Abstract
The current emergence of the carbapenemase OXA-48 among Enterobacteriaceae is related to the spread of a single IncL/M-type plasmid, pOXA-48a. This plasmid harbors the blaOXA-48 gene within a composite transposon, Tn1999, which is inserted into the tir gene, encoding a transfer inhibition protein. We showed that the insertion of Tn1999 into the tir gene was involved in a higher transfer frequency of plasmid pOXA-48a. This may likely be the key factor for the successful dissemination of this plasmid.
INTRODUCTION
Plasmids belonging to the IncL/M incompatibility group are commonly identified among environmental and clinical enterobacterial isolates (1–3). They have been increasingly identified as a source of multidrug resistance. Indeed, IncL/M-type plasmids are the main plasmids responsible for the dissemination of specific extended-spectrum β-lactamase genes such as the blaCTX-M-3 gene and have also been shown to harbor carbapenemase genes such as the blaNDM-1 and blaOXA-48 genes (4–6). OXA-48 carbapenemase is an Ambler class D β-lactamase that was first identified in 2001 from a multidrug-resistant Klebsiella pneumoniae isolate from Turkey (7). Many sporadic cases have been reported, first in Turkey and then in many other European countries and Israel (8–13). OXA-48 is now considered an endemic carbapenemase in Enterobacteriaceae, at least in Turkey and in North African countries such as Morocco, Algeria, and Tunisia, and a source of outbreak situations in France, Belgium, Spain, and the Netherlands (14–20). The spread of OXA-48 producers and of KPC producers represents the most important source of multidrug resistance in Europe and the United States. The spread of the blaOXA-48 gene is linked mostly to the dissemination of the single 62-kb IncL/M-type plasmid pOXA-48a (20). Most of the OXA-48-positive Enterobacteriaceae harbor this specific plasmid, which is spreading among various enterobacterial species (20). Sequence analysis of plasmid pOXA-48a showed that the blaOXA-48 gene is bracketed by two copies of IS1999, giving rise to a composite transposon named Tn1999 (8, 21). Tn1999 is inserted into the tir gene, encoding a transfer inhibition protein, which is therefore not functional (6). Interestingly, a gene similar to this tir gene has been shown to inhibit plasmid RP4 conjugal transfer (22). Therefore, the aim of this study was to evaluate whether the disruption of the tir gene consecutive to the Tn1999 insertion in pOXA-48a could play a role in the dissemination of this plasmid and, consequently, of the blaOXA-48 gene.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Clinical strains K. pneumoniae 11978 (blaOXA-48) and K. pneumoniae 601 (blaNDM-1) were previously described (7, 23). Escherichia coli TOP10(pOXA-48a) and E. coli TOP10(pNDM-OM), harboring the natural plasmids of K. pneumoniae 11978 and K. pneumoniae 601, respectively, were obtained by mating-out assays. Nalidixic acid-resistant E. coli JM109, nalidixic acid-resistant Enterobacter cloacae SB, and rifampin-resistant E. coli TOP10 were used for mating-out assays. The kanamycin-resistant plasmid pCRBluntII-TOPO (Invitrogen, Cergy-Pontoise, France) was used as a cloning vector. Bacterial strains were grown in Luria-Bertani (LB) broth at 37°C. When necessary, antibiotics were added at the following concentrations: ticarcillin at 50 μg · ml−1, temocillin at 50 μg · ml−1, kanamycin at 30 μg · ml−1, rifampin at 60 μg · ml−1, and nalidixic acid at 20 μg · ml−1.
PCR assays.
Phusion DNA polymerase (Thermo Fisher Scientific, Villebon-sur-Yvette, France) was used for PCR experiments according to the manufacturer's instructions. PCR primers are indicated in Table 1. On plasmid pOXA-48a, the tir gene is truncated by transposon Tn1999 harboring the blaOXA-48 gene (6). In order to construct a DNA fragment encompassing the entire tir gene, DNA fragments of 530 bp and 326 bp were amplified by PCR with primer pairs preTir-For/TirEnd-Rev and TirEnd-For/preTir-Rev, respectively (Table 1). Primer preTir-For was designed in order to replace the σ70 −35 promoter sequence (TCGGCA) of the tir gene by a −35 promoter sequence, TTGGCA, closer to the E. coli σ70 −35 promoter consensus sequence TTGACA in the corresponding amplicon (Fig. 1) (24). Primer TirEnd-For was designed as follows: the first 20 nucleotides of primer TirEnd-For were specific to the 3′ extremity of the first truncated fragment of tir, and the last 20 nucleotides were specific of the 5′ extremity of the second truncated fragment of tir (Fig. 1). Primer TirEnd-Rev was the reverse complement of primer TirEnd-For (Table 1). These DNA fragments were subsequently used as templates for a PCR experiment with primers preTir-For and preTir-Rev, giving rise to an 856-bp DNA fragment corresponding to the entire reconstructed sequence of the tir gene of plasmid pOXA-48a.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′–3′) | PCR product size (bp) | Use(s) |
|---|---|---|---|
| preTir-For | GCTAGCTTGGCAATCATTTTCTGTATC | 530 | Amplification and cloning |
| TirEnd-Rev | GTTGTACCTCGAACGGAAGACGTTCAGCATGACACCACGG | ||
| TirEnd-For | CCGTGGTGTCATGCTGAACGTCTTCCGTTCGAGGTACAAC | 326 | Amplification and cloning |
| preTir-Rev | GCTAGCGGTATGCATTTTCACCTCC | ||
| Tir-GSP1 | TCGTCATAAATGGCTCAGCG | 327a | 5′ RACE |
| Tir-GSP2 | GGTCCAGCACTTTACCCAGC | 303a | |
| Tir-GSP3 | GAAATTCACCGACATACACC | 282a | |
| RT-Tir-For | CTGCTCTATGGGCTGGG | 273 | RT-PCR |
| RT-Tir-Rev | GCAACGTCAGGATCACRTCG | ||
| RT-TraM-For | GGATCAAACTGACTCGGATC | 190 | RT-PCR |
| RT-TraM-Rev | GAAGCCAGTAACCCTGAAG | ||
| gapA-For | TATGACTGGTCCGTCTAAAGACAA | 192 | RT-PCR |
| gapA-Rev | GGTTTTCTGAGTAGCGGTAGTAGC |
Distance to the ATG start codon.
FIG 1.
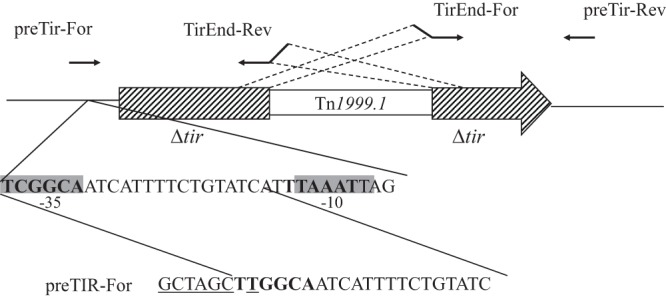
Design of primers for amplification of the tir gene. The −35 and −10 sequences of the promoter of the tir gene are indicated in boldface type and are shaded in gray. Nucleotides modified or added in primer preTIR-For are underlined. The orientation of the primers is indicated by arrows.
Cloning experiments and sequencing.
The reconstructed tir gene was subsequently cloned into the vector pCRBluntII-TOPO and electroporated into rifampin-resistant E. coli TOP10, giving rise to E. coli TOP10(pTOPO-Tir). As a control, a noncoding sequence of 683 bp was cloned into the vector pCRBluntII-TOPO and electroporated into rifampin-resistant E. coli TOP10, giving rise to E. coli TOP10(pTOPO-Nc). Sequences of the inserts of these recombinant plasmids were confirmed by sequencing.
Qualitative filter matings.
Plasmids pOXA-48a and pNDM-OM from K. pneumoniae 11978 and K. pneumoniae 601, respectively, were introduced into rifampin-resistant E. coli TOP10 by mixing 200 μl of donor cells with 800 μl of recipient cells and filtering 200 μl of the mating mix onto the surface of a 0.45-μm-pore-size filter (Millipore, Molsheim, France). The filter was placed onto the surface of a Trypticase soy plate, which was then incubated at 37°C for approximately 3 h. Following incubation, the bacterial lawn on the surface of the filter was streaked onto selective medium containing ticarcillin and rifampin. Those experiments gave rise to rifampin-resistant E. coli TOP10(pOXA-48a) and E. coli TOP10(pNDM-OM). Plasmid pOXA-48a was introduced into rifampin-resistant E. coli TOP10(pTOPO-Tir) and rifampin-resistant E. coli TOP10(pTOPO-Nc) according to the same protocol, using ticarcillin, kanamycin, and rifampin for selection. As a result, E. coli TOP10(pOXA-48a, pTOPO-Tir) and E. coli TOP10(pOXA-48a, pTOPO-Nc) were obtained.
Quantitative filter matings.
Donor and recipient cells were each grown overnight in LB broth supplemented with ticarcillin (50 μg · ml−1) for plasmid maintenance in donor cells. A 0.25-ml donor culture was mixed with 4.75 ml LB broth and incubated at 37°C for 5 h without shaking. Recipient cultures of E. coli JM109 or E. cloacae SB grown overnight were diluted 1:50 in LB broth and incubated at 37°C for 5 h with shaking. After incubation, 0.25 ml of the donor culture was gently mixed with 2.5 ml of the recipient culture, and 200 μl of this mating mix was filtered through a 0.45-μm filter (Millipore). Filters were incubated on prewarmed plates at 37°C for 2 h. Mating assays were ended by placement of filters into 4 ml of an ice-cold 0.9% NaCl solution, followed by vigorous agitation for 30 s. The number of transconjugants per donor cell was determined by plating dilutions of the mating mixture onto plates containing antibiotics. Strain donor cells were selected with ticarcillin (50 μg · ml−1) or temocillin (50 μg · ml−1) and rifampin (60 μg · ml−1). Transconjugants were selected with ticarcillin (50 μg.ml−1) or temocillin (50 μg · ml−1) and nalidixic acid (20 μg · ml−1). Transfer frequencies were calculated by dividing the number of transconjugants by the number of donor cells. For the measurement of conjugation frequencies, the standard deviation was calculated from three independent cultures. Statistical analysis was performed by using the Student t test; a P value of ≤0.05 was considered significant.
Primer extension experiments.
Total RNA was extracted from the E. coli TOP10(pOXA-48a) and E. coli TOP10(NDM-OM) strains with an RNeasy Midi kit (Qiagen, Courtaboeuf, France) by using RNAprotect (Qiagen) according to the recommendations of the manufacturer. RNA extracts were previously treated with DNase (Qiagen). The 5′ rapid amplification of cDNA ends (RACE) reactions were performed with 5 μg of total RNA of E. coli TOP10(pOXA-48a) and E. coli TOP10(pNDM-OM) and a 5′ RACE system kit (version 2.0; Invitrogen, Cergy-Pontoise, France), according to the recommendations of the manufacturer. The design of specific primers (Table 1) was performed as specified by the manufacturer for first-strand cDNA synthesis (primer Tir-GSP1), PCR of dC-tailed cDNA (primer Tir-GSP2), and nested amplification (primer Tir-GSP3) (Table 1). The 5′ RACE PCR products were then cloned into pCRBluntII-TOPO. Analysis of the cloned sequence allowed determination of the transcription initiation site and, subsequently, the promoter sequences.
Expression of the tir gene.
Real-time reverse transcription-PCR (RT-PCR) was performed to measure mRNA expression levels of the tir and the traM genes from plasmids pOXA-48a and pNDM-OM. A one-step RT-PCR was performed by using a Rotor-Gene instrument (Qiagen) with the Rotor-Gene SYBR green RT-PCR kit (Qiagen), with 100 ng of total RNA of E. coli TOP10(pOXA-48a) and E. coli TOP10(pNDM-OM) and 1 μM primer in a total volume of 25 μl (Table 1). Relative transcripts levels were calculated by using the 2−ΔΔCT method (25). Levels of gap gene (encoding d-glyceraldehyde-3-phosphate dehydrogenase) transcription were used as internal controls to normalize the data. At least three independent RNA samples isolated from three separate cultures were used to determine average transcript levels of each strain.
RESULTS AND DISCUSSION
Comparison of the transfer frequencies of plasmids pOXA-48a and pNDM-OM.
Transfer frequencies of two IncL/M plasmids, pOXA-48a, carrying the blaOXA-48 gene, and pNDM-OM, carrying the blaNDM-1 gene, were compared. Results of these experiments are shown in Table 2. The transfer efficiency of pOXA-48a was about 40-fold higher than that of pNDM-OM. Noteworthy, those two plasmids exhibit different sizes (61,881 bp for pOXA-48a and 87,185 bp for pNDM-OM) and display significant heterogeneity in several genes of the tra locus (traX, traY, and excA) that might be involved in transfer efficiency differences. Nevertheless, we hypothesized that the differences observed in terms of transfer frequency might also be related to the disruption of the tir gene resulting from the insertion of transposon Tn1999, leading to a lack of production of the inhibition protein for pOXA-48a transfer.
TABLE 2.
Transfer frequencies in E. coli JM109 and E. cloacae SBa
| Donor strain | Recipient strain | Mean transfer frequency ± SD |
|---|---|---|
| E. coli TOP10(pOXA-48a) | E. coli JM109 | 1.1 × 10−1 ± 0.02 |
| E. coli TOP10(pNDM-OM) | E. coli JM109 | 2.6 × 10−3 ± 0.016 |
| E. coli TOP10(pOXA-48a, pTOPO-Nc) | E. coli JM109 | 1.7 × 10−1 ± 0.03 |
| E. coli TOP10(pOXA-48a, pTOPO-TIR) | E. coli JM109 | 1.6 × 10−3 ± 0.0005 |
| E. coli TOP10(pOXA-48a, pTOPO-Nc) | E. cloacae SB | 4.9 × 10−2 ± 0.018 |
| E. coli TOP10(pOXA-48a, pTOPO-TIR) | E. cloacae SB | 1.2 × 10−3 ± 0.00004 |
In each case, three independent experiments were performed, and the means and standard deviations were calculated. Statistical analysis was performed by using the Student t test, and a P value of ≤0.05 of was considered significant.
Disruption of the tir gene and inhibition of the conjugative transfer of plasmid pOXA-48a.
The transfer frequencies of pOXA-48a in the presence or absence of the Tir protein were therefore evaluated by performing mating-out assays with E. coli TOP10(pOXA-48a, pTOPO-Tir) or E. coli TOP10(pOXA-48a, pTOPO-Nc) as the donor, respectively, and using E. coli JM109 or E. cloacae SB as the recipient (Table 2). Each of these donor strains harbored two plasmids, both having the natural plasmid pOXA-48a together with a recombinant plasmid, either pTOPO-Tir, encoding the transfer inhibition protein TIR, or pTOPO-Nc, possessing a noncoding DNA sequence, respectively. Interestingly, trans-complementation with the TIR protein resulted in approximately 100- and 50-fold decreases of the efficiency of pOXA-48a transfer in E. coli and E. cloacae, respectively (Table 2). These results confirm the role of TIR in the inhibition of plasmid transfer.
Characterization of tir gene promoter sequences in pOXA-48a and pNDM-OM.
Using the 5′ RACE technique, the +1 transcription initiation site and, subsequently, the promoter sequences of the tir gene were mapped for both pOXA-48a and pNDM-OM, respectively. In pOXA-48a, the +1 transcription site was located 53 bp upstream of the start codon of the tir gene. The promoter of the tir gene was made of a −35 box (TCGGCA) and a −10 box (TTAAAT) separated by 17 bp (Fig. 2). The promoter of the tir gene was made of a −35 box (TCGGTA) separated by 17 bp from the −10 box (TTAAAT) in pNDM-OM (Fig. 2). In comparison with the −35 box promoter sequences of E. coli TOP10(pNDM-OM) (TCGGTA), the −35 box promoter sequences of E. coli TOP10(pOXA-48a) (TCGGCA) displayed a single mutation, leading to a −35 box closer to the consensus sequence (24), suggesting a putative stronger promoter and, consequently, stronger expression of the tir gene in E. coli TOP10(pOXA-48a).
FIG 2.
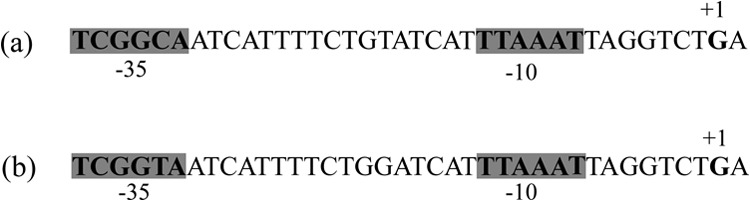
Promoter structures for the tir gene in plasmids pOXA-48a (a) and pNDM-OM (b). The −35 and −10 sequences of the promoter are indicated in boldface type and are shaded in gray. The +1 transcription start sites are indicated in boldface type.
Expression of tir genes in pOXA-48a and pNDM-OM.
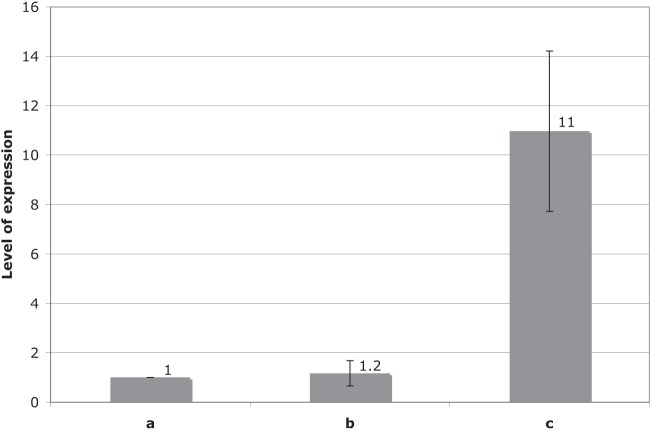
Quantitative RT-PCR was performed to measure the expression of the tir gene in E. coli TOP10(pOXA-48a) and in E. coli TOP10(pNDM-OM). The tir gene transcript levels were compared to those of the gap gene, which was taken as a chromosomal reference for gene expression, and to those of the traM gene, taken as a plasmid reference for gene expression for pOXA-48a and pNDM-OM. The traM genes of these two plasmids were very similar (only a single amino acid substitution in the corresponding protein), whereas the promoter sequences of these genes were strictly identical. As expected, the transcription levels of the traM genes of pOXA-48a and pNDM-OM, sharing identical promoter sequences, were almost identical (Fig. 3). Transcriptional profile analysis indicated that the tir genes were significantly expressed in both pOXA-48a and pNDM-OM. Furthermore, an 11-fold-increased expression level of the tir gene was observed in E. coli TOP10(pOXA-48a) compared to E. coli TOP10(pNDM-OM), in accordance with a stronger −35 promoter sequence of the tir gene in pOXA-48a (Fig. 3). However, the TIR protein was not functional when using pOXA-48a, with its corresponding gene being disrupted by Tn1999. Analyzing the sequence of another IncL/M-type plasmid, namely, pEL60 from Erwinia amylovora, considered to possess a typical IncL/M backbone (neither resistance gene nor insertion sequence), the −35 promoter sequence of the tir gene was found to be distantly related to that consensus, ACGGTA (1). Therefore, it appears that the expression of the tir gene was maintained at a low level in order to avoid the inhibition of plasmid transfer and therefore to increase the transfer frequency. In the case of pOXA-48a, the promoter of the tir gene was stronger; however, the gene was truncated and could not encode a functional protein. Altogether, these elements may constitute a negative regulation system contributing to the dissemination of IncL/M plasmids, which are known to be efficient vectors for resistance genes.
FIG 3.
Mean relative expression ratio of the traM (b) and the tir genes (c) for E. coli TOP10(pOXA-48a) compared to E. coli TOP10(pNDM-OM). The expression level of the gap gene (a) in E. coli TOP10 was used as a reference with a value of 1.
The current spread of the blaOXA-48 gene is largely the consequence of the dissemination of a single epidemic plasmid. In this study, we showed that the inactivation of the tir gene, encoding a transfer inhibition protein, by the insertion of Tn1999 may contribute to the efficient transfer of plasmid pOXA-48a among various genetic backgrounds. However, other genes of the operon transfer, namely, traX, traY, and excA, are very specific to the pOXA-48a plasmid, and further experiments will be necessary to evaluate their role in the transfer of pOXA-48a.
ACKNOWLEDGMENTS
This work was funded partially by a grant from the INSERM (U914), the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), and the Université Paris XI, France, and mostly by grants from the European Community (R-GNOSIS, FP7/HEALTH-F3-2011-282512, and MAGIC-BULLET, FP7/HEALTH-F3-2001-278232).
Footnotes
Published ahead of print 4 November 2013
REFERENCES
- 1.Foster GC, McGhee GC, Jones AL, Sundin GW. 2004. Nucleotide sequences, genetic organization, and distribution of pEU30 and pEL60 from Erwinia amylovora. Appl. Environ. Microbiol. 70:7539–7544. 10.1128/AEM.70.12.7539-7544.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnin RA, Nordmann P, Carattoli A, Poirel L. 2013. Comparative genomics of IncL/M-type plasmids: evolution by acquisition of resistance genes and insertion sequences. Antimicrob. Agents Chemother. 57:674–676. 10.1128/AAC.01086-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carattoli A. 2011. Plasmids in Gram negatives: molecular typing of resistance plasmids. Int. J. Med. Microbiol. 301:654–658. 10.1016/j.ijmm.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 4.Golebiewski M, Kern-Zdanowicz I, Zienkiewicz M, Adamczyk M, Zylinska J, Baraniak A, Gniadkowski M, Bardowski J, Ceglowski P. 2007. Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum β-lactamase gene blaCTX-M-3. Antimicrob. Agents Chemother. 51:3789–3795. 10.1128/AAC.00457-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I, Tong AH, Bao JY, Lok S, Lo JY. 2011. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One 6:e17989. 10.1371/journal.pone.0017989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirel L, Bonnin RA, Nordmann P. 2012. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 56:559–562. 10.1128/AAC.05289-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L, Héritier C, Tolün V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:15–22. 10.1128/AAC.48.1.15-22.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrër A, Poirel L, Eraksoy H, Cagatay AA, Badur S, Nordmann P. 2008. Spread of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Istanbul, Turkey. Antimicrob. Agents Chemother. 52:2950–2954. 10.1128/AAC.01672-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poirel L, Carbonnelle E, Bernabeu S, Gutmann L, Rotimi V, Nordmann P. 2012. Importation of OXA-48-producing Klebsiella pneumoniae from Kuwait. J. Antimicrob. Chemother. 67:2051–2052. 10.1093/jac/dks167 [DOI] [PubMed] [Google Scholar]
- 10.Adler A, Shklyar M, Schwaber MJ, Navon-Venezia S, Dhaher Y, Edgar R, Solter E, Benenson S, Masarwa S, Carmeli Y. 2011. Introduction of OXA-48-producing Enterobacteriaceae to Israeli hospitals by medical tourism. J. Antimicrob. Chemother. 66:2763–2766. 10.1093/jac/dkr382 [DOI] [PubMed] [Google Scholar]
- 11.Cuzon G, Naas T, Bogaerts P, Glupczynski Y, Huang TD, Nordmann P. 2008. Plasmid-encoded carbapenem-hydrolyzing β-lactamase OXA-48 in an imipenem-susceptible Klebsiella pneumoniae strain from Belgium. Antimicrob. Agents Chemother. 52:3463–3464. 10.1128/AAC.00543-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimou V, Dhanji H, Pike R, Livermore DM, Woodford N. 2012. Characterization of Enterobacteriaceae producing OXA-48-like carbapenemases in the UK. J. Antimicrob. Chemother. 67:1660–1665. 10.1093/jac/dks124 [DOI] [PubMed] [Google Scholar]
- 13.Pfeifer Y, Schlatterer K, Engelmann E, Schiller RA, Frangenberg HR, Stiewe D, Holfelder M, Witte W, Nordmann P, Poirel L. 2012. Emergence of OXA-48-type carbapenemase-producing Enterobacteriaceae in German hospitals. Antimicrob. Agents Chemother. 56:2125–2128. 10.1128/AAC.05315-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrër A, Poirel L, Yilmaz M, Akan OA, Feriha C, Cuzon G, Matar G, Honderlick P, Nordmann P. 2010. Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob. Agents Chemother. 54:1369–1373. 10.1128/AAC.01312-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuzon G, Ouanich J, Gondret R, Naas T, Nordmann P. 2011. Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob. Agents Chemother. 55:2420–2423. 10.1128/AAC.01452-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glupczynski Y, Huang TD, Bouchahrouf W, Rezende de Castro R, Bauraing C, Gérard M, Verbruggen AM, Deplano A, Denis O, Bogaerts P. 2012. Rapid emergence and spread of OXA-48-producing carbapenem-resistant Enterobacteriaceae isolates in Belgian hospitals. Int. J. Antimicrob. Agents 39:168–172. 10.1016/j.ijantimicag.2011.10.005 [DOI] [PubMed] [Google Scholar]
- 17.Pitart C, Solé M, Roca I, Fabrega A, Vila J, Marco F. 2011. First outbreak of a plasmid-mediated carbapenem-hydrolyzing OXA-48 β-lactamase in Klebsiella pneumoniae in Spain. Antimicrob. Agents Chemother. 55:4398–4401. 10.1128/AAC.00329-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Potron A, Kalpoe J, Poirel L, Nordmann P. 2011. European dissemination of a single OXA-48-producing Klebsiella pneumoniae clone. Clin. Microbiol. Infect. 17:E24–E26. 10.1111/j.1469-0691.2011.03669.x [DOI] [PubMed] [Google Scholar]
- 19.Voulgari E, Zarkotou O, Ranellou K, Karageorgopoulos DE, Vrioni G, Mamali V, Themeli-Digalaki K, Tsakris A. 2013. Outbreak of OXA-48 carbapenemase-producing Klebsiella pneumoniae in Greece involving an ST11 clone. J. Antimicrob. Chemother. 68:84–88. 10.1093/jac/dks356 [DOI] [PubMed] [Google Scholar]
- 20.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J. Antimicrob. Chemother. 67:1597–1606. 10.1093/jac/dks121 [DOI] [PubMed] [Google Scholar]
- 21.Aubert D, Naas T, Héritier C, Poirel L, Nordmann P. 2006. Functional characterization of IS1999, an IS4 family element involved in mobilization and expression of β-lactam resistance genes. J. Bacteriol. 188:6506–6514. 10.1128/JB.00375-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanimoto K, Lino T, Ohtsubo H, Ohtsubo E. 1985. Identification of a gene, tir of R100, functionally homologous to the F3 gene of F in the inhibition of RP4 transfer. Mol. Gen. Genet. 198:356–357. 10.1007/BF00383019 [DOI] [PubMed] [Google Scholar]
- 23.Poirel L, Al Maskari Z, Al Rashdi F, Bernabeu S, Nordmann P. 2011. NDM-1-producing Klebsiella pneumoniae isolated in the Sultanate of Oman. J. Antimicrob. Chemother. 66:304–306. 10.1093/jac/dkq428 [DOI] [PubMed] [Google Scholar]
- 24.Lisser S, Margalit H. 1993. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 21:1507–1516. 10.1093/nar/21.7.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan JS, Reed A, Chen F, Stewart CN., Jr 2006. Statistical analysis of real-time PCR data. BMC Bioinformatics 7:85. 10.1186/1471-2105-7-85 [DOI] [PMC free article] [PubMed] [Google Scholar]