Abstract
Two carbapenem-resistant Raoultella planticola clinical isolates were isolated from patients with pneumonia and Port-A catheter-related bacteremia, respectively, in Taiwan. These isolates remained susceptible to fluoroquinolone, aminoglycoside, and colistin. Though the two isolates had the same antibiogram, plasmidic carbapenemase blaIMP-8, class 1 integron cassette (dfrA12-orfF-aadA2), and qnrB2, they had different pulsed-field gel electrophoresis patterns, plasmid sizes, and outer membrane protein loss profiles. To our knowledge, this is the first report of blaIMP-8 found in R. planticola. Interestingly, blaIMP-8 is the most common carbapenemase found in Klebsiella pneumoniae in Taiwan. In the literature, carbapenemase genes in R. planticola in each country were also found in carbapenem-resistant Enterobacteriaceae in the same country.
TEXT
Raoultella planticola belongs to the Enterobacteriaceae family, is related to Klebsiella spp., and is mostly found in soil and water (1). Although the reports of R. planticola infection are limited, this bacterium has been reported in bloodstream infection, surgical site infection, and cystitis (2–4). Bacteremia was the most common clinical manifestation in six of the 11 R. planticola infection cases in the literature (3). To date, there are isolates of Raoultella ornithinolytica and R. planticola which are resistant to carbapenem and carry different carbapenemases, including blaKPC, blaOXA-48, and blaOXA-162 genes (2, 5, 6). However, blaIMP-8-producing R. planticola has not been reported in the literature.
Among 411 carbapenem-resistant Enterobacteriaceae isolates collected from a nationwide surveillance study in Taiwan in 2012, two carbapenem-resistant R. planticola clinical isolates were identified from patients in the National Taiwan University Hospital. One was isolated from a sputum specimen; the other was from a blood specimen. The 16S rRNA gene sequencing confirmed identification of R. planticola initially with the Vitek 2 system (7).
Case 1.
A 77-year-old male patient was a case of non-small-cell lung cancer. He was admitted with pneumonia in February 2012. After admission, respiratory failure and shock developed. Chest roentgenography revealed bronchopneumonia in the right lower lung field. Sputum culture yielded carbapenem-resistant R. planticola (isolate 139), and the patient received levofloxacin and cefepime. Due to progressive leukocytosis, unstable hemodynamics, and increased O2 demand, meropenem and colistin were prescribed instead of cefepime. The patient eventually died of pneumonia and shock.
Case 2.
A 57-year-old male patient had non-small-cell lung cancer with bilateral mediastinal lymph nodes and multiorgan metastasis. He received therapy with erlotinib and cisplatin. Port-A catheter-related bacteremia was suspected, and blood cultures grew Acinetobacter baumannii and carbapenem-resistant R. planticola (isolate 193). The patient received ceftazidime, levofloxacin, and gentamicin, and then the follow-up blood cultures were negative sterile 5 days later.
These two isolates were regarded as health care-associated pathogens and had the same antibiogram, where both were susceptible to aztreonam, piperacillin-tazobactam, aminoglycosides, fluoroquinolones, colistin, and tigecycline but resistant to imipenem, doripenem, ertapenem, ticarcillin-clavulanic acid, and trimethoprim-sulfamethoxazole (Table 1). Pulsed-field gel electrophoresis (PFGE) analysis revealed that these two isolates had more than 6 bands of difference and were regarded as having different banding patterns (Fig. 1A). Detection of carbapenemases (blaKPC, blaNDM, blaVIM, blaIMP, blaNMC, blaSME, blaSPM-1, blaGIM-1, blaSIM-1, blaIMI, blaGES, and blaOXA-48) and extended-spectrum β-lactamases (ESBLs) and AmpC genes (blaSHV, blaTEM, blaDHA, blaCMY, blaCTX-M-G1, blaCTX-M-G2, and blaCTX-M-G9) was performed, and only blaIMP-8 was found in these two isolates (8, 9). Both isolates contained two plasmids (ca. 204 and 239 kb in isolate 139; ca. 207 and 249 kb in isolate 193, respectively) which were identified by S1-nuclease PFGE analysis and calculated by BioNumerics GelCompar software package (version 5.0; Applied Mathematics, Sint-Martens-Latem, Belgium) (Fig. 1B).
TABLE 1.
Characterization of blaIMP-8-carrying R. planticola clinical isolates
| Drug or resistance profile | MICa (μg/ml) or characteristic of R. planticola clinical isolate |
|
|---|---|---|
| Isolate 139 (case 1) | Isolate 193 (case 2) | |
| Carbapenems | ||
| Imipenem | 4 (R) | 4 (R) |
| Meropenem | 2 (I) | 2 (I) |
| Doripenem | >2 (R) | >2 (R) |
| Ertapenem | 1 (R) | 2 (R) |
| Cephems | ||
| Cefepime | 16 (I) | >16 (R) |
| Cefoxitin | >16 (R) | >16 (R) |
| Ceftazidime | >16 (R) | >16 (R) |
| Cefazolin | >16 (R) | >16 (R) |
| Cefuroxime | >16 (R) | >16 (R) |
| Cefotaxime | >32 (R) | >32 (R) |
| Aztreonam | <1 (S) | <1 (S) |
| Ticarcillin-clavulanic acid | >64 (R) | >64 (R) |
| Piperacillin-tazobactam | 16 (S) | 8 (S) |
| Trimethoprim-sulfamethoxazole | >2 (R) | >2 (R) |
| Tigecycline | 0.5 | 0.5 |
| Amikacin | <4 (S) | <4 (S) |
| Gentamicin | 2 (S) | 2 (S) |
| Colistin | <0.5 (S) | 1 (S) |
| Nalidixic acid | >8 | >8 |
| Ciprofloxacin | 0.5 (S) | 1 (S) |
| Levofloxacin | 1 (S) | 1 (S) |
| Resistance profiles | ||
| Outer membrane porins | OmpK35 (−), OmpK36 (−) | OmpK35 (−), OmpK36 (+) |
| Class I integron | dfrA12-orfF-aadA2 | dfrA12-orfF-aadA2 |
| PMQRb genes | qnrB2 | qnrB2 |
MICs were determined by the agar dilution method. R, resistant; S, susceptible; I, intermediate.
PMQR, plasmid-mediated quinolone resistance genes, including qnrA, qnrB, qnrS, qepA, aac(6′)-Ib-cr, armA, and rmtB.
FIG 1.
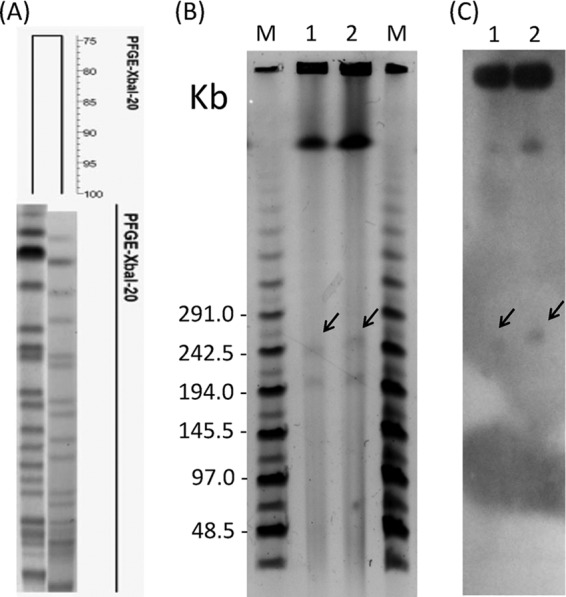
PFGE analysis of R. planticola. (A) XbaI-digested chromosome fragments were separated by PFGE, and the dendrogram was produced by BioNumerics software. (B) S1-nuclease-digested plasmid profiles separated by PFGE. (C) S1-nuclease-digested plasmid profiles hybridized with a blaIMP-8 probe. Lanes M, MidRange II PFG marker; lanes 1, isolate 139; lanes 2, isolate 193. The arrows show the locations of blaIMP-8 genes.
Although plasmid transfer assays included conjugation and electroporation, using Escherichia coli as the receptor (DH5α and J53) did not find the successful transformants. S1-nuclease PFGE combined with Southern blot hybridization showed that blaIMP-8 was located on the 239-kb plasmid of isolate 139 and the 249-kb plasmid of isolate 193 (Fig. 1C). This finding suggests that blaIMP-8-containing plasmids were likely to be nonconjugative, and blaIMP-8 was possibly acquired by mobile elements. Both isolates carried class 1 integron cassette arrays harboring dfrA12-orfF-aadA2 (1.8 kb). The dfrA12 gene, coding for dihydrofolate reductase, confers resistance to trimethoprim. The aadA2 gene, coding for aminoglycoside-3″-adenyltransferase, confers resistance to streptomycin and spectinomycin. This integron cassette array has been reported from different organisms and become globally disseminated (10). PCR and sequencing detection of plasmid-mediated quinolone resistance determinants, including qnrA, qnrB, qnrS, qepA, aac(6′)-Ib-cr, armA, and rmtB, found that both isolates contained only the qnrB2 gene (11). This gene conferred low-level resistance to all quinolones and was possibly transferred by a plasmid (12). SDS-PAGE analysis of outer membrane proteins found that loss of OmpK35 was found in isolate 193 and loss of OmpK35/36 was found in isolate 139. Previous study has shown that a double deletion of OmpK35 and OmpK36 reduced the susceptibilities of meropenem and cefepime in Klebsiella pneumoniae (13). This might be the reason why isolate 193 had a higher cefepime MIC than did isolate 139 (Table 1).
To our knowledge, this is the first report of blaIMP-8 in R. planticola. In the past decade, blaIMP-8-producing K. pneumoniae isolates have been reported in Taiwan (8, 9, 14). Previous studies have reported that carbapenemases (blaKPC, blaOXA-48, and blaOXA-162) could be found in R. planticola, R. ornithinolytica, and other Enterobacteriaceae (2, 5, 6, 9, 15). The carbapenemase genes in R. planticola in any specific country can be found in carbapenem-resistant Enterobacteriaceae, especially K. pneumoniae, in that country (Table 2). Our results identified a plasmid-located blaIMP-8 gene in R. planticola clinical isolates and suggested a possible association with blaIMP-8-harboring K. pneumoniae in Taiwan.
TABLE 2.
Correlation of carbapenem-resistant genes in Raoultella spp. and Enterobacteriaceae by country
| Report | Carbapenemase(s) | Microorganism(s) | Country |
|---|---|---|---|
| This study | blaIMP-8 | R. planticola | Taiwan |
| Ma et al. (9) | blaIMP-8 | K. pneumoniae | Taiwan |
| Castanheira et al. (2) | blaKPC-2, blaKPC-3 | R. planticola, R. ornithinolytica | United States |
| Chiang et al. (15) | blaKPC-2 | K. pneumoniae | United States |
| Österblad et al. (5) | blaOXA-48 | R. planticola, E. coli, K. pneumoniae | Finland |
| Pfeifer et al. (6) | blaOXA-162 | R. ornithinolytica, E. coli, Citrobacter freundii | Germany |
Footnotes
Published ahead of print 21 October 2013
REFERENCES
- 1.Bagley S, Seidler R, Brenner D. 1981. Klebsiella planticola sp. nov.: a new species of enterobacteriaceae found primarily in nonclinical environments. Curr. Microbiol. 6:105–109. 10.1007/BF01569013 [DOI] [Google Scholar]
- 2.Castanheira M, Deshpande LM, DiPersio JR, Kang J, Weinstein MP, Jones RN. 2009. First descriptions of blaKPC in Raoultella spp. (R. planticola and R. ornithinolytica): report from the SENTRY Antimicrobial Surveillance Program. J. Clin. Microbiol. 47:4129–4130. 10.1128/JCM.01502-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olson DS, Jr, Asare K, Lyons M, Hofinger DM. 2013. A novel case of Raoultella planticola urinary tract infection. Infection 41:259–261. 10.1007/s15010-012-0294-x [DOI] [PubMed] [Google Scholar]
- 4.Wolcott R, Dowd S. 2010. Molecular diagnosis of Raoultella planticola infection of a surgical site. J. Wound Care 19:329–332 [DOI] [PubMed] [Google Scholar]
- 5.Österblad M, Kirveskari J, Hakanen AJ, Tissari P, Vaara M, Jalava J. 2012. Carbapenemase-producing Enterobacteriaceae in Finland: the first years (2008–11). J. Antimicrob. Chemother. 67:2860–2864. 10.1093/jac/dks299 [DOI] [PubMed] [Google Scholar]
- 6.Pfeifer Y, Schlatterer K, Engelmann E, Schiller RA, Frangenberg HR, Stiewe D, Holfelder M, Witte W, Nordmann P, Poirel L. 2012. Emergence of OXA-48-type carbapenemase-producing Enterobacteriaceae in German hospitals. Antimicrob. Agents Chemother. 56:2125–2128. 10.1128/AAC.05315-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drancourt M, Bollet C, Carta A, Rousselier P. 2001. Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella gen. nov., with description of Raoultella ornithinolytica comb. nov., Raoultella terrigena comb. nov. and Raoultella planticola comb. nov. Int. J. Syst. Evol. Microbiol. 51:925–932. 10.1099/00207713-51-3-925 [DOI] [PubMed] [Google Scholar]
- 8.Lee CM, Liao CH, Lee WS, Liu YC, Mu JJ, Lee MC, Hsueh PR. 2012. Outbreak of Klebsiella pneumoniae carbapenemase-2-producing K. pneumoniae sequence type 11 in Taiwan in 2011. Antimicrob. Agents Chemother. 56:5016–5022. 10.1128/AAC.00878-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma L, Lu PL, Siu LK, Hsieh MH. 2013. Molecular typing and resistance mechanisms of imipenem-non-susceptible Klebsiella pneumoniae in Taiwan: results from the Taiwan surveillance of antibiotic resistance (TSAR) study, 2002–2009. J. Med. Microbiol. 62:101–107. 10.1099/jmm.0.050492-0 [DOI] [PubMed] [Google Scholar]
- 10.Gestal AM, Stokes HW, Partridge SR, Hall RM. 2005. Recombination between the dfrA12-orfF-aadA2 cassette array and an aadA1 gene cassette creates a hybrid cassette, aadA8b. Antimicrob. Agents Chemother. 49:4771–4774. 10.1128/AAC.49.11.4771-4774.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin CJ, Siu LK, Ma L, Chang YT, Lu PL. 2012. Molecular epidemiology of ciprofloxacin-resistant extended-spectrum β-lactamase-producing Klebsiella pneumoniae in Taiwan. Microb. Drug Resist. 18:52–58. 10.1089/mdr.2011.0060 [DOI] [PubMed] [Google Scholar]
- 12.Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, Hooper DC. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50:1178–1182. 10.1128/AAC.50.4.1178-1182.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai YK, Fung CP, Lin JC, Chen JH, Chang FY, Chen TL, Siu LK. 2011. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 55:1485–1493. 10.1128/AAC.01275-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan JJ, Ko WC, Wu JJ. 2001. Identification of a plasmid encoding SHV-12, TEM-1, and a variant of IMP-2 metallo-beta-lactamase, IMP-8, from a clinical isolate of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:2368–2371. 10.1128/AAC.45.8.2368-2371.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiang T, Mariano N, Urban C, Colon-Urban R, Grenner L, Eng RH, Huang D, Dholakia H, Rahal JJ. 2007. Identification of carbapenem-resistant Klebsiella pneumoniae harboring KPC enzymes in New Jersey. Microb. Drug Resist. 13:235–239. 10.1089/mdr.2007.767 [DOI] [PubMed] [Google Scholar]


