at codon 581. PLoS One 4:e4569. 10.1371/journal.pone.0004569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertin G, Briand V, Bonaventure D, Carrieu A, Massougbodji A, Cot M, Deloron P. 2011. Molecular markers of resistance to sulphadoxine-pyrimethamine during intermittent preventive treatment of pregnant women in Benin. Malar. J. 10:196. 10.1186/1475-2875-10-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basco LK, Le Bras J. 1993. In vitro activity of monodesethylamodiaquine and amopyroquine against African isolates and clones of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 48:120–125 [DOI] [PubMed] [Google Scholar]
- 41.Childs GE, Boudreau EF, Milhous WK, Wimonwattratee T, Pooyindee N, Pang L, Davidson DE., Jr 1989. A comparison of the in vitro activities of amodiaquine and desethylamodiaquine against isolates of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 40:7–11 [DOI] [PubMed] [Google Scholar]
- 42.Skinner TS, Manning LS, Johnston WA, Davis TM. 1996. In vitro stage-specific sensitivity of Plasmodium falciparum to quinine and artemisinin drugs. Int. J. Parasitol. 26:519–525. 10.1016/0020-7519(96)89380-5 [DOI] [PubMed] [Google Scholar]
- 43.Witkowski B, Khim N, Chim P, Kim S, Ke S, Kloeung N, Chy S, Duong S, Leang R, Ringwald P, Dondorp AM, Tripura R, Benoit-Vical F, Berry A, Gorgette O, Ariey F, Barale JC, Mercereau-Puijalon O, Menard D. 3 December 2012. Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in western Cambodia. Antimicrob. Agents Chemother.. 10.1128/AAC.01868-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basco LK, Bickii J, Ringwald P. 1998. In vitro activity of lumefantrine (benflumetol) against clinical isolates of Plasmodium falciparum in Yaounde, Cameroon. Antimicrob. Agents Chemother. 42:2347–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pradines B, Hovette P, Fusai T, Atanda HL, Baret E, Cheval P, Mosnier J, Callec A, Cren J, Amalvict R, Gardair JP, Rogier C. 2006. Prevalence of in vitro resistance to eleven standard or new antimalarial drugs among Plasmodium falciparum isolates from Pointe-Noire, Republic of the Congo. J. Clin. Microbiol. 44:2404–2408. 10.1128/JCM.00623-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duraisingh MT, Roper C, Walliker D, Warhurst DC. 2000. Increased sensitivity to the antimalarials mefloquine and artemisinin is conferred by mutations in the pfmdr1 gene of Plasmodium falciparum. Mol. Microbiol. 36:955–961. 10.1046/j.1365-2958.2000.01914.x [DOI] [PubMed] [Google Scholar]
- 47.Gadalla NB, Adam I, Elzaki SE, Bashir S, Mukhtar I, Oguike M, Gadalla A, Mansour F, Warhurst D, El-Sayed BB, Sutherland CJ. 2011. Increased pfmdr1 copy number and sequence polymorphisms in Plasmodium falciparum isolates from Sudanese malaria patients treated with artemether-lumefantrine. Antimicrob. Agents Chemother. 55:5408–5411. 10.1128/AAC.05102-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baliraine FN, Rosenthal PJ. 2011. Prolonged selection of pfmdr1 polymorphisms after treatment of falciparum malaria with artemether-lumefantrine in Uganda. J. Infect. Dis. 204:1120–1124. 10.1093/infdis/jir486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stepniewska K, White NJ. 2008. Pharmacokinetic determinants of the window of selection for antimalarial drug resistance. Antimicrob. Agents Chemother. 52:1589–1596. 10.1128/AAC.00903-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White N. 1999. Antimalarial drug resistance and combination chemotherapy. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354:739–749. 10.1098/rstb.1999.0426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malmberg M, Ferreira PE, Tarning J, Ursing J, Ngasala B, Björkman A, Mårtensson A, Gil JP. 5 December 2012. Plasmodium falciparum drug resistance phenotype as assessed by patient antimalarial drug levels and its association with pfmdr1 polymorphisms. J. Infect. Dis. 10.1093/infdis/jis747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. 2000. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403:906–909. 10.1038/35002615 [DOI] [PubMed] [Google Scholar]
- 53.Okombo J, Ohuma E, Picot S, Nzila A. 2011. Update on genetic markers of quinine resistance in Plasmodium falciparum. Mol. Biochem. Parasitol. 177:77–82. 10.1016/j.molbiopara.2011.01.012 [DOI] [PubMed] [Google Scholar]
- 54.Ferdig MT, Cooper RA, Mu J, Deng B, Joy DA, Su XZ, Wellems TE. 2004. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol. Microbiol. 52:985–997. 10.1111/j.1365-2958.2004.04035.x [DOI] [PubMed] [Google Scholar]
- 55.Dai D, Zeldin DC, Blaisdell JA, Chanas B, Coulter SJ, Ghanayem BI, Goldstein JA. 2001. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics 11:597–607. 10.1097/00008571-200110000-00006 [DOI] [PubMed] [Google Scholar]
- 56.Li XQ, Bjorkman A, Andersson TB, Ridderstrom M, Masimirembwa CM. 2002. Amodiaquine clearance and its metabolism to N-desethylamodiaquine is mediated by CYP2C8: a new high affinity and turnover enzyme-specific probe substrate. J. Pharmacol. Exp. Ther. 300:399–407. 10.1124/jpet.300.2.399 [DOI] [PubMed] [Google Scholar]
- 57.Aubouy A, Mayombo J, Keundjian A, Bakary M, Le Bras J, Deloron P. 2004. Short report: lack of prediction of amodiaquine efficacy in treating Plasmodium falciparum malaria by in vitro tests. Am. J. Trop. Med. Hyg. 71:294–296 [PubMed] [Google Scholar]
- 58.Smith DL, Klein EY, McKenzie FE, Laxminarayan R. 2010. Prospective strategies to delay the evolution of anti-malarial drug resistance: weighing the uncertainty. Malar. J. 9:217. 10.1186/1475-2875-9-217 [DOI] [PMC free article] [PubMed] [Google Scholar]