Abstract
Bacterial pathogens commonly associated with chronic periodontitis are the spirochete Treponema denticola and the Gram-negative, proteolytic species Porphyromonas gingivalis and Tannerella forsythia. These species rely on complex anaerobic respiration of amino acids, and the anthelmintic drug oxantel has been shown to inhibit fumarate reductase (Frd) activity in some pathogenic bacteria and inhibit P. gingivalis homotypic biofilm formation. Here, we demonstrate that oxantel inhibited P. gingivalis Frd activity with a 50% inhibitory concentration (IC50) of 2.2 μM and planktonic growth of T. forsythia with a MIC of 295 μM, but it had no effect on the growth of T. denticola. Oxantel treatment caused the downregulation of six P. gingivalis gene products and the upregulation of 22 gene products. All of these genes are part of a regulon controlled by heme availability. There was no large-scale change in the expression of genes encoding metabolic enzymes, indicating that P. gingivalis may be unable to overcome Frd inhibition. Oxantel disrupted the development of polymicrobial biofilms composed of P. gingivalis, T. forsythia, and T. denticola in a concentration-dependent manner. In these biofilms, all three species were inhibited to a similar degree, demonstrating the synergistic nature of biofilm formation by these species and the dependence of T. denticola on the other two species. In a murine alveolar bone loss model of periodontitis oxantel addition to the drinking water of P. gingivalis-infected mice reduced bone loss to the same level as the uninfected control.
INTRODUCTION
Periodontal diseases range from the relatively mild form, gingivitis, to the more aggressive forms, periodontitis, which are characterized by the destruction of the tooth's supporting structures that can lead to tooth loss. The most common form of periodontitis is chronic periodontitis. Chronic periodontitis is a major public health problem in all societies and is estimated to affect around 30 to 47% of the adult population, with severe forms affecting 5 to 10% (1, 2).
Over the last decade, there has been a dramatic increase in the number of studies describing relationships between chronic periodontitis and systemic diseases. Epidemiological surveys have shown that clinical indicators of periodontal disease such as tooth loss and bleeding gums are associated with a greater risk of certain cancers and systemic diseases and disorders such as cardiovascular disease as well as preterm and underweight birth (3, 4). Clinical markers of periodontitis have recently been associated with an increased risk of cancer of the head, neck, and esophagus (5), tongue (6), pancreas (7–9), and also inflammation in solid organ transplant recipients (10).
The bacterial etiology of chronic periodontitis is acknowledged to be polymicrobial in nature, and while the concepts of the roles of particular oral bacterial species in disease have changed over the past 2 decades, there is wide consensus that anaerobic, proteolytic, amino-acid-fermenting species, including Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia, play a crucial role (11–13). Based on animal model data, P. gingivalis has recently been proposed to be a “keystone pathogen” that manipulates the host response to favor proliferation of other oral bacterial species, which then results in disease progression (12). This proposal has more recently been modified to emphasize the importance of synergistic interactions between potentially pathogenic bacterial species that result in the formation of a pathogenic polymicrobial plaque that is composed of one or more primary and a number of accessory bacterial pathogens expressing community virulence factors that elicit a nonresolving and tissue-destructive host response (12). We have previously demonstrated in a longitudinal human study that the imminent progression of chronic periodontitis could be predicted by increases in the relative proportions of P. gingivalis and T. denticola in subgingival plaque (14), which is consistent with other clinical studies demonstrating that the proportion of P. gingivalis in the subgingival plaque bacterial load is predictive of human disease progression (15, 16). When coinoculated in small animal models of periodontitis, P. gingivalis and T. denticola exhibit a synergistic virulence (17–19). These species also display synergistic biofilm formation (20–22) and respond to each other's presence by modulating the abundance of a range of proteins (22).
The surface of the tooth is a unique microbial habitat in the human body, as it is the only hard, permanent, nonshedding surface of the orogastrointestinal tract. This allows the accretion of a substantial bacterial biofilm over a lengthy period, as opposed to mucosal surfaces, where epithelial cell shedding limits development of the biofilm. Bacterial biofilms are defined as matrix-enclosed bacterial populations that are adherent to each other and/or to surfaces or interfaces (23). Sessile bacterial cells such as P. gingivalis can release antigens, toxins, endotoxin, and hydrolytic enzymes, such as proteinases as well as hemagglutinins—either directly into the surrounding milieu or associated with outer membrane vesicles that stimulate an inflammatory immune response. However, the host response is not very effective at killing bacteria within biofilms, and a chronic response can cause damage to host tissues. The presence of biofilms therefore often complicates treatment of chronic infections, including periodontitis, by protecting bacteria from the immune system, decreasing antibiotic/antimicrobial efficacy, and by allowing dispersal of planktonic cells to distant sites spreading the infection (24, 25).
Fumarate respiration is the most widespread type of anaerobic respiration, and fumarate is the only metabolic intermediate known to serve as an electron acceptor, yielding ∼0.5 ATP/electron to form succinate as the end product (26). The P. gingivalis fumarate reductase (Frd) is a trimeric enzyme complex that belongs to the succinate:quinone oxidoreductase (SQOR) family.
Anthelmintics such as oxantel, pyrantel, thiabendazole, and morantel have been used effectively since the 1970s to eradicate intestinal parasites, such as the whipworm, Trichocephalus trichiurus, in animals and humans (27–29). The main target of some of the anthelmintics appears to be the Frd complex of parasites that rely on fumarate as the terminal electron acceptor in the regeneration of NAD+, although other effects have also been reported (30–34).
We have previously shown that the anthelmintic oxantel not only kills P. gingivalis but at sublethal concentrations causes it to disperse from biofilms (35). To cause disease, P. gingivalis must grow as part of a polymicrobial subgingival biofilm, so the use of oxantel to remove or exclude P. gingivalis and other periodontal pathogens from the biofilm may help to prevent development of chronic periodontitis. In this study, we determined the effect of oxantel on other periodontal pathogens, characterized the P. gingivalis transcriptome in the presence of oxantel, investigated the effect of oxantel on polymicrobial biofilm development, and determined its efficacy in an animal model of disease. Collectively, these data indicate that oxantel may have potential for the treatment of human chronic periodontitis.
MATERIALS AND METHODS
Bacterial strains and growth media.
Porphyromonas gingivalis W50, Treponema denticola ATCC 35405, and Tannerella forsythia ATCC 43037 were obtained from the culture collection of the Oral Health CRC, The University of Melbourne. The bacteria were maintained in an anaerobic workstation (MG500; Don Whitley Scientific) at 37°C. Planktonic P. gingivalis cultures were routinely grown in brain heart infusion (BHI) medium containing 37 g/liter brain heart infusion (Oxoid), l-cysteine hydrochloride (5.0 mg/ml), hemin (5.0 μg/ml), and vitamin K (5.0 μg/ml). T. forsythia was cultured in Trypticase soy (15 g/liter), BHI (18.5 g/liter) supplemented with yeast extract (10 g/liter), hemin (5 mg/liter), menadione (0.4 mg/liter), N-acetyl muramic acid (10 mg/liter), cysteine (0.5 g/liter), and fetal bovine serum (5% [vol/vol]). T. denticola was grown in oral bacterium growth medium (OBGM), a modified and adapted version of new oral spirochete medium (NOS) containing N-acetyl muramic acid (10 mg/liter) (36) and GM-1 (17, 19, 37). Growth of bacterial cultures was monitored by measuring absorbance at a wavelength of 650 nm (AU650), and cells were harvested by centrifugation. Culture purity was routinely checked by Gram staining and colony morphology.
Fumarate reductase assay.
P. gingivalis W50 crude lysates were prepared by repeated passage through a French pressure cell at 138 MPa, as previously described (38). Benzyl viologen-linked reductase assays were carried out with crude cell lysates in a 1-ml assay. The reaction mixture contained 10 mM Tris-HCl buffer (pH 7.5) and a 0.1 mM concentration of the electron donor benzyl viologen (Sigma). Following the addition of bacterial lysate, 20 mM sodium dithionite (Merck) was added to the cuvette to achieve an absorbance of between 1.0 and 1.1 at 585 nm. The assay was initiated by the addition of a 5 mM concentration of the electron acceptor sodium fumarate (Sigma), and the oxidation of benzyl viologen (extinction coefficient 8.65 cm−1 mM−1) was spectrophotometrically measured at 585 nm. Oxantel pamoate (Sigma) was added to the assay at a range of concentrations up to 5 μM. All reagents were maintained anaerobically. Frd activity was expressed as nmol of reduced benzyl viologen oxidized min−1 mg−1 of protein.
MICs.
T. forsythia and T. denticola were resuspended in fresh growth medium to give a final cell density of 2.5 × 107 cells/ml. Oxantel pamoate (Sigma) was dissolved in dimethyl sulfoxide (DMSO) to achieve stock concentrations of up to 250 mM, and the MIC of oxantel was determined essentially as described previously (35). Cell density was monitored over a period of 110 h by measuring the absorbance of the culture at 650 nm (A650) using a UV-visible spectrophotometer (Varian). The MIC was calculated by linear regression of the growth data after 50 h of incubation with increasing concentrations of the inhibitor (39).
Polymicrobial biofilm assay.
Cultures of T. forsythia, T. denticola, and P. gingivalis W50 were individually grown in OBGM, which was prereduced under anaerobic conditions for more than 24 h before use. The cultures were grown under anaerobic conditions at 37°C until they reached A650 values of 0.6 for T. forsythia and P. gingivalis and 0.15 for T. denticola. Under these conditions, their viability was ∼90%, as determined by flow cytometry (described below).
For the polymicrobial biofilms, individual T. denticola, T. forsythia, and P. gingivalis planktonic cultures were diluted with fresh OBGM and combined in equal volumes to give a total cell density equivalent to an A650 of 0.15. This was immediately used as the inoculum for the polymicrobial biofilm assay. Two milliliters of each of the combined cultures was aliquoted into each well of a 2-cm-diameter 12-well flat-well plate (Nunc). A stock solution of 250 mM oxantel pamoate dissolved in DMSO or DMSO alone to a volume of 2 μl was added to achieve final concentrations of 0, 125, and 250 μM oxantel, and the plates were incubated at 37°C anaerobically for 48 h. After incubation, the medium in the well was decanted. Each well was then gently rinsed with 1 ml of OBGM to remove loosely attached and planktonic cells. The biofilm was then mechanically removed into 1 ml of OBGM, and the bacterial cells were enumerated by flow cytometry and real-time PCR (described below).
Flow cytometry.
Bacterial cells were enumerated, and viability was determined by flow cytometery on a Cell Lab Quanta (Beckman Coulter) using the LIVE/DEAD BacLight bacterial viability and counting kit (Invitrogen) according to the manufacturer's instructions. Bacterial cell suspensions were diluted in saline, and 20-μl aliquots were mixed with 180 μl cytometry medium containing 1 μl/ml Syto9 and 1 μl/ml propidium iodide in saline and applied to the flow cytometer.
Real-time PCR.
DNA from 0.5 ml of each biofilm sample was extracted using the PowerBiofilm DNA isolation kit (Mo Bio Laboratories) following the manufacturer's instructions. Real-time PCR was performed as previously described by Byrne et al. (14). Briefly, all oligonucleotide primers targeted the 16S rRNA gene (Table 1). Real-time PCRs, previously optimized for the Corbett Rotor-Gene (Qiagen), were carried out in triplicate, in a 25-μl reaction volume consisting of 12.5 μl Platinum SYBR green quantitative PCR (qPCR) Supermix UDG (Invitrogen), 9.5 μl DNase-free deionized water, a 200 nM final concentration of forward and reverse primers, and 2 μl template. Real-time PCR conditions for all primer pairs consisted of an initial heating step at 50°C for 2 min and initial denaturation at 95°C for 2 min, followed by 35 cycles of denaturation at 95°C for 15 s, annealing at 58°C for 30 s, and extension at 72°C for 30 s. Fluorescence data were collected immediately following the extension step of each cycle. Specificity of the primer pairs was confirmed by melt curve analysis by heating from 72°C to 95°C in 0.2°C increments according to the manufacturer's instructions.
TABLE 1.
Sequence of the 16S rRNA oligonucleotide primers used for real-time PCR enumeration of bacteria in the polymicrobial biofilms
| Species | Primer |
Product size (bp) | GenBank accession no. | Reference | |
|---|---|---|---|---|---|
| Forward | Reverse | ||||
| P. gingivalis | AGGCAGCTTGCCATACTGCG | ACTGTTAGTAACTACCGATGT | 404 | AB035456 | 14 |
| T. denticola | TAATACCGAATGTGCTCATTTACAT | TCAAAGAAGCATTCCCTCTTCTTCTTA | 316 | AF139203 | 40 |
| T. forsythia | AAAACAGGGGTTCCGCATGG | TTCACCGCGGACTTAACAGC | 426 | AB035460 | 41 |
Tenfold serial dilutions of DNA of known concentration as determined using a Nanodrop fluorospectrometer ND 1000 (Thermo Scientific) were used to construct standard curves for quantification of each species. P. gingivalis has four copies of the 16S rRNA gene per genome, while T. denticola and T. forsythia each have two. The total cell number and percentage of each bacterial species were then calculated for each biofilm sample.
Continuous culture and oxantel treatment of P. gingivalis.
P. gingivalis W50 was grown in continuous culture using a Bioflo 110 fermentor/bioreactor (New Brunswick Scientific) with an 800-ml working volume essentially as described previously (38). The growth medium was a dilute modified heart infusion broth (9.25 g/liter heart infusion medium [Oxoid], supplemented with 0.5 g/liter cysteine hydrochloride, 0.25 mg/liter vitamin K, 5 g/liter NaCl, 2.5 g/liter Na2HPO4, 1.25 mg/liter hemin, and 5 mg/liter resazurin). The dilution rate was 0.1 h−1, resulting in a mean generation time of 6.9 h. The temperature of the vessel was maintained at 37°C, and the culture was continuously gassed with 5% CO2 in 95% N2. Cultures reached and maintained a stable cell density of ∼6 × 108 CFU/ml, as determined by measuring the optical density at a wavelength of 600 nm. Oxantel treatment consisted of initially removing samples from the bioreactor for time zero (untreated controls). This was followed by the addition of powdered oxantel pamoate directly to the bioreactor and the medium supply reservoir to a final concentration of 125 μM. After the addition of the oxantel pamoate, cells were still under conditions of continuous culture. Samples were taken at 30-min intervals for 2 h. Separate bioreactors were used for individual experiments to give four biological replicates.
Extraction of RNA for transcriptomic analyses.
Extraction of total RNA was performed as previously described (38).
Microarray hybridization and analyses.
P. gingivalis W83 microarray slides, version 1, were obtained from the Pathogen Functional Genomics Resource Centre of the J. Craig Venter Institute. P. gingivalis W83 microarrays have previously been shown to be suitable to determine the effects of environmental conditions on the P. gingivalis W50 transcriptome due to the nearly identical genome sequences of the two strains (42). cDNA synthesis, labeling, and microarray hybridization were all performed as previously described, except that 5 μg total RNA was reverse transcribed instead of 10 μg (38). Paired samples were compared on a single microarray using a two-color system. Time zero control samples were paired with each of the 30-, 60-, 90-, and 120-min samples to give a total of 4 paired microarray hybridizations for each biological replicate (16 microarrays in total). A balanced dye design was used, with the analysis for each biological replicate, including two microarrays in which P. gingivalis W50 control samples were labeled with Cy3 and the paired oxantel-treated samples were labeled with Cy5, and two other microarrays where samples were labeled with the opposite combination of fluorophores. Image analysis was also performed as previously described, except that print tip loess normalization was used (38).
Therapeutic murine alveolar bone loss model.
BALB/c mice were obtained from the animal facility of the Melbourne Dental School at The University of Melbourne, and animal experimentation was approved by the University of Melbourne Animal Ethics Committee. Six groups of 6- to 8-week-old mice (12 animals per group) were housed in microisolators and given kanamycin (Sigma-Aldrich) at 0.34 mg/ml in deionized drinking water ad libitum for 7 days to suppress the oral microbiota. After a 3-day wash out period with no kanamycin in the drinking water, three groups were intraorally inoculated with 1 × 1010 viable cells of P. gingivalis in 5% carboxymethyl cellulose (CMC) four times over a 7-day period, while the other three groups received CMC alone (19). Three days after the last inoculation, oxantel pamoate (0.50 mg/ml [830 μM]) or amoxicillin (0.50 mg/ml) was added to the drinking water of two of the inoculated groups and two of the uninoculated groups, while the third group had no additions to the drinking water. A second round of P. gingivalis inoculation occurred 2 weeks after the first inoculation. After 6 weeks, the mice were euthanized, the right-half maxillae were defleshed, and the periodontal bone level was assessed by computer-assisted image analysis, by determining the mean area in mm2 from the cementoenamel junction to the alveolar bone crest of the buccal aspect of each molar, essentially as previously described (43).
RESULTS
Fumarate reductase activity.
Cellular extracts of P. gingivalis oxidized benzyl viologen upon the addition of 5 mM fumarate at a rate of 398.3 ± 94.4 nmol min−1 mg−1 protein. Addition of oxantel pamoate resulted in a dose-dependent reduction in Frd activity with an IC50 of 2.2 μM (Table 2).
TABLE 2.
Inhibition of P. gingivalis fumarate reductase by oxantela
| Fumarate reductase activity (nmol/min/mg)b | Oxantel concn (μM) | Inhibition (%) |
|---|---|---|
| 398.3 ± 94.4 | 0–0.5 | 0 |
| 282.8 ± 84.0 | 1.0 | 24 |
| 185.7 ± 139.7 | 2.5 | 50 |
| 27.6 ± 6.5 | 5.0 | 100 |
P. gingivalis cytoplasmic extracts were incubated with fumarate and a range of oxantel concentations. Absorbance at 585 nm was monitored, and all assays contained 0.1 mM benzyl viologen. The negative control was P. gingivalis cytoplasmic extract with no fumarate or oxantel. The rate obtained with 5 μM oxantel was not significantly different from the rate obtained without substrate (fumarate) or enzyme. A plot of inhibition versus log10 oxantel concentration gave an IC50 of 2.2 μM (R = 0.9925).
Mean ± standard deviation (n = 3 to 5).
Effect of oxantel on planktonic growth.
Oxantel had negligible effects on the growth of T. denticola at concentrations of up to 1 mM (Table 3). The MIC of oxantel against T. forsythia was calculated to be 295 μM (R2 = 0.9342). The MIC of oxantel against P. gingivalis W50 has previously been shown to be 112 μM (35). Oxantel addition at the sub-MICs of 31.25 and 62.5 μM substantially slowed T. forsythia growth in a similar manner to its effect on P. gingivalis growth (35). The effect of DMSO at the concentrations used in this assay on T. forsythia and T. denticola planktonic growth was negligible.
TABLE 3.
Effect of oxantel on planktonic growth of T. forsythia and T. denticola
| Species | MIC (μM) | Mean generation time (h−1) with: |
||
|---|---|---|---|---|
| DMSO only | Oxantel |
|||
| 31.25 μM | 62.5 μM | |||
| T. forsythia | 295 | 20 | 28 | 35 |
| T. denticola | >1,000a | 33 | 27 | 28 |
The highest concentration tested.
The effect of oxantel on the P. gingivalis transcriptome.
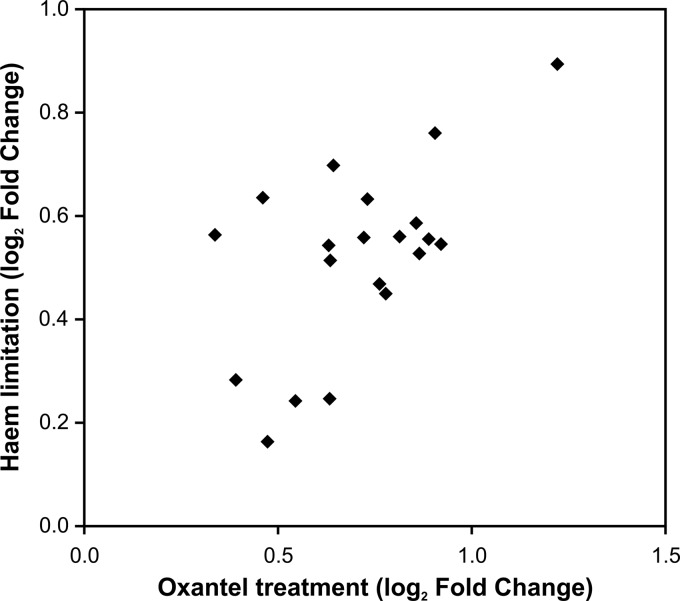
To determine the effect of sub-MIC oxantel treatment on P. gingivalis gene expression, the bacterium was grown in continuous culture in heme excess until steady state was achieved. The expression of 22 genes was significantly upregulated, with a 1.4-fold or higher change for at least one of the four time points in the 2 h after addition of 125 μM oxtanel pamoate to P. gingivalis growing in continuous culture at a cell density of ∼6 × 108 cells/ml. Sixteen of these 22 genes were in six predicted operons, and 13 of these genes are predicted to encode hypothetical proteins with no known function. The expression of six genes was significantly downregulated after oxantel treatment using the same criteria (Table 4). Compared with the heme regulon of P. gingivalis, the 22 genes upregulated after oxantel treatment were found to be largely a subset of the 152 genes whose expression was upregulated during heme limitation (38). Only two genes (PG1314 and PG0218) that were upregulated after oxantel treatment were not detected to be significantly upregulated during heme limitation. PG1314 is in a three-gene operon (PG1314 to -6), and PG1315 expression was significantly upregulated under heme-limited conditions. Similarly, PG0218 is part of a four-gene operon (PG0214 to -8), and the other three genes in the operon were consistently upregulated under heme limitation. The expression of both of PG1314 and PG0218 was higher under heme limitation, but neither met the cutoff criteria for significance. A comparison of the upregulated genes indicated a significant correlation between the effect of oxantel and heme limitation on gene expression, (y = 0.507x + 0.162; R = 0.5958, P < 0.05) (Fig. 1).
TABLE 4.
Effect of oxantel pamoate treatment on the transcriptome of P. gingivalis growing in continuous culturea
| Gene designation no.b | Log2 fold change at: |
Adjusted P value | Gene name | Product descriptionc | |||
|---|---|---|---|---|---|---|---|
| 30 min | 60 min | 90 min | 120 min | ||||
| Upregulated genes | |||||||
| PG0192 | 0.38 | 0.40 | 0.58 | 0.64 | 4.44E−12 | ompH-1 | Cationic outer membrane protein OmpH |
| PG0193 | 0.40 | 0.45 | 0.68 | 0.86 | 1.42E−22 | ompH-2 | Cationic outer membrane protein OmpH |
| PG0214 | 0.36 | 0.35 | 0.31 | 0.63 | 3.24E−04 | RNA polymerase σ70 factor, ECF subfamily | |
| PG0215 | 0.37 | 0.51 | 0.52 | 0.63 | 9.50E−07 | Hypothetical protein | |
| PG0217 | 0.59 | 0.66 | 0.76 | 0.89 | 1.51E−09 | Hypothetical protein | |
| PG0218 | 0.79 | 1.10 | 1.15 | 1.22 | 2.00E−12 | Hypothetical protein | |
| PG0419 | 0.42 | 0.85 | 0.98 | 0.92 | 1.89E−24 | Hypothetical protein | |
| PG0593 | 0.43 | 0.69 | 0.87 | 0.90 | 2.84E−18 | htrA | HtrA protein |
| PG0613 | 0.29 | 0.27 | 0.57 | 0.34 | 1.90E−05 | Hypothetical protein | |
| PG0614 | 0.30 | 0.55 | 0.45 | 0.86 | 9.37E-08 | Hypothetical protein | |
| PG0927 | 0.23 | 0.63 | 0.36 | 0.54 | 4.52E−03 | Hypothetical protein | |
| PG0985 | 0.20 | 0.25 | 0.36 | 0.72 | 9.22E−05 | RNA polymerase σ70 factor, ECF subfamily | |
| PG0986 | 0.42 | 0.51 | 0.78 | 0.76 | 7.86E−16 | Hypothetical protein | |
| PG0987 | 0.48 | 0.68 | 1.00 | 1.22 | 1.22E−22 | Hypothetical protein | |
| PG1314 | 0.14 | 0.49 | 0.63 | 0.51 | 9.26E−06 | aroC | Chorismate synthase (EC 4.2.3.5) (5-enolpyruvylshikimate-3-phosphate phospholyase) |
| PG1315 | 0.39 | 0.46 | 0.68 | 0.81 | 4.33E−13 | slyD | Peptidyl-prolyl cis-trans isomerase SlyD, FKBP-type |
| PG1316 | 0.38 | 0.61 | 0.55 | 0.63 | 1.42E−22 | Hypothetical protein | |
| PG1579 | 0.30 | 0.51 | 0.58 | 0.39 | 1.81E−05 | ATPase, MoxR family | |
| PG1584 | 0.21 | 0.46 | 0.83 | 0.47 | 7.92E−10 | batC | BatC protein |
| PG1625 | 0.37 | 0.48 | 0.68 | 0.78 | 3.18E−15 | Hypothetical protein | |
| PG1626 | 0.28 | 0.46 | 0.63 | 0.73 | 1.02E−18 | Hypothetical protein | |
| PG1715 | 0.27 | 0.57 | 0.38 | 0.46 | 5.45E−08 | Hypothetical protein | |
| Downregulated genes | |||||||
| PG0045 | −0.33 | −0.48 | −0.64 | −0.56 | 1.33E−04 | htpG | Chaperone protein HtpG (heat shock protein HtpG [high-temp protein G]) |
| PG0520 | −0.23 | −0.40 | −0.41 | −0.60 | 1.00E−08 | groEL | 60-kDa chaperonin (protein Cpn60; GroEL protein) |
| PG0707 | −0.38 | −0.30 | −0.56 | −0.33 | 2.06E−05 | Hypothetical TonB-linked outer membrane receptor PG50 (TonB-dependent receptor, putative) | |
| PG1129 | −0.30 | −0.60 | −0.85 | −0.77 | 1.44E−06 | nrd | Ribonucleotide reductase |
| PG1208 | −0.33 | −0.40 | −0.41 | −0.57 | 7.92E−10 | dnaK | Chaperone protein DnaK (heat shock protein 70; heat shock 70-kDa protein [HSP70]) |
| PG1844 | −0.22 | −0.48 | −0.38 | −0.60 | 1.58E−05 | hagD | Hemagglutinin protein HagD |
Gene transcription was considered to be significantly regulated if there was a change of 1.4-fold and a P value of < 0.05.
Gene designations that are underlined or in boldface represent genes predicted to be part of the same operon.
ECF, extracytoplasmic function; FKBP, FK506-binding protein.
FIG 1.
Correlation of fold changes for the genes upregulated by oxantel treatment and heme limitation. Both heme limitation and oxantel treatment were achieved by using P. gingivalis cells growing in anaerobic, planktonic continuous culture at identical growth rates. The symbols represent the genes shown as upregulated in Table 4.
The effect of oxantel on polymicrobial biofilm formation.
Polymicrobial biofilms composed of P. gingivalis, T. forsythia, and T. denticola were cultured in a static well assay in OBGM, a growth medium we have developed for the culture of these three bacterial species (19, 22). The starting inoculum for each biofilm in the 2-ml well was composed of an average of 6.9 × 106 P. gingivalis cells, 6.7 × 106 T. forsythia cells, or 8.8 × 106 T. denticola cells, with a combined viability of >97%, as determined by flow cytometry.
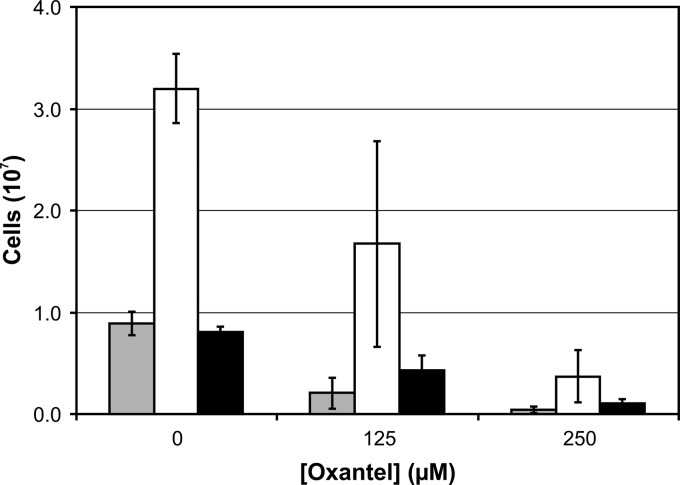
The biofilm model was initially used to culture T. denticola, P. gingivalis W50, and T. forsythia individually as single-species biofilms (homotypic) in OBGM. Forty-eight hours after inoculation, there were (1.11 ± 0.18) × 107 P. gingivalis cells, (1.40 ± 0.51) × 106 T. forsythia cells, and (2.08 ± 1.56) × 105 T. denticola cells in these homotypic biofilms. In the polymicrobial biofilm model containing all three species, there was a total of (4.87 ± 0.51) × 107 cells (Fig. 2), a 4-fold increase compared with the sum of the bacterial cells in the three individual homotypic biofilms. Treatment of the homotypic biofilms with 125 μM oxantel resulted in significant decreases in P. gingivalis ([8.64 ± 0.85] × 106) and T. forsythia ([3.21 ± 0.62] × 105) cell numbers but had no significant effect on T. denticola cell numbers ([1.58 ± 0.14] × 105).
FIG 2.
Effect of oxantel on polymicrobial biofilm development. Polymicrobial biofilms composed of P. gingivalis (gray bars), T. denticola (white bars), and T. forsythia (black bars) were cultured in OBGM in 12-well flat-well plates for 48 h in the presence or absence of oxantel. The polymicrobial biofilm was removed from the substratum after being washed and resuspended in OBGM prior to bacterial cell numbers being determined by real-time PCR. The data points represent the mean and standard deviation of five replicates. A one-way ANOVA with Dunnett's test showed that the numbers of bacterial cells for each species were significantly different (P < 0.05) between the three oxantel concentrations.
The polymicrobial biofilms in this static model biofilm system were dominated by T. denticola, which accounted for 65.7% of the total bacterial cells in the biofilm, as determined by real-time PCR analysis. P. gingivalis (17.7%) and T. forsythia (16.6%) were present in approximately equal proportions in the biofilm (Fig. 2). Incorporation of oxantel pamoate into the growth medium at concentrations of 125 and 250 μM resulted in a significant decrease in the numbers of all three species in the biofilm, with total cell numbers decreasing from 4.87 × 107 to 2.31 × 107 to 0.52 × 107, respectively. The relative proportions of the three species in the biofilms remained relatively stable with increasing oxantel concentrations, with T. denticola making up 72.3% and 71.2% of the total cells at 125 and 250 μM concentrations, respectively. There was a decrease in the relative proportion of P. gingivalis in the biofilm, which declined from 17.7% of the total cell number to 9.1% and 9.6% at oxantel concentrations of 125 and 250 μM, respectively (Fig. 2).
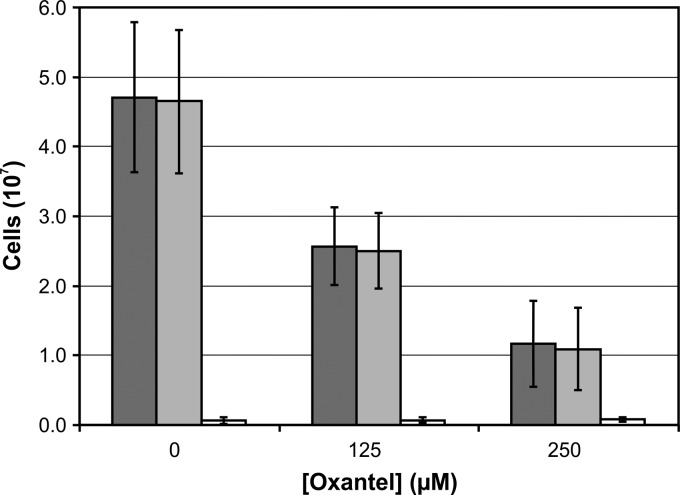
The incorporation of oxantel pamoate into the growth medium did not result in a significant increase in the number of dead bacterial cells in the polymicrobial biofilms, as determined by flow cytometric analysis of LIVE/DEAD-stained bacteria (Fig. 3).
FIG 3.
Effect of oxantel on a polymicrobial biofilm composed of P. gingivalis, T. forsythia, and T. denticola cultured in OBGM in a static assay for 48 h. Total (dark gray bars), live (light gray bars), and dead (white bars) cells were determined by flow cytometric analysis of LIVE/DEAD-stained cells. A one-way ANOVA with Dunnett's test showed that the numbers of total and live bacterial cells were significantly different (P < 0.05) between the three oxantel concentrations. There was no significant difference in dead bacterial cell numbers for the three oxantel concentrations.
Murine periodontitis model.
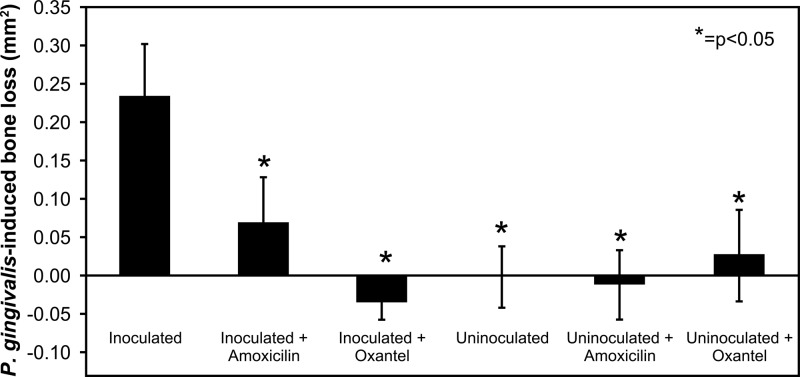
To evaluate the effect of oxantel on P. gingivalis-induced alveolar bone loss, three groups of BALB/c mice were orally infected with two rounds of inoculation of four doses of 1 × 1010 viable P. gingivalis cells 2 weeks apart. Oxantel pamoate or amoxicillin was incorporated into the drinking water 3 days after the end of the first round of inoculation in two groups, and a control group had no additions to the drinking water. The P. gingivalis W50-inoculated control group with no additions to the drinking water had significantly higher alveolar bone loss than all of the other groups, including the inoculated groups treated with oxantel or amoxicillin (Fig. 4). The P. gingivalis-inoculated, oxantel-treated group did not have significantly different bone loss compared with the uninoculated untreated control and the uninoculated groups that received oxantel or amoxicillin treatment. A one-way analysis of variance (ANOVA) with a post hoc Dunnett's T3 test showed that the oxantel-treated group had significantly less bone loss than the amoxicillin-treated group.
FIG 4.
Effect of oxantel and amoxicillin treatment on alveolar bone loss in mice orally infected with P. gingivalis. Three groups of mice were orally infected with P. gingivalis W50 prior to addition of amoxicillin or oxantel to the drinking water, while the remaining three groups were sham inoculated. Mice were killed 29 days after the last oral inoculation and 46 days after the start of amoxicillin or oxantel treatment. The maxillae were removed and defleshed, and alveolar bone loss was determined in the right-hand maxillae by computer-assisted image analysis (43). The data are presented as means plus standard deviations (n = 12) and were analyzed by one-way ANOVA with Dunnett's test. Values that were significantly different (P < 0.05) from the value for the group inoculated with the P. gingivalis W50 wild-type strain are indicated by an asterisk.
DISCUSSION
Fumarate reductase (Frd) has been proposed to play a key role in the fermentation of amino acids by the oral bacterium P. gingivalis, as well as other anaerobic bacterial pathogens, such as Helicobacter pylori and Campylobacter jejuni (38, 45). A search of the sequenced oral bacterial genomes available from the NCBI genome database (http://www.ncbi.nih.gov/genome) showed that some species linked to the initiation and progression of various forms of periodontal disease, including P. gingivalis, T. forsythia, Fusobacterium nucleatum, and Aggregatibacter actinomycetemcomitans, have genes predicted to encode the components of the trimeric SQOR family fumarate reductase (Frd). The ability to selectively inhibit keystone and accessory pathogens like P. gingivalis, T. forsythia, and F. nucleatum using an Frd inhibitor could make this approach more attractive than the use of broad-spectrum antibiotics. The Frd inhibitor may allow commensal species that do not rely on fumarate reduction for energy production to establish and thereby help prevent the reemergence of the pathogenic species and the development of a dysbiotic polymicrobial biofilm (12).
Anthelmintics that have been used to cure helminthic infections in animals and humans have bacteriostatic and bactericidal effects against some bacteria by inhibiting the Frd enzyme complex (44, 46–48). In the present study, we demonstrated that P. gingivalis Frd was inhibited by oxantel in a dose-dependent manner with an IC50 of 2.2 μM, and at 5 μM, oxantel effectively abolished Frd activity in a cytoplasmic extract (Table 2). This is a much lower concentration than the previously determined MIC of 125 μM, suggesting that entry into the cell is the limiting factor in growth inhibition (35). In addition, the P. gingivalis Frd was much more sensitive to oxantel inhibition than recombinantly expressed and purified C. jejuni and H. pylori Frd (48). Oxantel also inhibited the growth of the two other selected oral bacterial pathogens that are predicted to rely on Frd for catabolism. The MIC of oxantel for T. forsythia was 295 μM, which was approximately 2.5 times higher than the P. gingivalis MIC but still substantially lower than the MICs for H. pylori and C. jejuni (44, 46, 48). Oxantel, as expected, had no effect on the planktonic growth of T. denticola, as its genome is not predicted to encode the components of the trimeric SQOR family Frd. Similar to the effects seen with P. gingivalis, oxantel at sub-MICs lowered the growth rate of T. forsythia (Table 3) (35).
We hypothesized that inhibition of P. gingivalis Frd activity by oxantel would cause a shift in its catabolic activity if the bacterium was able to utilize other catabolic pathways. P. gingivalis grown in heme excess in continuous culture to a steady-state cell density of 6 × 108 cells/ml was treated with a concentration of oxantel (125 μM) that did not affect cell density (i.e., the sub-MIC). This resulted in significant upregulation of the expression of 22 genes that were a subset of the 152 genes upregulated during heme-limited growth of P. gingivalis (Table 4 and Fig. 1) (38). P. gingivalis is auxotrophic for protoporphyrin IX, and as such it is unable to synthesize heme de novo, relying on the uptake of environmental heme (49, 50). As the Frd enzyme complex contains two heme cofactors that are essential for activity, Frd inhibition by oxantel to some extent may mimic the effects of heme limitation. We have shown using a quantitative differential proteomics approach that both cytoplasmic fumarate reductase (Frd) subunit proteins decreased in abundance in response to growth in continuous culture under heme limitation (38). In addition, we have shown that the expression of these genes was downregulated in a P. gingivalis ferric uptake regulator (Fur) orthologue knockout mutant (unpublished observation). We have also shown that heme-limited P. gingivalis has reduced biofilm-forming capacity compared with P. gingivalis grown under excess heme conditions (51). It is therefore possible that at the sub-MIC used in this study, the oxantel inhibition of Frd activity reduced energy levels in the cell, which then acted as a weak inducer of a subset of genes of this regulon. There was no obvious shift in the expression of genes encoding catabolic pathways, suggesting that P. gingivalis is unable to modulate its catabolism and avoid the effects of Frd inhibition, unlike some bacteria, such as Actinomyces spp., which have Frd but have alternative catabolic pathways and are able to avoid the growth-limiting consequences of Frd inhibition.
P. gingivalis, T. denticola, and T. forsythia have been strongly associated with chronic periodontitis and have been found to coexist in subgingival plaque in deep periodontal pockets (13–15, 52, 53). Synergistic interactions between these pathogens and other species have been proposed to be a major contributor toward dysbiosis and the progression of the disease (12). In vitro, P. gingivalis and T. denticola display a mutualistic symbiotic relationship in nutrient utilization and growth promotion (52, 54). P. gingivalis and T. denticola coaggregation has been demonstrated (54–57), which would contribute to their colocalization and synergism in biofilm formation (20, 21, 37). When cultured together as part of a polymicrobial biofilm, significant changes occur in the abundance of P. gingivalis and T. denticola peptidases and enzymes involved in glutamate and glycine catabolism, supporting previous reports of syntrophy, as well as changes in abundance of proteins involved in iron/heme acquisition (22).
In the present study, these three species were used as a model dysbiotic pathogenic polymicrobial biofilm (12, 22). As oxantel has no direct effect on T. denticola due to its lack of Frd, there is the possibility that T. denticola could benefit from the suppression of T. forsythia and P. gingivalis. We have previously shown that an oxantel concentration of 125 μM had significant inhibitory effects on P. gingivalis ATCC 33277 homotypic biofilm formation in a static biofilm assay (35), and because polymicrobial biofilms may be more resistant to oxantel, due in part to the higher MIC against T. forsythia, we tested oxantel at 125 and 250 μM.
In the current static assay, 48 h after inoculation with approximately equal numbers of P. gingivalis, T. denticola, and T. forsythia, all three species could be recovered in the biofilm. This indicates a degree of synergy in biofilm formation, as under planktonic conditions, P. gingivalis and T. forsythia have a much higher growth rate than T. denticola and could be expected to outgrow the spirochete. However, 65.7% of cells in the polymicrobial biofilm were T. denticola, as determined by real-time PCR, which is similar to results obtained recently in a polymicrobial flow cell model (22). Addition of oxantel to the growth medium resulted in a concentration-dependent inhibition of polymicrobial biofilm development, which affected all species in the biofilm. In addition to the decrease in the total number of bacterial cells in the biofilm, T. denticola cell numbers fell by a similar proportion to P. gingivalis and T. forsythia cell numbers. This demonstrates the dependence of T. denticola on P. gingivalis and/or T. forsythia for polymicrobial biofilm formation and suggests that T. denticola may be unable to proliferate in subgingival plaque if P. gingivalis and T. forsythia are suppressed (Fig. 2). Oxantel treatment caused a larger reduction in P. gingivalis numbers in the polymicrobial biofilm relative to T. forsythia, which is consistent with the higher MIC of oxantel against T. forsythia. The data obtained from the LIVE/DEAD staining of bacterial cells from the polymicrobial biofilms indicated that the majority of cells were still intact, which is consistent with the oxantel inhibitory mechanism not being associated with cell lysis but energy production (Fig. 3). Frd is an essential and major energy-transducing enzyme in this bacterium, and its inhibition will result in a decrease in available energy to the cell. It is possible that the cellular signaling systems interpret this inhibition as an unfavorable environment to colonize, and so the cell does not adopt a sessile lifestyle, preferring to remain planktonic until more favorable environmental conditions are encountered.
To determine the efficacy of oxantel in vivo, a therapeutic murine alveolar bone loss model, based on a prophylactic model previously reported, was used (43, 58). In this therapeutic model, the addition of 0.50 mg/ml of oxantel pamoate to the drinking water of mice infected with P. gingivalis prevented alveolar bone loss and was superior to 0.50 mg/ml amoxicillin (Fig. 4). These in vivo results provide confirmation that oxantel can inhibit pathogen-associated disease in an animal model.
In conclusion, these combined data indicate that oxantel may have potential for the treatment of periodontitis in humans.
ACKNOWLEDGMENT
This research was partially supported by an International Association for Dental Research/Glaxo-Smith Kline Innovation in Oral Care Award.
Footnotes
Published ahead of print 28 October 2013
REFERENCES
- 1.Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ. 2012. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J. Dent. Res. 91:914–920. 10.1177/0022034512457373 [DOI] [PubMed] [Google Scholar]
- 2.Oliver RC, Brown LJ, Loe H. 1998. Periodontal diseases in the United States population. J. Periodontol. 69:269–278. 10.1902/jop.1998.69.2.269 [DOI] [PubMed] [Google Scholar]
- 3.Beck JD, Offenbacher S, Williams R, Gibbs P, Garcia R. 1998. Periodontitis: a risk factor for coronary heart disease? Ann. Periodontol. 3:127–141. 10.1902/annals.1998.3.1.127 [DOI] [PubMed] [Google Scholar]
- 4.Spahr A, Klein E, Khuseyinova N, Boeckh C, Muche R, Kunze M, Rothenbacher D, Pezeshki G, Hoffmeister A, Koenig W. 2006. Periodontal infections and coronary heart disease: role of periodontal bacteria and importance of total pathogen burden in the Coronary Event and Periodontal Disease (CORODONT) study. Arch. Intern. Med. 166:554–559. 10.1001/archinte.166.5.554 [DOI] [PubMed] [Google Scholar]
- 5.Guha N, Boffetta P, Wunsch Filho V, Eluf Neto J, Shangina O, Zaridze D, Curado MP, Koifman S, Matos E, Menezes A, Szeszenia-Dabrowska N, Fernandez L, Mates D, Daudt AW, Lissowska J, Dikshit R, Brennan P. 2007. Oral health and risk of squamous cell carcinoma of the head and neck and esophagus: results of two multicentric case-control studies. Am. J. Epidemiol. 166:1159–1173. 10.1093/aje/kwm193 [DOI] [PubMed] [Google Scholar]
- 6.Tezal M, Sullivan MA, Reid ME, Marshall JR, Hyland A, Loree T, Lillis C, Hauck L, Wactawski-Wende J, Scannapieco FA. 2007. Chronic periodontitis and the risk of tongue cancer. Arch. Otolaryngol. Head Neck Surg. 133:450–454. 10.1001/archotol.133.5.450 [DOI] [PubMed] [Google Scholar]
- 7.Hujoel PP, Drangsholt M, Spiekerman C, Weiss NS. 2003. An exploration of the periodontitis-cancer association. Ann. Epidemiol. 13:312–316. 10.1016/S1047-2797(02)00425-8 [DOI] [PubMed] [Google Scholar]
- 8.Stolzenberg-Solomon RZ, Dodd KW, Blaser MJ, Virtamo J, Taylor PR, Albanes D. 2003. Tooth loss, pancreatic cancer, and Helicobacter pylori. Am. J. Clin. Nutr. 78:176–181 http://ajcn.nutrition.org/content/78/1/176.long [DOI] [PubMed] [Google Scholar]
- 9.Michaud DS, Joshipura K, Giovannucci E, Fuchs CS. 2007. A prospective study of periodontal disease and pancreatic cancer in US male health professionals. J. Natl. Cancer Inst. 99:171–175. 10.1093/jnci/djk021 [DOI] [PubMed] [Google Scholar]
- 10.Ioannidou E, Kao D, Chang N, Burleson J, Dongari-Bagtzoglou A. 2006. Elevated serum interleukin-6 (IL-6) in solid-organ transplant recipients is positively associated with tissue destruction and IL-6 gene expression in the periodontium. J. Periodontol. 77:1871–1878. 10.1902/jop.2006.060014 [DOI] [PubMed] [Google Scholar]
- 11.Darveau RP. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8:481–490. 10.1038/nrmicro2337 [DOI] [PubMed] [Google Scholar]
- 12.Hajishengallis G, Lamont RJ. 2012. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 27:409–419. 10.1111/j.2041-1014.2012.00663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134–144. 10.1111/j.1600-051X.1998.tb02419.x [DOI] [PubMed] [Google Scholar]
- 14.Byrne SJ, Dashper SG, Darby IB, Adams GG, Hoffmann B, Reynolds EC. 2009. Progression of chronic periodontitis can be predicted by the levels of Porphyromonas gingivalis and Treponema denticola in subgingival plaque. Oral Microbiol. Immunol. 24:469–477. 10.1111/j.1399-302X.2009.00544.x [DOI] [PubMed] [Google Scholar]
- 15.Haffajee AD, Socransky SS, Smith C, Dibart S. 1991. Relation of baseline microbial parameters to future periodontal attachment loss. J. Clin. Periodontol. 18:744–750. 10.1111/j.1600-051X.1991.tb00066.x [DOI] [PubMed] [Google Scholar]
- 16.Brown LF, Beck JD, Rozier RG. 1994. Incidence of attachment loss in community-dwelling older adults. J. Periodontol. 65:316–323. 10.1902/jop.1994.65.4.316 [DOI] [PubMed] [Google Scholar]
- 17.Kesavalu L, Holt SC, Ebersole JL. 1998. Virulence of a polymicrobic complex, Treponema denticola and Porphyromonas gingivalis, in a murine model. Oral Microbiol. Immunol. 13:373–377. 10.1111/j.1399-302X.1998.tb00694.x [DOI] [PubMed] [Google Scholar]
- 18.Kesavalu L, Sathishkumar S, Bakthavatchalu V, Matthews C, Dawson D, Steffen M, Ebersole JL. 2007. Rat model of polymicrobial infection, immunity, and alveolar bone resorption in periodontal disease. Infect. Immun. 75:1704–1712. 10.1128/IAI.00733-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orth RK, O'Brien-Simpson NM, Dashper SG, Reynolds EC. 2011. Synergistic virulence of Porphyromonas gingivalis and Treponema denticola in a murine periodontitis model. Mol. Oral Microbiol. 26:229–240. 10.1111/j.2041-1014.2011.00612.x [DOI] [PubMed] [Google Scholar]
- 20.Kuramitsu HK, Chen W, Ikegami A. 2005. Biofilm formation by the periodontopathic bacteria Treponema denticola and Porphyromonas gingivalis. J. Periodontol. 76:2047–2051. 10.1902/jop.2005.76.11-S.2047 [DOI] [PubMed] [Google Scholar]
- 21.Yamada M, Ikegami A, Kuramitsu HK. 2005. Synergistic biofilm formation by Treponema denticola and Porphyromonas gingivalis. FEMS Microbiol. Lett. 250:271–277. 10.1016/j.femsle.2005.07.019 [DOI] [PubMed] [Google Scholar]
- 22.Zainal-Abidin Z, Veith PD, Dashper SG, Zhu Y, Catmull DV, Chen YY, Heryanto DC, Chen D, Pyke JS, Tan K, Mitchell HL, Reynolds EC. 2012. Differential proteomic analysis of a polymicrobial biofilm. J. Proteome Res. 11:4449–4464. 10.1021/pr300201c [DOI] [PubMed] [Google Scholar]
- 23.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745. 10.1146/annurev.mi.49.100195.003431 [DOI] [PubMed] [Google Scholar]
- 24.Cvitkovitch DG, Li YH, Ellen RP. 2003. Quorum sensing and biofilm formation in streptococcal infections. J. Clin. Invest. 112:1626–1632. 10.1172/JCI20430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167–193. 10.1128/CMR.15.2.167-193.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kroger A, Geisler V, Lemma E, Theis F, Lenger R. 1992. Bacterial fumarate respiration Arch. Microbiol. 158:311–314 [Google Scholar]
- 27.Choi WY, Lee OR, Lee WK, Kim WK, Chung CS, Ough BO. 1979. A clinical trial of oxantel and pyrantel against intestinal nematodes infections. Kisaengch'ung Hak Chapchi 17:60–66 [DOI] [PubMed] [Google Scholar]
- 28.Garcia EG. 1976. Treatment for trichuriasis with oxantel. Am. J. Trop. Med. Hyg. 25:914–915 [DOI] [PubMed] [Google Scholar]
- 29.Horton L. 2003. The efficacy of anthelmintics: past, present, and future, p 143–155 In Crompton D, Montresor A, Nesheim M, Savioli L. (ed), Controlling disease due to helminth infections. World Health Organization, Geneva, Switzerland [Google Scholar]
- 30.Bossche VD. 1985. Handbook of experimental pharmacology. Springer, New York, NY [Google Scholar]
- 31.Fornelio C, Caabeiro R, Gonzalez J. 1987. The mode of action of some benzimidazole drugs on Trichinella spiralis. Parasitology 95:61–70. 10.1017/S0031182000057541 [DOI] [PubMed] [Google Scholar]
- 32.Harrow ID, Gration KA, Evans NA. 1991. Neurobiology of arthropod parasites. Parasitology 102(Suppl):S59–S69 [DOI] [PubMed] [Google Scholar]
- 33.Martin RJ, Murray I, Robertson AP, Bjorn H, Sangster N. 1998. Anthelmintics and ion-channels: after a puncture, use a patch. Int. J. Parasitol. 28:849–862. 10.1016/S0020-7519(98)00048-4 [DOI] [PubMed] [Google Scholar]
- 34.Prichard RK. 1970. Mode of action of the anthelminthic thiabendazole in Haemonchus contortus. Nature 228:684–685. 10.1038/228684a0 [DOI] [PubMed] [Google Scholar]
- 35.Dashper S, Ang CS, Liu SW, Paolini R, Veith P, Reynolds E. 2010. Inhibition of Porphyromonas gingivalis biofilm by oxantel. Antimicrob. Agents Chemother. 54:1311–1314. 10.1128/AAC.00946-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leschine SB, Canale-Parola E. 1980. Rifampin as a selective agent for isolation of oral spirochetes. J. Clin. Microbiol. 12:792–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Y, Dashper SG, Chen Y-Y, Crawford S, Slakeski N, Reynolds EC. 2013. Porphyromonas gingivalis and Treponema denticola synergistic polymicrobial biofilm development. PLoS One 8:e71727. 10.1371/journal.pone.0071727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dashper SG, Ang CS, Veith PD, Mitchell HL, Lo AW, Seers CA, Walsh KA, Slakeski N, Chen D, Lissel JP, Butler CA, O'Brien-Simpson NM, Barr IG, Reynolds EC. 2009. Response of Porphyromonas gingivalis to heme limitation in continuous culture. J. Bacteriol. 191:1044–1055. 10.1128/JB.01270-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dashper SG, O'Brien-Simpson NM, Cross KJ, Paolini RA, Hoffmann B, Catmull DV, Malkoski M, Reynolds EC. 2005. Divalent metal cations increase the activity of the antimicrobial peptide kappacin. Antimicrob. Agents Chemother. 49:2322–2328. 10.1128/AAC.49.6.2322-2328.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakamoto M, Takeuchi Y, Umeda M, Ishikawa I, Benno Y. 2001. Rapid detection and quantification of five periodontopathic bacteria by real-time PCR. Microbiol. Immunol. 45:39–44 [DOI] [PubMed] [Google Scholar]
- 41.Fujise O, Hamachi T, Inoue K, Miura M, Maeda K. 2002. Microbiological markers for prediction and assessment of treatment outcome following non-surgical periodontal therapy. J. Periodontol. 73:1253–1259. 10.1902/jop.2002.73.11.1253 [DOI] [PubMed] [Google Scholar]
- 42.Lo AW, Seers CA, Boyce JD, Dashper SG, Slakeski N, Lissel JP, Reynolds EC. 2009. Comparative transcriptomic analysis of Porphyromonas gingivalis biofilm and planktonic cells. BMC Microbiol. 9:18. 10.1186/1471-2180-9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pathirana RD, O'Brien-Simpson NM, Brammar GC, Slakeski N, Reynolds EC. 2007. Kgp and RgpB, but not RgpA, are important for Porphyromonas gingivalis virulence in the murine periodontitis model. Infect. Immun. 75:1436–1442. 10.1128/IAI.01627-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendz GL, Hazell SL, Srinivasan S. 1995. Fumarate reductase: a target for therapeutic intervention against Helicobacter pylori. Arch. Biochem. Biophys. 321:153–159. 10.1006/abbi.1995.1380 [DOI] [PubMed] [Google Scholar]
- 45.Takahashi N, Sato T, Yamada T. 2000. Metabolic pathways for cytotoxic end product formation from glutamate- and aspartate-containing peptides by Porphyromonas gingivalis. J. Bacteriol. 182:4704–4710. 10.1128/JB.182.17.4704-4710.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mendz GL, Meek DJ, Hazell SL. 1998. Characterization of fumarate transport in Helicobacter pylori. J. Membr. Biol. 165:65–76. 10.1007/s002329900421 [DOI] [PubMed] [Google Scholar]
- 47.Smith MA, Mendz GL, Jorgensen MA, Hazell SL. 1999. Fumarate metabolism and the microaerophily of Campylobacter species. Int. J. Biochem. Cell Biol. 31:961–975. 10.1016/S1357-2725(99)00062-X [DOI] [PubMed] [Google Scholar]
- 48.Mileni M, MacMillan F, Tziatzios C, Zwicker K, Haas AH, Mantele W, Simon J, Lancaster CR. 2006. Heterologous production in Wolinella succinogenes and characterization of the quinol:fumarate reductase enzymes from Helicobacter pylori and Campylobacter jejuni. Biochem. J. 395:191–201. 10.1042/BJ20051675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roper JM, Raux E, Brindley AA, Schubert HL, Gharbia SE, Shah HN, Warren MJ. 2000. The enigma of cobalamin (vitamin B12) biosynthesis in Porphyromonas gingivalis. Identification and characterization of a functional corrin pathway. J. Biol. Chem. 275:40316–40323. 10.1074/jbc.M007146200 [DOI] [PubMed] [Google Scholar]
- 50.Schifferle RE, Shostad SA, Bayers-Thering MT, Dyer DW, Neiders ME. 1996. Effect of protoporphyrin IX limitation on Porphyromonas gingivalis. J. Endod. 22:352–355. 10.1016/S0099-2399(96)80216-0 [DOI] [PubMed] [Google Scholar]
- 51.Dashper SG, Pan Y, Veith PD, Chen YY, Toh EC, Liu SW, Cross KJ, Reynolds EC. 2012. Lactoferrin inhibits Porphyromonas gingivalis proteinases and has sustained biofilm inhibitory activity. Antimicrob. Agents Chemother. 56:1548–1556. 10.1128/AAC.05100-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kigure T, Saito A, Seida K, Yamada S, Ishihara K, Okuda K. 1995. Distribution of Porphyromonas gingivalis and Treponema denticola in human subgingival plaque at different periodontal pocket depths examined by immunohistochemical methods. J. Periodontal Res. 30:332–341. 10.1111/j.1600-0765.1995.tb01284.x [DOI] [PubMed] [Google Scholar]
- 53.Simonson LG, Robinson PJ, Pranger RJ, Cohen ME, Morton HE. 1992. Treponema denticola and Porphyromonas gingivalis as prognostic markers following periodontal treatment. J. Periodontol. 63:270–273. 10.1902/jop.1992.63.4.270 [DOI] [PubMed] [Google Scholar]
- 54.Grenier D. 1992. Nutritional interactions between two suspected periodontopathogens, Treponema denticola and Porphyromonas gingivalis. Infect. Immun. 60:5298–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ito R, Ishihara K, Shoji M, Nakayama K, Okuda K. 2010. Hemagglutinin/adhesin domains of Porphyromonas gingivalis play key roles in coaggregation with Treponema denticola. FEMS Immunol. Med. Microbiol. 60:251–260. 10.1111/j.1574-695X.2010.00737.x [DOI] [PubMed] [Google Scholar]
- 56.Kolenbrander PE. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413–437. 10.1146/annurev.micro.54.1.413 [DOI] [PubMed] [Google Scholar]
- 57.Yao ES, Lamont RJ, Leu SP, Weinberg A. 1996. Interbacterial binding among strains of pathogenic and commensal oral bacterial species. Oral Microbiol. Immunol. 11:35–41. 10.1111/j.1399-302X.1996.tb00334.x [DOI] [PubMed] [Google Scholar]
- 58.O'Brien-Simpson NM, Pathirana RD, Paolini RA, Chen YY, Veith PD, Tam V, Ally N, Pike RN, Reynolds EC. 2005. An immune response directed to proteinase and adhesin functional epitopes protects against Porphyromonas gingivalis-induced periodontal bone loss. J. Immunol. 175:3980–3989 http://www.jimmunol.org/content/175/6/3980.long [DOI] [PubMed] [Google Scholar]