Abstract
Four out of 143 phenotypically isoniazid-resistant but rifampin-susceptible Mycobacterium tuberculosis strains that were isolated from patients in Germany in 2011 had mutations in the rifampin resistance-determining region of rpoB. After performing drug susceptibility testing (DST) with two methods, the proportion method on Löwenstein-Jensen medium and using the Bactec 960 Mycobacteria Growth Indicator Tube system, we conclude that the two methods are equally reliable for phenotypic DST and MIC determination.
TEXT
The development of multidrug-resistant tuberculosis (MDR-TB), defined as resistance against at least isoniazid (INH) and rifampin (RMP), is assumed to occur as resistance to drugs serially acquired due to inadequate treatment. This includes the use of single drugs or single, usually effective drugs in the treatment of patients that are infected with resistant strains. INH-resistant but RMP-susceptible Mycobacterium tuberculosis strains may represent a source for the development of MDR strains in the case of nonadapted treatment.
The molecular basis of RMP resistance is localized mainly in a specific region of the rpoB gene, the RMP resistance-determining region (RRDR), which enables rapid detection by molecular analysis (1). Extensive use of molecular assays consistently found, although rarely, isolates harboring rpoB mutations but which did not test resistant by phenotypical drug susceptibility testing (DST) methods (2, 3).
In this study, we investigated the presence of rpoB mutations in all 143 INH-resistant and RMP-susceptible M. tuberculosis isolates from patients living in Germany that were submitted to the German National Reference Laboratory for Mycobacteria in 2011. In addition, RMP susceptibility was evaluated with two different DST methods. Primary detection and DST had been performed earlier by using the Bactec 960 Mycobacterial Growth Indicator Tube (MGIT) system (Becton, Dickinson, USA). The processing of the specimens, the DNA preparation, and the sequencing of katG and the ribosome binding site (RBS) of inhA for INH resistance and of the RRDR were carried out as described previously (4–6).
Of the 143 M. tuberculosis strains, 139 strains showed no mutation, while four strains (2.8%) had at least one rpoB mutation within the RRDR. This is consistent with two other studies reporting 4 of 94 (4.3%) and 4 of 202 (2.0%) strains with at least one rpoB mutation within the RRDR (7) (8). Two of the four strains had the mutation L533P. The other two strains had the mutation D516Y in combination with a second mutation within the RRDR (N518D or E510H) (Table 1). None of the sequencing profiles showed underlying wild-type peaks indicating heteroresistance. All mutations found have been described previously (9). The mutation N518D has been described in combination with L533P in a strain with high-level RMP resistance (10). Therefore, this mutation does not seem to have a reverse effect on a resistant phenotype in general. The second double mutation within rpoB, resulting in E510H D516Y, has been described in an RMP-resistant strain (11). A possible explanation for the discrepancy could be different genetic backgrounds of the strains. An influence of the M. tuberculosis lineage on the resistance level has been described for INH (12). Isolates with the mutations D516Y and L533P, as well as isolates with the mutations L511P and H526L, have been reported independently as susceptible by standard Bactec MGIT methods. It has been debated whether these “disputed” mutations are of clinical relevance, in spite of test results indicating susceptibility (7, 13–15). We did not find any of the mutations that frequently confer high-level RMP resistance (e.g., S531L or H526D) (16, 17).
TABLE 1.
Resistance profiles, mutations, and MICs of four phenotypically INH-resistant but RMP-susceptible strainsa
| Strain | Resistance to first-line agents | inhA and katG mutationsb | INH susceptibility determined by MGIT 960 at MIC (μg/ml) of: |
rpoB mutation(s) | RMP susceptibility at indicated MIC (μg/ml) determined by: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LJ medium |
MGIT 960 |
|||||||||||||||
| 0.1 | 1 | 3 | 10 | 1.25 | 2.5 | 5 | 10 | 20 | 40 | 0.25 | 0.5 | 1 | ||||
| 1 | INH, SM | −15 C/T, S315T | R | R | R | R | L533P | R | R | S | S | S | S | R | S | S |
| 2 | INH, SM | WT, S315T | R | R | S | S | D516Y, N518D | B | S | S | S | S | S | S | S | S |
| 3 | INH, EMB | −15 C/T, WT | R | S | S | S | L533P | R | R | R | S | S | S | R | S | S |
| 4 | INH | −8 T/A, S315T | R | R | R | R | E510H, D516Y | R | R | R | R | R | S | R | R | S |
Abbreviations: INH, isoniazid; SM, streptomycin; EMB, ethambutol; R, resistant; S, susceptible; B, borderline.
Designations such as −15 C/T indicate a C/T exchange at position −15. WT, wild type.
Three of the four strains showed high-level INH resistance at ≥1.0 μg/ml and an S315T mutation in katG, while one of the strains with an L533P mutation had low-level INH resistance at 0.1 μg/ml, no mutation within katG, but a mutation within the RBS of inhA. A relation between RMP-susceptible strains with rpoB mutations and INH high-level resistance has been described by Williamson et al. (7), who, as a consequence, proposed to test all isolates with high-level INH resistance for rpoB mutations.
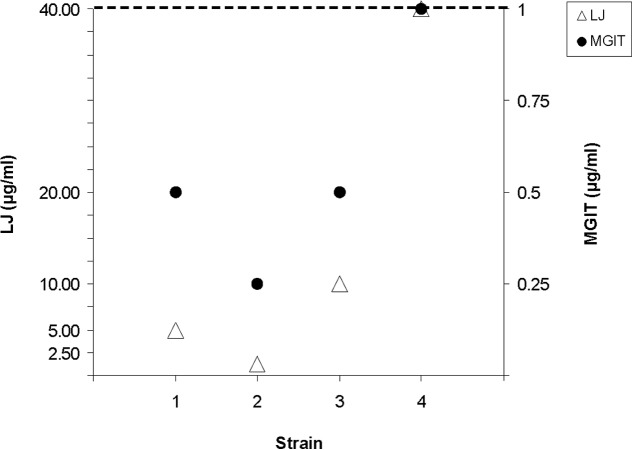
RMP MICs below the standard critical concentrations were determined by the proportion method on Löwenstein-Jensen (LJ) medium according to German guidelines (18, 19) and by using the Bactec 960 MGIT system (for concentrations, see Table 1). While three strains had MICs below the critical concentrations, one strain had MICs identical to both critical concentrations (40 and 1 μg/ml) (Table 1 and Fig. 1). However, these values still remain below the critical concentration that defines resistance of clinical importance. Since this slightly increased MIC level compared to that of strain H37Rv has been discussed as possibly relevant to treatment failure, knowledge of the MICs might be helpful.
FIG 1.
RMP MICs below the critical concentrations determined by the Bactec 960 MGIT system and the proportion method on LJ medium. Only the concentrations tested, beginning with 2.5 μg/ml, are given on the y axes. The adjusted critical concentrations for the two methods (LJ medium, 40 μg/ml; Bactec 960 MGIT, 1 μg/ml) are represented by a dashed line.
In contrast to our study, a multicenter study has described different results for the two methods (2). Of 12 strains selected on the basis of discordant results in earlier drug proficiency testing and classified as resistant or probably resistant, all were susceptible by the Bactec 960 MGIT method but only one was susceptible by the proportion method on LJ medium. In a more recent study, the two DST methods were compared using strains with different rpoB mutations (20). The authors described a high discrepancy of DST results for strains with the mutations L511P, D516Y, and L533P and several mutations at residue 526. Strains with the mutations D516Y and L533P were associated with high RMP MICs on LJ medium but tested sensitive for RMP by the Bactec 960 MGIT system. In contrast, the four strains of our study, which were detected by molecular sequencing, were classified as susceptible at the current critical concentrations independent of the technique used.
A major limitation of this study is the low number of strains, only four, with a disputed rpoB mutation. In addition, we have no relevant clinical data on, e.g., treatment regimen or outcome. Since we did not receive any additional specimens from the patients, we can only speculate that the treatment might have been successful. Poor treatment outcome for four of four patients with phenotypically INH-resistant but RMP-susceptible isolates has been described by Williamson et al. (7). With similar isolates, Ho et al. found one TB relapse case among four patients (8). Recently, among selected first-failure and relapse patients, equally poor first-line retreatment outcomes were reported for strains with common or disputed mutations (21).
So far there is little knowledge on the consequences of an infection with phenotypically INH-resistant but RMP-susceptible strains that have disputed rpoB mutations. Larger studies including clinical data and based on strains which are not preselected on clinical outcome are needed. Yet, from our data, we conclude that the liquid Bactec 960 MGIT system and the proportion method on LJ medium are equally reliable for phenotypic RMP DST and MIC determination.
ACKNOWLEDGMENTS
This research project was supported by the Bundesministerium für Gesundheit (German Federal Ministry of Health).
We thank Kirsten Ott, Daniela Stephan, Margit Kernbach, and Ilse Radzio for technical assistance.
Footnotes
Published ahead of print 21 October 2013
REFERENCES
- 1.Zhang Y, Yew WW. 2009. Mechanisms of drug resistance in Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 13:1320–1330 [PubMed] [Google Scholar]
- 2.Van Deun A, Barrera L, Bastian I, Fattorini L, Hoffmann H, Kam KM, Rigouts L, Rüsch-Gerdes S, Wright A. 2009. Mycobacterium tuberculosis strains with highly discordant rifampin susceptibility test results. J. Clin. Microbiol. 47:3501–3506. 10.1128/JCM.01209-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drobniewski F, Nikolayevskyy V, Balabanova Y, Bang D, Papaventsis D. 2012. Diagnosis of tuberculosis and drug resistance: what can new tools bring us? Int. J. Tuberc. Lung Dis. 16:860–870. 10.5588/ijtld.12.0180 [DOI] [PubMed] [Google Scholar]
- 4.Hillemann D, Rüsch-Gerdes S, Richter E. 2007. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J. Clin. Microbiol. 45:2635–2640. 10.1128/JCM.00521-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hillemann D, Richter E, Rüsch-Gerdes S. 2006. Use of the BACTEC Mycobacteria Growth Indicator Tube 960 automated system for recovery of mycobacteria from 9,558 extrapulmonary specimens, including urine samples. J. Clin. Microbiol. 44:4014–4017. 10.1128/JCM.00829-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Telenti A, Honoré N, Bernasconi C, March J, Ortega A, Heym B, Takiff HE, Cole ST. 1997. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J. Clin. Microbiol. 35:719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williamson DA, Roberts SA, Bower JE, Vaughan R, Newton S, Lowe O, Lewis CA, Freeman JT. 2012. Clinical failures associated with rpoB mutations in phenotypically occult multidrug-resistant Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 16:216–220. 10.5588/ijtld.11.0178 [DOI] [PubMed] [Google Scholar]
- 8.Ho J, Jelfs P, Sintchencko V. 2013. Phenotypically occult multidrug-resistant Mycobacterium tuberculosis: dilemmas in diagnosis and treatment. J. Antimicrob. Chemother. 68:2915–2920. 10.1093/jac/dkt284 [DOI] [PubMed] [Google Scholar]
- 9.Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. 2009. Tuberculosis drug resistance mutation database. PLoS Med. 6:e1000002. 10.1371/journal.pmed.1000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan RCY, Hui M, Chan EWC, Au TK, Chin ML, Yip CK, AuYeang CKW, Yeung CYL, Kam KM, Yip PCW, Cheng AFB. 2007. Genetic and phenotypic characterization of drug-resistant Mycobacterium tuberculosis isolates in Hong Kong. J. Antimicrob. Chemother. 59:866–873. 10.1093/jac/dkm054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sajduda A, Brzostek A, Poplawska M, Augustynowicz-Kopec E, Zwolska Z, Niemann S, Dziadek J, Hillemann D. 2004. Molecular characterization of rifampin- and isoniazid-resistant Mycobacterium tuberculosis strains isolated in Poland. J. Clin. Microbiol. 42:2425–2431. 10.1128/JCM.42.6.2425-2431.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenner L, Egger M, Bodmer T, Altpeter E, Zwahlen M, Jaton K, Pfyffer GE, Borrell S, Dubuis O, Bruderer T, Siegrist HH, Furrer H, Calmy A, Fehr J, Stalder JM, Ninet B, Böttger EC, Gagneux S. 2012. Effect of mutation and genetic background on drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 56:3047–3053. 10.1128/AAC.06460-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feuerriegel S, Oberhauser B, George AG, Dafae F, Richter E, Rüsch-Gerdes S, Niemann S. 2012. Sequence analysis for detection of first-line drug resistance in Mycobacterium tuberculosis strains from a high-incidence setting. BMC Microbiol. 12:90. 10.1186/1471-2180-12-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Traore H, Ogwang S, Mallard K, Joloba ML, Mumbowa F, Narayan K, Kayes S, Jones-Lopez EC, Smith PG, Ellner JJ, Mugerwa RD, Eisenach KD, McNerney R. 2007. Low-cost rapid detection of rifampicin resistant tuberculosis using bacteriophage in Kampala, Uganda. Ann. Clin. Microbiol. Antimicrob. 6:1. 10.1186/1476-0711-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohno H, Koga H, Kohno S, Tashiro T, Hara K. 1996. Relationship between rifampin MICs for and rpoB mutations of Mycobacterium tuberculosis strains isolated in Japan. Antimicrob. Agents Chemother. 40:1053–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hillemann D, Kubica T, Rüsch-Gerdes S, Niemann S. 2005. Disequilibrium in distribution of resistance mutations among Mycobacterium tuberculosis Beijing and non-Beijing strains isolated from patients in Germany. Antimicrob. Agents Chemother. 49:1229–1231. 10.1128/AAC.49.3.1229-1231.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huitric E, Werngren J, Juréen P, Hoffner S. 2006. Resistance levels and rpoB gene mutations among in vitro-selected rifampin-resistant Mycobacterium tuberculosis mutants. Antimicrob. Agents Chemother. 50:2860–2862. 10.1128/AAC.00303-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deutsche Institut für Normung E. V 2009. DIN 58943–8, Medical microbiology—diagnosis of tuberculosis. Part 8: Methods for the determination of susceptibility of tubercle bacilli to chemotherapeutic agents. [Google Scholar]
- 19.Canetti G, Fox W, Khomenko A, Mahler HT, Menon NK, Mitchison DA, Rist N, Smelev NA. 1969. Advances in techniques of testing mycobacterial drug sensitivity, and the use of sensitivity tests in tuberculosis control programmes. Bull. World Health Organ. 41:21–43 [PMC free article] [PubMed] [Google Scholar]
- 20.Rigouts L, Gumusboga M, de Rijk WB, Nduwamahoro E, Uwizeye C, de Jong B, Deun AV. 2013. Rifampicin resistance missed in automated liquid culture system for Mycobacterium tuberculosis with specific rpoB mutations. J. Clin. Microbiol. 51:2641–2645. 10.1128/JCM.02741-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Deun A, Aung KJM, Bola V, Lebeke R, Hossain MA, de Rijk WB, Rigouts L, Gumusboga A, Torrea G, de Jong BC. 2013. Rifampin drug resistance tests for tuberculosis: challenging the gold standard. J. Clin. Microbiol. 51:2633–2640. 10.1128/JCM.00553-13 [DOI] [PMC free article] [PubMed] [Google Scholar]