Abstract
Letermovir is a novel antiviral compound currently in clinical development for the prevention of human cytomegalovirus (HCMV) infections. In contrast to all currently approved anti-HCMV drugs that target the viral DNA polymerase, letermovir acts via a distinct mode of action involving the viral terminase subunit pUL56. To extend our understanding of potential letermovir resistance mechanisms, we used marker transfer to characterize mutations identified in letermovir-resistant HCMV variants that were selected in cell culture.
TEXT
Human cytomegalovirus (HCMV) disease continues to be a serious and life-threatening condition in immunocompromised patients such as transplant recipients. Currently approved anti-HCMV drugs, including (val)ganciclovir, cidofovir, and foscarnet, are associated with profound toxicity as well as drug resistance that restricts their long-term clinical benefit. All these substances ultimately target the viral DNA polymerase (pUL54), though ganciclovir requires prior activation by the viral pUL97 kinase. Accordingly, mutations in open reading frame (ORF) UL97 are associated with (val)ganciclovir resistance whereas mutations in ORF UL54 can confer cross-resistance to all approved anti-HCMV drugs, an increasingly frequent scenario representing a serious threat for specific patient populations (1–3).
Letermovir (AIC246) is a new anti-HCMV drug with a distinct but well-characterized mechanism of action targeting the viral terminase subunit pUL56 (4, 5). The drug has proven to be well tolerated in numerous (pre)clinical studies and has demonstrated clinical efficacy in a recent phase IIb trial, meeting all primary endpoints as a prophylactic drug (6).
In line with its distinct mode of action, in vitro studies have shown that letermovir is active against HCMV strains resistant to current anti-HCMV drugs (4, 5, 7). In vivo confirmation of resistance-breaking potential was demonstrated in a patient infected with a multidrug-resistant HCMV strain (8). So far, no letermovir-resistant HCMV strains have been reported from clinical trials. Nonetheless, as letermovir is a direct-acting antiviral, the emergence of resistance is ultimately to be expected.
HCMV resistance testing previously required virus cultivation and phenotypic characterization of virus isolates; this is slow, labor-intensive, and nonstandardized and often has a poor success rate (3, 9). Current standard clinical practice uses genotyping and virtual phenotyping based, e.g., on mutation databases (10). Once a database is established, this genetic approach is faster, more sensitive, and more cost-effective and provides detailed and quantitative resistance information to the physician (3). However, genetic monitoring of drug resistance requires detailed knowledge of potential resistance mutations in order to develop virtual phenotyping profiles for screening databases (6).
To date, 2 letermovir resistance mutations leading to amino acid substitutions have been identified in vitro, both within the coding region for the pUL56 subunit of the viral terminase complex (4). This study was intended to (i) identify and characterize additional amino acid substitutions associated with letermovir resistance, (ii) extend our understanding of letermovir resistance mechanisms, and (iii) create a basis for genotypic resistance testing in patients.
Two different approaches were used to generate letermovir-resistant mutants of the HCMV laboratory strain AD169: (i) a single-step selection method, using virus-infected fibroblast cells incubated in the presence of letermovir (or a chemically closely related predecessor compound) at ∼10-fold 50% effective concentration (EC50) (50 nM for letermovir) as described in reference 4 (4 experiments; each using 30 96-well plates/assay; multiplicity of infection [MOI] of 0.03), or (ii) classical resistance induction using virus-infected fibroblasts (MOI, 0.03 to 0.01) initially incubated at subinhibitory concentrations (∼1× EC50) of antiviral compound. Each time that virus breakthrough (100% cytopathic effect [CPE]) was observed, virus supernatant was subcultured and the drug concentration in the medium was doubled until either CPE formation was suppressed or drug concentration reached ≥128-fold EC50. Resistant virus mutants were plaque purified by limiting dilution in the presence of 10× EC50 of letermovir followed by serial passage 8 to 10 times in drug-free cell culture medium to ensure the stability of the resistance mutation. It is of note that the applied procedures were intended to generate drug-resistant strains for mechanistic studies and not to examine, e.g., the frequency of the emergence of drug resistance or the barrier to resistance.
Letermovir susceptibility (EC50) of the selected viruses was assessed using a cytopathic effect assay (4, 5). As summarized in Table 1, all strains were letermovir resistant, with resistance indices (RI; EC50 for mutant/EC50 for wild type) ranging from ∼13 to ∼6,000. As expected, no cross-resistance to ganciclovir was evident. In this way, 10 letermovir-resistant AD169 isolates were obtained (8 via single-step selection, 2 using induction), of which two have been described previously (4).
TABLE 1.
Pheno- and genotypic characterization of letermovir-resistant HCMV mutants obtained in vitro
| HCMV strain | EC50 (μM)a |
RIb | Amino acid substitutionc |
||||
|---|---|---|---|---|---|---|---|
| Letermovir | GCVj | UL56d | UL89d | UL104d | UL51d | ||
| AD169 | 0.0046 ± 0.0019 | 3.6 ± 1.4 | 1 | NAe | NA | NA | NA |
| Selected mutantsf | |||||||
| rAIC246-1g | 1.23 ± 0.32 | 1.2 ± 0.2 | 268 | L241P | —h | — | — |
| rAIC246-2g | 0.37 ± 0.07 | 4.0 ± 0.9 | 81 | R369S | A345Si | — | — |
| rAIC246-3 | 27.23 ± 3.27 | 3.0 ± 2.4 | 5,870 | C325Y | — | — | — |
| rAIC246-4 | 0.13 ± 0.01 | 4.2 ± 1.3 | 28 | V231L | — | — | — |
| rAIC246-5 | 0.11 ± 0.01 | 5.0 ± 0.4 | 23 | R369M | — | — | — |
| rAIC246-6 | 0.08 ± 0.02 | 2.9 ± 0.9 | 17 | R369M | — | — | — |
| rAIC246-7 | 0.92 ± 0.12 | 2.2 ± 0.6 | 200 | L241P | — | — | — |
| rAIC246-8 | 25.01 ± 5.53 | 2.2 ± 1.2 | 5,413 | C325Y | — | — | — |
| rAIC246-9 | 0.06 ± 0.04 | 1.7 ± 0.2 | 13 | R369G | — | — | — |
| rAIC246-10 | 0.09 ± 0.02 | 1.4 ± 0.4 | 19 | V236M | A345Si | — | — |
EC50s were determined by a CPE reduction assay. Data are means from at least three independent experiments and are expressed with standard deviations.
The resistance index (RI) is the letermovir EC50 for mutant virus divided by the letermovir EC50 for wild-type virus.
Amino acid substitution identified by HCMV genotyping.
HCMV gene involved in cleavage/packaging of viral progeny DNA.
NA, not applicable.
HCMV strain AD169 virus mutants obtained in vitro under selective pressure with letermovir.
Previously published (4).
—, no amino acid substitution.
Interstrain variation (12) not associated with letermovir resistance.
GCV, ganciclovir.
In order to identify amino acid substitutions that account for the observed drug resistance, 4 HCMV open reading frames (ORFs) encoding proteins involved in cleavage/packaging of progeny DNA were sequenced (4, 11). A sequence comparison with the parental AD169 reference strain revealed 7 unique single amino acid substitutions; all were point mutations, and all were clustered in the region between amino acid (aa) 230 and 370 of ORF UL56 (Table 1; Fig. 1). Of these, 2 have been previously reported (L241P and R369S) (4) and 5 were novel substitutions (V231L, V236M, C325Y, R369M, and R369G). Interestingly, 3 of the 7 detected substitutions were located at position 369. No substitutions with significance for letermovir resistance were identified in UL89, UL51, and UL104. The known UL89 A345S interstrain variation (12, 13) was apparent in 2 resistant strains, likely because the AD169 strain used in our laboratory included both the UL89 A345 and S345 genotypes. An association between this polymorphism and letermovir resistance was excluded since letermovir susceptibility to AD169 UL89 A345 variants (e.g., AD169-GFP) is comparable with that of AD169 UL89 S345 variants (e.g., RV-HG) (reference 5 and unpublished data).
FIG 1.
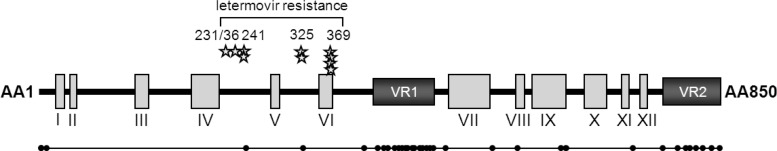
Schematic representation of the UL56 domain organization, showing known wild-type polymorphisms. (Top) Schematic representation of the UL56 domain organization according to data from reference 19. Conserved regions are indicated as gray boxes (I to XII); variable regions (VR1 and VR2) are indicated as black boxes. Positions of amino acids associated with in vitro resistance to letermovir are indicated. Each star represents an amino acid mutation identified in an independently selected resistant virus variant. (Bottom) Schematic representation of a UL56 consensus sequence alignment derived from 61 unique HCMV UL56 amino acid sequences. Each dot indicates a naturally occurring amino acid sequence polymorphism.
To confirm that letermovir resistance resulted from the identified UL56 substitutions, a two-step, markerless red recombination system was used to introduce the respective single point mutations into the enhanced green fluorescent protein (EGFP)-expressing, AD169-derived HCMV bacterial artificial chromosome (BAC) pHG (4, 14) (see Table S1 in the supplemental material). Sequence fidelity of the recombinant BACs was confirmed by sequencing and restriction analysis as described previously (4, 15). Following virus reconstitution, viral growth curves were generated exactly as described in reference 4 in order to confirm the viability of all recombinant virus strains. None of the amino acid substitutions had any discernible effect upon the growth properties of these laboratory strains in vitro (data not shown). However, the fitness of these variants in natural isolates in patients remains to be determined.
To confirm that the identified UL56 substitutions were indeed responsible for letermovir resistance and to examine possible cross-resistance, we proceeded to phenotypically characterize the recombinant viruses using a fluorescence reduction assay as a measure of drug susceptibility (5) (Table 2). Drug resistance profiles were comparable between recombinant viruses (UL89 S345 background) and their corresponding original isolates (UL89 background indicated in Table 1), confirming that solely the identified UL56 amino acid substitutions were sufficient to confer the observed resistance.
TABLE 2.
Susceptibility of reconstituted wild-type virus and constructed UL56 mutant viruses to letermovir, approved polymerase inhibitors, and drugs targeting the viral terminase
| Drug | EC50a (μM) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RV-HG | RV-HG UL56-V231L | RIb | RV-HG UL56-V236M | RI | RV-HG UL56-L241Pc | RI | RV-HG UL56-C325Y | RI | RV-HG UL56-R369M | RI | RV-HG UL56-R369G | RI | RV-HG UL56-R369Sc | RI | |
| Letermovir | 0.0030 ± 0.0011 | 0.02 ± 0.00 | 5 | 0.13 ± 0.01 | 45 | 0.65 ± 0.24 | 218 | 26.4 ± 6.5 | 8,796 | 0.04 ± 0.01 | 13 | 0.13 ± 0.03 | 44 | 0.14 ± 0.07 | 48 |
| Ganciclovird | 1.84 ± 0.92 | 0.72 ± 0.24 | —f | 1.92 ± 0.35 | — | 2.11 ± 0.77 | — | 2.11 ± 0.71 | — | 1.25 ± 0.61 | — | 1.22 ± 0.11 | — | 1.96 ± 1.09 | — |
| Cidofovird | 0.18 ± 0.13 | 0.09 ± 0.02 | — | 0.40 ± 0.11 | — | 0.21 ± 0.09 | — | 0.35 ± 0.05 | — | 0.18 ± 0.11 | — | 0.14 ± 0.03 | — | 0.16 ± 0.05 | — |
| Foscarnetd | 95 ± 49 | 67 ± 18 | — | 128 ± 13 | — | 115 ± 71 | — | 126 ± 20 | — | 68 ± 24 | — | 55 ± 0.00 | — | 61 ± 21 | — |
| Aciclovird | 53 ± 33 | 29 ± 9 | — | 75 ± 14 | — | 53 ± 12 | — | 49 ± 16 | — | 61 ± 25 | — | 102 ± 0.00 | — | 45 ± 10 | — |
| BAY38-4766e | 0.39 ± 0.14 | 0.36 ± 0.09 | — | 0.61 ± 0.03 | — | 0.34 ± 0.14 | — | 1.07 ± 0.14 | — | 1.75 ± 0.16 | 5 | 0.43 ± 0.18 | — | 0.58 ± 0.13 | — |
| BDCRBe | 0.30 ± 0.11 | 0.21 ± 0.04 | — | 0.32 ± 0.05 | — | 0.27 ± 0.04 | — | 0.30 ± 0.04 | — | 0.11 ± 0.05 | — | 0.12 ± 0.07 | — | 0.19 ± 0.02 | — |
EC50s were determined by a fluorescence reduction assay. Data are means from at least three independent experiments and are expressed with standard deviations.
The resistance index (RI) is the EC50 for the indicated RV-HG mutant divided by the EC50 of the RV-HG wild type.
Previously published (4).
Approved polymerase inhibitor.
—, no antiviral resistance (RI < 3).
The lack of cross-resistance to all approved anti-HCMV drugs is consistent with letermovir's alternate mechanism of action. Interestingly, although all mutants were susceptible to the chemically unrelated cleavage/packaging inhibitor BDCRB (16, 17), a low-grade cross-resistance (∼5-fold) was observed against another unrelated terminase inhibitor named BAY38-4766 (18) for one UL56 mutant strain (R369M) but not for strains carrying other amino acid substitutions at the same position (369G and 369S).
It is conceivable that the observed high-grade drug resistance conferred by the detected resistance mutations is due to the steep dose-response curve that characterizes the activity of letermovir in cell culture (5). However, the magnitude of drug resistance is not the sole measure of clinical relevance since, e.g., even mutations mediating low-level ganciclovir resistance (>3-fold EC50) are often therapeutically prohibitive due to dose-limiting toxicities and/or physicochemical properties of the drug (2, 3). Moreover, an “all-or-nothing” inhibition kinetics, as observed with letermovir in vitro (5), might be advantageous in vivo since it renders low-level virus replication unlikely and may thus limit the emergence of drug resistance. However, resistance emergence at clinical doses remains to be determined, as it will depend on additional factors not captured in cell culture studies, including human pharmacokinetics (PK) and unbound drug concentrations in human plasma.
Based on these results, we defined the region between aa 230 and 370 as that involved in letermovir resistance. A sequence alignment comprising 61 UL56 sequences available at NCBI GenBank, including 41 sequences from clinical isolates previously compared by Champier et al. (19), was generated (see Table S2 in the supplemental material). A schematic representation of the results (Fig. 1) shows 34 naturally occurring polymorphisms within ORF UL56 relative to the HCMV reference strain Merlin. The letermovir resistance region (aa 230 to 370) appears to be within a conserved area of pUL56, with only 2 naturally occurring polymorphisms (D242G and A327V) which were not selected by letermovir. These data suggest that preexisting natural resistance to letermovir is unlikely, although compound activity against D242G and A327V is yet to be examined.
In conclusion, the work presented here identifies the likely location of letermovir resistance substitutions within an ∼140-aa region of pUL56 and therefore should provide a useful basis for resistance monitoring in clinical trials. No letermovir resistance-associated substitutions were uncovered in the coding regions of other viral proteins known to be involved in genome cleavage/packaging (pUL89, pUL104, and pUL51), suggesting a highly specific interaction of the inhibitor with the pUL56 terminase subunit. Additional structural information on pUL56 and biophysical/biochemical characterization of compound binding will shed more light on the mechanism of action of this novel inhibitor, which has been shown to be fully active against HCMV strains resistant to currently approved antivirals.
Supplementary Material
ACKNOWLEDGMENTS
Plasmid pEPKan-S was provided by Klaus Osterrieder, Cornell University, Ithaca, NY, USA, and the HCMV Bac pHG was a kind gift of Martin Messerle, MHH Hannover, Germany. We thank Rob Saunders, biomed context, for his assistance preparing the draft manuscript and H.C. Huang (Merck & Co., Inc.) for reviewing the final draft. The excellent technical assistance of Marion Heidtmann, Kerstin Pixberg, and Bernd Schulz is gratefully acknowledged.
All authors are employees of AiCuris GmbH & Co. KG.
Footnotes
Published ahead of print 4 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01794-13.
REFERENCES
- 1.Harter G, Michel D. 2012. Antiviral treatment of cytomegalovirus infection: an update. Expert Opin. Pharmacother. 13:623–627. 10.1517/14656566.2012.658775 [DOI] [PubMed] [Google Scholar]
- 2.Schreiber A, Harter G, Schubert A, Bunjes D, Mertens T, Michel D. 2009. Antiviral treatment of cytomegalovirus infection and resistant strains. Expert Opin. Pharmacother. 10:191–209. 10.1517/14656560802678138 [DOI] [PubMed] [Google Scholar]
- 3.Lurain NS, Chou S. 2010. Antiviral drug resistance of human cytomegalovirus. Clin. Microbiol. Rev. 23:689–712. 10.1128/CMR.00009-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldner T, Hewlett G, Ettischer N, Ruebsamen-Schaeff H, Zimmermann H, Lischka P. 2011. The novel anticytomegalovirus compound AIC246 (letermovir) inhibits human cytomegalovirus replication through a specific antiviral mechanism that involves the viral terminase. J. Virol. 85:10884–10893. 10.1128/JVI.05265-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lischka P, Hewlett G, Wunberg T, Baumeister J, Paulsen D, Goldner T, Ruebsamen-Schaeff H, Zimmermann H. 2010. In vitro and in vivo activities of the novel anticytomegalovirus compound AIC246. Antimicrob. Agents Chemother. 54:1290–1297. 10.1128/AAC.01596-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chemaly RF, Ehninger G, Champlin R, Richard MP, Zimmermann H, Lischka P, Stoelben S, McCormick D, Ruebsamen-Schaeff H. 2012. Letermovir (AIC246) for the prevention of CMV infections meets primary endpoint in phase 2b trial in human blood precursor cell transplant (HBPCT) recipients, abstr. T-356 Abstr. 52nd Intersci. Conf. Antimicrob. Agents Chemother [Google Scholar]
- 7.Marschall M, Stamminger T, Urban A, Wildum S, Ruebsamen-Schaeff H, Zimmermann H, Lischka P. 2012. In vitro evaluation of the activities of the novel anti-cytomegalovirus compound AIC246 (letermovir) against herpesviruses and other human pathogenic viruses. Antimicrob. Agents Chemother. 56:1135–1137. 10.1128/AAC.05908-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaul DR, Stoelben S, Cober E, Ojo T, Sandusky E, Lischka P, Zimmermann H, Rubsamen-Schaeff H. 2011. First report of successful treatment of multidrug-resistant cytomegalovirus disease with the novel anti-CMV compound AIC246. Am. J. Transplant. 11:1079–1084. 10.1111/j.1600-6143.2011.03530.x [DOI] [PubMed] [Google Scholar]
- 9.Boeckh M, Boivin G. 1998. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin. Microbiol. Rev. 11:533–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chevillotte M, von EJ, Meier BM, Lin FM, Kestler HA, Mertens T. 2010. A new tool linking human cytomegalovirus drug resistance mutations to resistance phenotypes. Antiviral Res. 85:318–327. 10.1016/j.antiviral.2009.10.004 [DOI] [PubMed] [Google Scholar]
- 11.Borst EM, Kleine-Albers J, Gabaev I, Babic M, Wagner K, Binz A, Degenhardt I, Kalesse M, Jonjic S, Bauerfeind R, Messerle M. 2013. The human cytomegalovirus UL51 protein is essential for viral genome cleavage-packaging and interacts with the terminase subunits pUL56 and pUL89. J. Virol. 87:1720–1732. 10.1128/JVI.01955-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradley AJ, Lurain NS, Ghazal P, Trivedi U, Cunningham C, Baluchova K, Gatherer D, Wilkinson GW, Dargan DJ, Davison AJ. 2009. High-throughput sequence analysis of variants of human cytomegalovirus strains Towne and AD169. J. Gen. Virol. 90:2375–2380. 10.1099/vir.0.013250-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Champier G, Hantz S, Couvreux A, Stuppfler S, Mazeron MC, Bouaziz S, Denis F, Alain S. 2007. New functional domains of human cytomegalovirus pUL89 predicted by sequence analysis and three-dimensional modelling of the catalytic site DEXDc. Antivir. Ther. 12:217–232 [PubMed] [Google Scholar]
- 14.Tischer BK, von EJ, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. 10.2144/000112096 [DOI] [PubMed] [Google Scholar]
- 15.Tavalai N, Kraiger M, Kaiser N, Stamminger T. 2008. Insertion of an EYFP-pp71 (UL82) coding sequence into the human cytomegalovirus genome results in a recombinant virus with enhanced viral growth. J. Virol. 82:10543–10555. 10.1128/JVI.01006-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reefschlaeger J, Bender W, Hallenberger S, Weber O, Eckenberg P, Goldmann S, Haerter M, Buerger I, Trappe J, Herrington JA, Haebich D, Ruebsamen-Waigmann H. 2001. Novel non-nucleoside inhibitors of cytomegaloviruses (BAY 38-4766): in vitro and in vivo antiviral activity and mechanism of action. J. Antimicrob. Chemother. 48:757–767. 10.1093/jac/48.6.757 [DOI] [PubMed] [Google Scholar]
- 17.Underwood MR, Harvey RJ, Stanat SC, Hemphill ML, Miller T, Drach JC, Townsend LB, Biron KK. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buerger I, Reefschlaeger J, Bender W, Eckenberg P, Popp A, Weber O, Graeper S, Klenk HD, Ruebsamen-Waigmann H, Hallenberger S. 2001. A novel nonnucleoside inhibitor specifically targets cytomegalovirus DNA maturation via the UL89 and UL56 gene products. J. Virol. 75:9077–9086. 10.1128/JVI.75.19.9077-9086.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Champier G, Couvreux A, Hantz S, Rametti A, Mazeron MC, Bouaziz S, Denis F, Alain S. 2008. Putative functional domains of human cytomegalovirus pUL56 involved in dimerization and benzimidazole D-ribonucleoside activity. Antivir. Ther. 13:643–654 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


