Abstract
Among 220 clinical isolates of Gram-negative bacilli collected in India during 2000, 22 strains showing elevated imipenem MICs were evaluated for carbapenemase production. One DIM-1-producing Pseudomonas stutzeri isolate was detected, and no other carbapenemase-encoding genes were identified. This detection of a DIM-1-producing P. stutzeri isolate from India predating the finding of this gene in the index Dutch strain and the very recent detection of DIM-1 in Africa suggest an unidentified environmental source of this metallo-β-lactamase gene.
TEXT
Numerous types of metallo-β-lactamases (MBL) have been discovered and characterized to date, and the IMP and VIM types and a few NDM variants seem to possess the most global distribution. In contrast, some other MBL-encoding genes are limited to a single report or to certain geographic locations, including genes encoding SPM-1, GIM-1, SIM-1, AIM-1, KHM-1, DIM-1, BIC-1, and TMB-1 (1). The NDM-1 (New Delhi Metallo-β-lactamase)-encoding gene was detected in a diabetic Swedish patient of Indian origin who traveled to New Delhi and acquired a urinary tract infection (2). Since this initial report, NDM-producing isolates have been reported in several countries across nearly all continents, and in many instances these reports were related to patient travel to the Indian subcontinent. Although much has been studied about NDM producers, the timeline for the emergence of the NDM-encoding gene was not clearly established before 2006 (3).
(This study was presented at the 22nd European Congress of Clinical Microbiology and Infectious Diseases, London, United Kingdom, 31 March to 3 April 2012.)
This study was initiated to determine the presence of blaNDM and other carbapenemase genes among a collection of Gram-negative bacilli cultured during 2000 at hospitals located in India (4). Among 220 Gram-negative clinical isolates available from five hospitals (one each in the cities Mumbai, Vellore, New Delhi, Lucknow, and Indore), 10.0% (22 strains) demonstrated imipenem MICs of ≥0.5 μg/ml when tested by reference broth microdilution methods (5). These organisms included five Enterobacter cloacae isolates, four Pseudomonas aeruginosa isolates, four Pseudomonas fluorescens isolates, two isolates each of Klebsiella pneumoniae, Acinetobacter baumannii, Citrobacter freundii, and Pseudomonas stutzeri, and one Proteus vulgaris isolate. Enterobacteriaceae strains exhibited the lowest imipenem MICs (0.5 to 1 μg/ml), and higher carbapenem MIC results were noted for Pseudomonas spp. (1 to 8 μg/ml). The two A. baumannii isolates displayed imipenem MICs of 1 μg/ml (Table 1).
TABLE 1.
Susceptibility profiles of the Gram-negative isolates displaying elevated imipenem MICs collected in Indian hospitals in 2000a
| Isolate | Organism | City of isolation | MIC (μg/ml) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Imipenem | Ceftazidime | Ceftriaxone | Piperacillin- tazobactam | Ciprofloxacin | Gentamicin | Tetracycline | Tigecycline | Trimethoprim- sulfamethoxazole | |||
| 10627 | P. stutzeri | New Delhi | 2 | 16 | >8 | 64 | >4 | >8 | 8 | 0.12 | >4 |
| 10532 | P. stutzeri | Lucknow | 1 | 0.25 | 2 | 16 | 0.12 | ≤1 | 1 | 0.25 | >4 |
| 10526 | P. aeruginosa | Lucknow | 8 | 2 | >8 | 4 | 0.12 | ≤1 | >8 | 4 | 4 |
| 10417 | P. aeruginosa | Vellore | 2 | 2 | >8 | 16 | >4 | >8 | >8 | >4 | >4 |
| 10550 | P. aeruginosa | Lucknow | 8 | 2 | >8 | 4 | 0.12 | ≤1 | >8 | 4 | 4 |
| 10552 | P. aeruginosa | Lucknow | 8 | 2 | >8 | 4 | 0.12 | ≤1 | >8 | 4 | 4 |
| 10528 | P. fluorescens | Lucknow | 8 | 4 | >8 | 4 | 0.06 | ≤1 | 1 | 0.25 | ≤0.5 |
| 10540 | P. fluorescens | Lucknow | 8 | 4 | >8 | 4 | 0.12 | >8 | 2 | 0.12 | 2 |
| 10543 | P. fluorescens | Lucknow | 8 | 4 | >8 | 8 | 0.12 | >8 | 2 | 0.12 | 4 |
| 10545 | P. fluorescens | Lucknow | 4 | 4 | >8 | 4 | ≤0.03 | ≤1 | 1 | 0.12 | 1 |
| 10430 | A. baumannii | Vellore | 1 | >32 | >8 | ≤0.5 | >4 | >8 | >8 | 0.25 | >4 |
| 10456 | A. baumannii | Indore | 1 | 32 | >8 | >64 | >4 | >8 | >8 | 0.5 | ≤0.5 |
| 10606 | C. freundii | New Delhi | 0.5 | >32 | >8 | >64 | >4 | >8 | 1 | 0.25 | ≤0.5 |
| 10431 | C. freundii | Indore | 1 | >32 | >8 | 2 | 2 | 4 | >8 | 0.5 | >4 |
| 10525 | E. cloacae | Lucknow | 0.5 | >32 | >8 | >64 | 2 | >8 | >8 | 0.5 | >4 |
| 10418 | E. cloacae | Vellore | 0.5 | >32 | >8 | 32 | ≤0.03 | >8 | 2 | 1 | >4 |
| 10433 | E. cloacae | Indore | 0.5 | >32 | >8 | >64 | ≤0.03 | ≤1 | 2 | 0.25 | ≤0.5 |
| 10476 | E. cloacae | Mumbai | 0.5 | >32 | >8 | >64 | 0.06 | >8 | >8 | 0.5 | >4 |
| 10488 | E. cloacae | New Delhi | 0.5 | >32 | >8 | 32 | 0.5 | >8 | >8 | 0.25 | ≤0.5 |
| 10447 | K. pneumoniae | Indore | 1 | >32 | >8 | >64 | >4 | >8 | 4 | 0.25 | >4 |
| 10480 | K. pneumoniae | Mumbai | 0.5 | 0.06 | ≤0.06 | 2 | ≤0.03 | ≤1 | 1 | 0.25 | ≤0.5 |
| 10432 | P. stuartii | Vellore | 1 | >32 | >8 | 2 | 2 | 4 | >8 | 0.5 | >4 |
The data for the DIM-1-producing strain are set in boldface.
The candidate isolates were further evaluated for the presence of genes encoding carbapenemases by the use of PCRs targeting the following genes/groups: blaIMP, blaVIM, blaSPM-1, blaKPC, blaSME, blaIMI, blaNMC-A, blaGES, blaKHM-1, blaDIM-1, blaBIC-1, blaNDM, and blaOXA-48 by multiplex PCR using custom primers (3, 6). Acinetobacter spp. were also screened for the genes encoding OXA-23, OXA-24/OXA-40, OXA-51, and OXA-58 (7). Enterobacteriaceae isolates were screened by the modified Hodge test (MHT) using imipenem and meropenem as the substrates (8).
None of the isolates were positive for blaNDM, but one P. stutzeri strain yielded positive PCR results for blaDIM primers. No other carbapenemase genes were detected, and all Enterobacteriaceae strains yielded negative MHT phenotypic results. The two A. baumannii strains were positive only for blaOXA-51-like genes.
DIM-1 was first identified in a Pseudomonas stutzeri strain collected in 2007 from a Dutch patient (9). This enzyme shares 45 to 52% amino acid identity with IMP types and GIM-1 and only 30 to 38% identity with SPM-1, VIM, and NDM types. The greatest amino acid sequence identity (63.7%) is shared with TMB-1, a new MBL identified in Achromobacter spp. (10). Similar to other MBLs, DIM-1 hydrolyzes broad-spectrum cephalosporins and carbapenems but not monobactams such as aztreonam.
The MBL-positive P. stutzeri was nonsusceptible to cephalosporins, gentamicin, ciprofloxacin, piperacillin-tazobactam, and tetracycline, but the imipenem MIC result was only marginally elevated (MIC, 2 μg/ml) (Table 1) and this isolate was inhibited by 0.25 μg/ml of tigecycline. The MBL genetic environment was amplified using primers annealing to the 5′ and 3′ conserved sequence (CS) regions of class 1 integrons in combination with the blaDIM-1 primers. Sequences were analyzed using the Lasergene software (DNASTAR, Madison, WI) and compared to others available via internet sources (http://www.ncbi.nlm.nih.gov/blast/). P. stutzeri carried blaDIM-1, and integron sequencing identified the MBL gene located in the second position of a class 1 integron with aadB located upstream. No 3′-end CS was detected using multiple primers. The comparison of the genetic environment of this strain and that of the index DIM-1-producing strain demonstrated different arrays of genes in the integron structures. The index strain carried blaDIM-1 in the first position, followed by aadB and qacH, and no 3′-end CS (9).
Clonality among the DIM-1-producing P. stutzeri isolate from India and the index strain from the same bacterial species detected in the Netherlands (kindly supplied by L. Poirel, Bicetre Hospital, France) was evaluated by pulsed-field gel electrophoresis (PFGE), as described elsewhere (9). The Indian DIM-1-producing isolate was genetically distinct from the Dutch P. stutzeri index strain (Fig. 1). Southern blot and hybridization of S1 endonuclease and I-CeuI digests of agarose-embedded DNA using a digoxigenin-labeled (Roche Diagnostics GmbH, Mannheim, Germany) blaDIM-1-specific probe showed one copy of the DIM-1-encoding gene located on a plasmid of approximately 240 kb, in contrast to the index strain, which carried blaDIM-1 on a much smaller plasmid (70 kb). Plasmid sizes were estimated by linear regression using a high-molecular-weight lambda ladder (Lonza, Rockland, ME).
FIG 1.
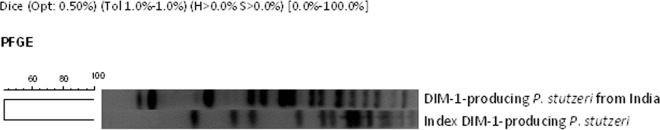
PFGE comparison of the DIM-1-producing P. stutzeri isolate from India and the DIM-1-producing P. stutzeri index isolate from the Netherlands.
P. stutzeri is considered a low-frequency opportunistic pathogen that causes infections mainly in patients with serious underlying diseases (11). The most frequent infection sites have included the bloodstream, wounds, the respiratory tract, and urine. However, in recent years this species has emerged as a pathogen capable of causing serious invasive infections, such as indolent endocarditis, community-acquired (CA) meningitis, neonatal septicemia, and community-acquired pneumonia. Furthermore, P. stutzeri has been sporadically reported to carry MBLs, primarily of the VIM types (12–14).
Genes encoding NDM or other carbapenemases (VIM types among P. aeruginosa and OXA-181 among Enterobacteriaceae) that have been reported from India (3, 15) were not detected in this collection dating from 2000. This suggests that these enzymes emerged in India after 2000 and before 2006, when NDM-1-, OXA-181-, and VIM-producing isolates became more commonly reported. The finding of DIM-1 in two P. stutzeri isolates separated geographically and temporally may imply that the DIM-1-encoding gene has emerged in the Indian subcontinent; however, DIM-1 has not been detected in any other carbapenem-resistant isolate of Indian origin screened to date in our collection (2006-2007 and 2010-2011 sample years; data not shown) or cited in the literature. Additionally, DIM-1 has been very recently detected in an Enterobacter species from Sierra Leone, West Africa (16), and the emergence of the DIM-1-encoding gene on different continents with no apparent worldwide dissemination might indicate that blaDIM-1 was acquired independently from an unknown natural reservoir. The protein similarity of DIM-1 and TMB-1 characterized from an environmental sample of Achromobacter species (10) also suggests that the possible origin of DIM-1 is as an intrinsic β-lactamase in an environmental species.
Although the potential of the DIM-1-encoding gene to proliferate in clinical isolates seems low, its appearance on three continents warrants screening for the corresponding resistance genotype among isolates with elevated carbapenem MICs.
ACKNOWLEDGMENTS
We thank L. Poirel and P. Nordmann (Bicetre Hospital, France) for kindly sharing the DIM-1-producing P. stutzeri index strain with our group. We also acknowledge the technical assistance of R. R. Dietrich and the contribution of Indian investigators, mainly Dilip Mathai, who coordinated the 2000 collection of isolates tested in this investigation.
JMI Laboratories, Inc. has received research and educational grants in 2010 to 2013 from the following: Achaogen, Aires, American Proficiency Institute (API), Anacor, Astellas, AstraZeneca, bioMérieux, Cempra, Cerexa, Contrafect, Cubist, Dipexium, Enanta, Furiex, GlaxoSmithKline, Johnson & Johnson, LegoChem Biosciences Inc., Meiji Seika Kaisha, Nabriva, Novartis, Pfizer, PPD Therapeutics, Premier Research Group, Rempex, Melinta (Rib-X) Pharmaceuticals, Seachaid, Shionogi, The Medicines Co., Theravance, and ThermoFisher. Some JMI employees are advisors/consultants for Astellas, Cubist, Pfizer, Cempra, Cerexa-Forest, and Theravance.
Footnotes
Published ahead of print 21 October 2013
REFERENCES
- 1.Queenan AM, Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20:440–458. 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. 2011. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob. Agents Chemother. 55:1274–1278. 10.1128/AAC.01497-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathai D, Rhomberg PR, Biedenbach DJ, Jones RN. 2002. Evaluation of the in vitro activity of six broad-spectrum beta-lactam antimicrobial agents tested against recent clinical isolates from India: a survey of ten medical center laboratories. Diagn. Microbiol. Infect. Dis. 44:367–377. 10.1016/S0732-8893(02)00466-2 [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute 2012. M07-A9. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard: ninth edition. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70:119–1123. 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 7.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, Amyes SG, Livermore DM. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351–353. 10.1016/j.ijantimicag.2006.01.004 [DOI] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute 2013. M100-S23. Performance standards for antimicrobial susceptibility testing: 23rd informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 9.Poirel L, Rodriguez-Martinez JM, Al Naiemi N, Debets-Ossenkopp YJ, Nordmann P. 2010. Characterization of DIM-1, an integron-encoded metallo-beta-lactamase from a Pseudomonas stutzeri clinical isolate in the Netherlands. Antimicrob. Agents Chemother. 54:2420–2424. 10.1128/AAC.01456-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Salabi A, Borra PS, Toleman MA, Samuelsen O, Walsh TR. 2012. Genetic and biochemical characterization of a novel metallo-beta-lactamase, TMB-1, from an Achromobacter xylosoxidans strain isolated in Tripoli, Libya. Antimicrob. Agents Chemother. 56:2241–2245. 10.1128/AAC.05640-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noble RC, Overman SB. 1994. Pseudomonas stutzeri infection. A review of hospital isolates and a review of the literature. Diagn. Microbiol. Infect. Dis. 19:51–56. 10.1016/0732-8893(94)90051-5 [DOI] [PubMed] [Google Scholar]
- 12.Yan JJ, Hsueh PR, Ko WC, Luh KT, Tsai SH, Wu HM, Wu JJ. 2001. Metallo-beta-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob. Agents Chemother. 45:2224–2228. 10.1128/AAC.45.8.2224-2228.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee NY, Yan JJ, Lee HC, Liu KH, Huang ST, Ko WC. 2004. Clinical experiences of bacteremia caused by metallo-beta-lactamase-producing gram-negative organisms. J. Microbiol. Immunol. Infect. 37:343–349 [PubMed] [Google Scholar]
- 14.Amudhan MS, Sekar U, Kamalanathan A, Balaraman S. 2012. bla(IMP) and bla(VIM) mediated carbapenem resistance in Pseudomonas and Acinetobacter species in India. J. Infect. Dev. Ctries. 6:757–762. 10.3855/jidc.2268 [DOI] [PubMed] [Google Scholar]
- 15.Castanheira M, Bell JM, Turnidge JD, Mathai D, Jones RN. 2009. Carbapenem resistance among Pseudomonas aeruginosa strains from India: evidence for nationwide endemicity of multiple metallo-beta-lactamase clones (VIM-2, -5, -6, and -11 and the newly characterized VIM-18). Antimicrob. Agents Chemother. 53:1225–1227. 10.1128/AAC.01011-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leski TA, Bangura U, Jimmy DH, Ansumana R, Lizewski SE, Li RW, Stenger DA, Taitt CR, Vora GJ. 2013. Identification of blaOXA-51-like, blaOXA-58, blaDIM-1, and blaVIM carbapenemase genes in hospital Enterobacteriaceae isolates from Sierra Leone. J. Clin. Microbiol. 51:2435–2438. 10.1128/JCM.00832-13 [DOI] [PMC free article] [PubMed] [Google Scholar]


