Abstract
Human fungal infections represent a therapeutic challenge. Although effective strategies for treatment are available, resistance is spreading, and many therapies have unacceptable side effects. A clear need for novel antifungal targets and molecules is thus emerging. Here, we present the identification and characterization of the plant-derived diyne-furan fatty acid EV-086 as a novel antifungal compound. EV-086 has potent and broad-spectrum activity in vitro against Candida, Aspergillus, and Trichophyton spp., whereas activities against bacteria and human cell lines are very low. Chemical-genetic profiling of Saccharomyces cerevisiae deletion mutants identified lipid metabolic processes and organelle organization and biogenesis as targets of EV-086. Pathway modeling suggested that EV-086 inhibits delta-9 fatty acid desaturation, an essential process in S. cerevisiae, depending on the delta-9 fatty acid desaturase OLE1. Delta-9 unsaturated fatty acids—but not saturated fatty acids—antagonized the EV-086-mediated growth inhibition, and transcription of the OLE1 gene was strongly upregulated in the presence of EV-086. EV-086 increased the ratio of saturated to unsaturated free fatty acids and phosphatidylethanolamine fatty acyl chains, respectively. Furthermore, EV-086 was rapidly taken up into the lipid fraction of the cell and incorporated into phospholipids. Together, these findings demonstrate that EV-086 is an inhibitor of delta-9 fatty acid desaturation and that the mechanism of inhibition might involve an EV-086–phospholipid. Finally, EV-086 showed efficacy in a guinea pig skin dermatophytosis model of topical Trichophyton infection, which demonstrates that delta-9 fatty acid desaturation is a valid antifungal target, at least for dermatophytoses.
INTRODUCTION
Healthy individuals are remarkably resistant to fungal infections. However, predisposing factors, such as age, immunosuppression, and underlying disease, weaken the immune system and render the host susceptible to fungal infections. Humid conditions and occlusive clothing further favor fungal growth on the skin. Fungal disease is therefore widespread and manifests in a spectrum from relatively mild dermatophytoses to serious systemic infections (1).
Chemotherapy has proven to be a highly effective strategy for the treatment of fungal infections. The majority of agents applied for the treatment of dermatophytoses are from the allylamine and azole family. Triazoles, polyenes, and echinocandins are currently the main classes used to treat systemic fungal infections. Both the allylamines and the azoles inhibit the biosynthesis of ergosterol, an essential membrane component crucial for the physico-chemical properties of the membrane. Polyenes do not target the ergosterol biosynthetic pathway but directly bind to membrane sterols. The echinocandins target beta-glucan synthesis, which is essential for cell wall biosynthesis (2–4).
Even though current chemotherapeutic strategies for the treatment of human fungal infections are effective, the widespread use of antifungal compounds has generated resistance, and many therapies are now losing efficacy (5, 6). A clear need for novel antifungal targets and molecules is therefore emerging. Ideally, antimicrobial molecules act multifactorially, inhibiting different essential processes. This ensures fast and efficient clearance of the pathogen while minimizing the risk of resistance development. Remarkably, the azoles, polyenes, and allylamines all target either ergosterol directly or its biosynthesis, which eventually impairs membrane-associated processes and adaptation to the environment. Membrane components such as ergosterol are therefore attractive antifungal targets (7).
Here, we present the identification of the plant derived diyne-furan fatty acid EV-086 as a novel and specific antifungal compound. Chemical-genetic profiling and transcript analysis revealed that EV-086 targets delta-9 fatty acid desaturation, an essential metabolic step in the biosynthesis of unsaturated fatty acids. EV-086 thus interferes with the biosynthesis of membrane components similar as do the azoles, polyenes, and allylamines, but via a different mechanism. The efficacy of EV-086 was validated in vivo in a guinea pig skin dermatophytosis model of fungal infection.
MATERIALS AND METHODS
Isolation of metabolites with antifungal activity from plant cell cultures.
A library of purified plant metabolites was screened for compounds with antifungal activity. The library was prepared from plant cell cultures as described previously (8, 9). Briefly, callus cultures were established on agar and then grown in suspension culture. The suspension cultures were subjected to a series of treatments to activate secondary metabolic pathways. About 40,000 different metabolites were purified from more than 2,000 plant cell cultures by using preparative high-performance liquid chromatography. The isolated compounds were then screened for antifungal activity by broth microdilution at final concentrations of 200, 100, 50, 10, and 1 μg/ml as described previously (10). In each well of a 96-well plate, 5 μl of test sample in dimethyl sulfoxide (DMSO) was combined with 95 μl Candida albicans SC5314 (11) suspension in RPMI medium (10.4 g/liter; R6504 buffered at pH 7 with 0.165 M morpholinepropanesulfonic acid; Sigma), resulting in a final suspension of 5 × 103 cells/ml. The plates were shaken and incubated at 35°C for 24 h. A 25-μl aliquot of 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide salt (XTT) stock solution was added to each well, and the plate was incubated on a shaker for another 2 h at 35°C. Absorption was measured at 470 nm in a spectrometer, and the 50% inhibitory concentation (IC50) was determined by using nonlinear least square fitting.
A metabolite with an IC50 below 1 μg/ml was identified from a plant cell culture of Anarrhinum bellidifolium and designated EV-086. Detailed culture conditions and extraction procedures for the isolation of EV-086 were as follows. Plant seeds were sterilized, and seeds were placed on seed germination medium (modified after Gamborg's B5 recipe [12]; 1% sucrose, no hormones, and 10 mg/liter propiconazole). Seedlings were chopped into small pieces and placed on solidified callus induction medium (modified after Gamborg's B5 recipe, containing 1 mg/liter 2,4-dichlorophenoxyacetic acid, 0.1 mg/liter kinetin, 100 ml/liter coconut water, and 2% sucrose). Portions of seedling explant and callus were placed in suspension medium (modified after Gamborg's B5 recipe containing 1 mg/liter 2,4-dichlorophenoxyacetic acid, 0.1 mg/liter kinetin, 100 ml/liter coconut water, and 3% sucrose) and replenished at 14-day intervals. Conical flasks containing 750 ml of suspension medium were inoculated with 250 ml of 14-day-old suspension culture. After 7 days of growth, filter-sterilized methyl jasmonate (250 μM, final concentration) and an autoclaved C. albicans preparation (50 mg/liter, final concentration) were added for defense system induction. Cell cultures were incubated for 6 days, harvested by vacuum filtration, and freeze-dried. Freeze-dried cell biomass was extracted with methylene chloride-methanol (1:1). The dried extract was absorbed and fractionated on a Diaion HP-20 column (catalog number 13606; inner diameter of 2.5 cm, 25 cm deep; Sigma) and eluted with H2O, 20% methanol in H2O, methanol, and acetone. The structure of EV-086 was elucidated by mass spectrometry and nuclear magnetic resonance.
Susceptibility testing of yeasts and filamentous fungi.
MICs were determined by broth microdilution according to the Clinical and Laboratory Standards Institute (CLSI) reference methods for antifungal susceptibility testing of yeasts and filamentous fungi (13, 14). Briefly, EV-086 was dissolved in ethanol, and serial 2-fold dilutions of 100× concentrated compound were further diluted 1/50 in RPMI medium. Aliquots of 100 μl of cell or conidia suspensions (2,000 cells or conidia/ml) were added to 96-well plates, and 100 μl of test compound diluted in RPMI medium was added. The endpoint was complete growth inhibition (optically clear), determined by visual inspection of growth wells. Saccharomyces cerevisiae strains were tested in synthetic complete (SC) medium (15). Checkerboard assays combining test compounds with fatty acids were set up accordingly. EV-086 and fatty acids were prepared as 4-fold dilutions in RPMI medium. Fifty-microliter aliquots were combined in 96-well plates and inoculated with 100 μl cell or conidia suspension.
Viability of C. albicans in response to EV-086 was determined in a time-kill kinetic analysis. C. albicans ATCC 24433 was inoculated at 1,000 cells/ml in RPMI medium. Viability of the cells was analyzed before compound addition and after 1 h, 2 h, 4 h, 8 h, 24 h, and 48 h with compound by plating 100 μl of culture on potato dextrose agar plates (PDA; Difco catalog number 213300). Dilutions in saline were made when cultures reached higher densities. Plates were incubated for 2 days, and culture cell densities were calculated based on the CFU. A carryover effect was negligible up to 500 ng/ml EV-086 in culture.
Susceptibility testing of bacteria.
The MICs were determined according to the CLSI reference method for dilution antimicrobial susceptibility tests for bacteria (16). Briefly, EV-086 was dissolved in ethanol, and serial 2-fold dilutions of 100× concentrated compounds were further diluted 1/50 in Mueller-Hinton broth. Cell suspension aliquots (100 μl; 106 cells/ml) were added to 96-well plates, and 100 μl of diluted test compound was added. The endpoint was complete growth inhibition (optically clear), determined by visual inspection of growth wells.
Mammalian cytotoxicity assay.
Cell viability was tested with a colorimetric assay that measured metabolic activity (17). Briefly, HepG2 and HeLa cells were grown at 37°C in 95% oxygen and 5% CO2 in Dulbecco's modified Eagle medium (Sigma) supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin (Pen/Strep). Cells were seeded into 96-well plates in medium without Pen/Strep at a density of 2 × 104 (HepG2) or 104 (HeLa) cells per well and incubated for 1 day without test compound. Serial 3-fold dilutions of EV-086 were prepared as DMSO stock solutions, diluted in medium without Pen/Strep, and added to the cell layers (1% DMSO, final concentration). Cells were incubated for 2 days with compound. Fifty microliters of XTT stock solution was added to each well. After 4 h, absorption at 470 nm was determined as the measure of viability. Absorption of control wells was subtracted from all measurements. Nonlinear least square fitting was applied to determine the 50% cytotoxic concentration.
Chemical-genetic profiling of Saccharomyces cerevisiae against EV-086.
A collection of 4,917 homozygous diploid BY4743-derived S. cerevisiae knockout strains served as the screening library (catalog number YSC1056; Thermo Fisher Scientific, Open Biosystems). The library strains were cultured in YPD medium (1% Bacto yeast extract, 2% Bacto peptone, 2% dextrose) in a 96-well format, and aliquots were spotted onto YPD agar (2% agar) containing 0, 0.5, 2, or 8 μg/ml EV-086 by using a 96-pin replicator. Susceptibility to EV-086 was determined after 2 days by visual inspection of the colony diameters in comparison to those of the parent strain, BY4743 (18). Primary hit strains were recultured and verified in a secondary drop spot assay. The biological processes associated with the deleted genes were determined for the identified strains (19, 20). A hypergeometric distribution was calculated to determine significantly overrepresented biological processes in the sample compared to the population, and a Bonferroni correction was applied to correct for the family-wise error rate (21).
Transcript profiling of the OLE1 gene.
S. cerevisiae BY4743 (18) was cultured in SC medium (15). RNA was isolated by glass bead extraction from 2 × 107 yeast cells (RNeasy minikit; Qiagen). cDNA was prepared by two-step reverse transcription-PCR (RT-PCR; QuantiTect reverse transcription kit; Qiagen). cDNA levels were determined quantitatively on a Rotor-Gene Q 2plex platform (Rotor-Gene SYBR green PCR kit; Qiagen). Reactions were performed in triplicate, and the normalized relative expression was calculated using the ΔΔCT method (22). The alpha-tubulin gene (TUB1) served for normalization of cDNA. Oligonucleotides 5′-TGGGACAAACAAACCTT-3′ and 5′-CACCACCTGGATGTTCAGAG-3′ were used for amplification of the S. cerevisiae OLE1 gene, and oligonucleotides 5′-CCAGATGGTCAAGTGTGATCC-3′ and 5′-CAACCAATTGGACGGTCTTC-3′ were used for the amplification of the TUB1 gene (23–25). One-way analysis of variance (ANOVA) with Dunnett's post hoc test was performed using the Prism software (version 5.00 for Windows; GraphPad Software, San Diego CA).
S. cerevisiae lipid extraction and analysis.
To analyze relative levels of unsaturated and saturated free fatty acids and molecular species of phospholipids, lipids were extracted and analyzed as described previously (26). Briefly, cultured S. cerevisiae cells were collected by centrifugation, resuspended in 150 mM ammonium bicarbonate (pH 8), and disrupted with 0.5-mm zirconia beads (catalog number D1032-05; Benchmark Scientific, Edison, NJ). Disrupted yeast cells were extracted with chlorophorm-methanol (17:1, vol/vol) and analyzed by mass spectrometry on a Brucker Esquire HCT ion trap mass spectrometer (electrospray ionization) with a flow rate of 250 μl/h and a capillary tension of −250 V. Ion fragmentation was induced with argon as the collision gas at a pressure of 800 Pa. To compare uptake and incorporation of EV-086 to those of palmitic acid, cells were cultivated in the presence of 5 μg/ml of either [14C]EV-086 or [14C]palmitic acid, both with a specific activity of 0.08 μCi/ml. At each time point, 10 optical density (OD) units of cells were withdrawn. Lipids were extracted with chlorophorm-methanol (1:1) from the cell pellet and the culture supernatant and subjected to scintillation counting. Aliquots were dried and separated by thin-layer chromatography (TLC) on silica gel 60 plates (Merck, Darmstadt, Germany) by using the solvents CHCl-methanol-acetic acid (65/25/8, vol/vol/vol). The TLC was dried and exposed to a phosphorimager screen, and the screen was developed using a Typhoon FLA 9500 imager (GE Healthcare, GmbH, Europe).
Guinea pig skin dermatophytosis model of infection.
The efficacy of EV-086 was determined in guinea pig model of Trichophyton menagrophytes skin dermatophytosis as previously described (27). Briefly, T. mentagrophytes ATCC 24953 was prepared as a suspension of 2 × 108 conidia/ml. Male albino guinea pigs (Harlan-Sprague-Dawley, San Diego, CA) were randomly assigned to five different experimental groups with 10 animals in each group. Square areas (3 cm by 3 cm) of skin were shaved on the animals' backs and abraded using sterile emery paper. A total of 1 × 107 T. mentagrophytes conidia were applied. The five experimental groups were treated as follows: untreated control, vehicle control (Captex 350; Abitec, Columbus, OH), 0.1% EV-086 (in Captex 350), 1% EV-086 (in Captex 350), or 1% terbinafine (Lamisil commercial formulation; Novartis) as the positive control. For mycological evaluations of the test compounds, hair samples were removed with a sterile forceps from the infected areas. Ten hair samples from each animal were inoculated, and the mycological efficacy was expressed as the percentage of mycologically positive hairs compared to the untreated control. Clinical assessments of local changes of the infected skin were scored from 0 (no findings) to 5 (extensive damage), and the clinical efficacy was expressed as a percentage relative to the result for the untreated control group. One-way ANOVA with Dunnett's post hoc test was performed using Prism software (version 5.00 for Windows; GraphPad Software, San Diego CA). All procedures in the protocol were in compliance with the Animal Welfare Act, the NRC Guide for the Care and Use of Laboratory Animals (56), and the Office of Laboratory Animal Welfare. Guinea pigs were used upon review and approval of an addendum to our existing protocol by the Institutional Animal Care and Use Committee (IACUC).
RESULTS
A plant cell culture screen for natural antifungal compounds identified the diyne-furan fatty acid EV-086.
In our search for novel natural compounds with antifungal activity, a library of purified plant cell culture metabolites was screened for compounds with activity against C. albicans. More than 40,000 purified compounds were tested for antifungal activity in an XTT-based broth microdilution assay. This screen identified a metabolite from Anarrhinum bellidifolium with a MIC of 2.5 ng/ml against the C. albicans strain ATCC 24433. The compound had a Mw of 284.35 and was designated EV-086. Spectroscopic analyses identified EV-086 as a diyne-furan substituted (Z)-9 unsaturated fatty acid [chemical name, (Z)-14-(furan-2-yl)tetradeca-9-en-11,13-diynoic acid] (Fig. 1a). Chemically synthesized EV-086 (as the potassium salt, Mw 322.44) confirmed the activity of the original molecule and was used for all further studies unless otherwise indicated. The thermodynamic aqueous solubility determined for EV-086 (potassium salt) was above 96.5 mg/ml (28). The calculated average logP of EV-086 was 4.98 ± 0.92 (29).
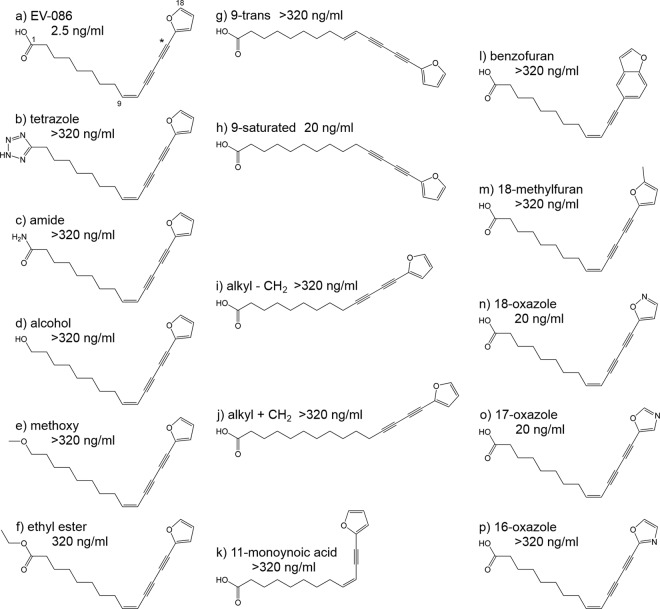
FIG 1.
Structures and activities of EV-086 and analogues. (a) EV-086 as the free acid. Numbering of the carbon atoms starts at the carboxylic acid carbon, with 1, and ends in the furan ring, with 18. The position of the [14C]carbon in the labeled EV-086 is indicated with an asterisk at position 14. (b to p) Analogues of EV-086. Activities are given as MICs against the C. albicans ATCC 24433 reference strain.
EV-086 is a selective, broad-spectrum antifungal compound with potent activity in vitro.
The MIC of EV-086 was determined against a panel of reference yeasts and filamentous fungi relevant for systemic and topical human fungal infections by using CLSI reference methods (13, 14). Fluconazole, amphotericin B, and terbinafine served as comparators (Table 1). EV-086 was highly potent against Candida spp., with MICs of 2.5 ng/ml for C. albicans and 80 ng/ml for C. glabrata. Similar high potency was observed for Trichophyton spp., with an MIC of 8 ng/ml for T. mentagrophytes. EV-086 was also active against Aspergillus fumigatus, with a MIC of 640 ng/ml. EV-086 showed only low activities against Candida parapsilosis, Aspergillus flavus, and Aspergillus niger. There was no major difference in activity between the potassium salt and the free acid of EV-086 as determined for five different Candida species. The MIC of EV-086 against the diploid S. cerevisiae laboratory strain BY4743 was 64 ng/ml, and it was 16 ng/ml for the haploid BY4742 and BY4741 strains.
TABLE 1.
MICs of EV-086 and comparators against different fungal reference strains and S. cerevisiae laboratory strains
| Species and strain(s) | Incubation temp and time | MIC (ng/ml) |
||||
|---|---|---|---|---|---|---|
| EV-086 potassium salt | EV-086 free acid | FLC | AMB | TRB | ||
| C. albicans | ||||||
| ATCC 24433 | 35°C, 2 days | 2.5 | 2.5 | 400 | 1,000 | — |
| SC5314 | 35°C, 2 days | 2.5 | — | — | — | — |
| C. glabrata ATCC 90030 | 35°C, 2 days | 80 | 40 | 200 | 1,000 | — |
| C. krusei ATCC 6258 | 35°C, 2 days | 40 | 40 | 1,600 | 2,000 | — |
| C. tropicalis ATCC 750 | 35°C, 2 days | 5 | 5 | 200 | 2,000 | — |
| C. parapsilosis ATCC 22019 | 35°C, 2 days | 10,240 | 10,240 | 50 | 1,000 | — |
| A. fumigatus ATCC MYA-3626 | 35°C, 2 days | 640 | — | — | 1,000 | — |
| A. flavus ATCC MYA-3631 | 35°C, 2 days | >5,120 | — | — | 1,000 | — |
| A. niger ATCC 10578 | 35°C, 2 days | >5,120 | — | — | 1,000 | — |
| A. terreus ATCC MYA-3633 | 35°C, 2 days | 80 | — | — | — | |
| T. rubrum ATCC 4438 | 35°C, 4 days | 4 | — | — | >512 | |
| T. mentagrophytes strains | ||||||
| ATCC 4439 | 35°C, 4 days | 8 | — | — | 8 | |
| ATCC 24953 | 35°C, 4 days | 4 | — | — | — | |
| S. cerevisiae strains | ||||||
| BY 4741 | 30°C, 1 day | 16 | — | — | — | |
| BY 4742 | 30°C, 1 day | 16 | — | — | — | |
| BY 4743 | 30°C, 1 day | 64 | — | — | — | |
FLC, fluconazole; AMB, amphotericin B; TRB, terbinafine; —, not determined.
The effect of EV-086 on the viability of C. albicans ATCC 24433 was investigated in a time-kill kinetic assay. Cell suspensions were incubated in shaking flasks, and CFUs were determined during a 48-h incubation period with agar plating (Fig. 2). The reference MICs against the C. albicans assay strain ATCC 24433 were 2.5 ng/ml for EV-086, 400 ng/ml for fluconazole, and 1000 ng/ml for amphotericin B. A reduction of CFUs was observed after ≥4 h and at ≥5 ng/ml EV-086. After an incubation period of 24 h, EV-086 reduced the CFUs by >10-fold and >100-fold at 20 and 320 ng/ml, respectively. Fluconazole at 16 μg/ml showed an increase in the CFUs of >137-fold after 24 h, while the CFUs for amphotericin B were below the limit of detection at this concentration.
FIG 2.
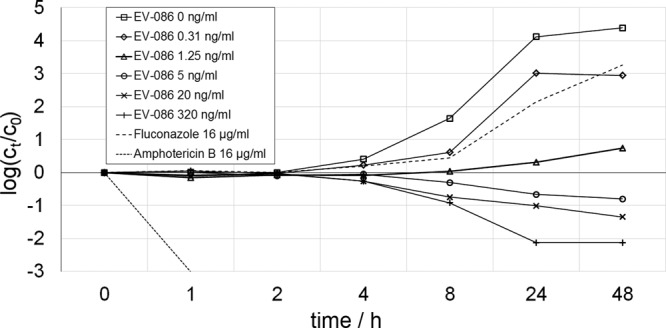
Time-kill kinetics of EV-086 and comparators against C. albicans ATCC 24433. C. albicans growth is given as the log CFU at time t (ct) versus the CFU at time zero (c0). Compound concentrations tested in cultures and corresponding symbols are shown in the key.
The activity of EV-086 was selective against fungi. Only very high concentrations inhibited the growth of mammalian cells or bacteria. The cytotoxic concentration of EV-086 against HeLa and HepG2 cells was >40 μg/ml. The MICs against Staphylococcus aureus, Enterococcus faecalis, and Escherichia coli were 50, 12.5, and >200 μg/ml, respectively.
In summary, EV-086 showed a broad-spectrum activity that was selective against fungi. EV-086 was equally potent or more potent compared to the reference compounds fluconazole, amphotericin B, and terbinafine for most of the tested fungi, and EV-086 reduced viability of the cells in the time-kill kinetic assay.
Evaluation of EV-086 analogues revealed a narrow structure-activity relationship.
A primary structure-activity relationship (SAR) was determined to characterize the functional groups in EV-086. Fifteen analogues of EV-086 were synthesized, and the activities were determined as a measure of C. albicans in vitro susceptibility (Fig. 1).
Modifications of the carboxylic acid group were not tolerated. EV-086–tetrazole, which mimics the electron distribution of a carboxylic acid, EV-086–amide, –alcohol, –methoxy, or –ethylester were inactive, with >100-fold decreased potency compared to the parent compound EV-086 (Fig. 1a to f). A trans conformation at carbon C-9 abolished the activity, whereas a saturation of the C-9–C-10 bond was tolerated, resulting in a MIC of 20 ng/ml for the EV-086–9–saturated analogue (Fig. 1g and h). Alkyl chain length modifications at the carboxylic acid terminus were not tolerated. Similarly, the removal of one triple bond inactivated the molecule (Fig. 1i to k). Mimicking the diyne-furan moiety with a benzofuran also inactivated the molecule (Fig. 1l). Certain modifications in the furan ring were tolerated. Oxazole analogues with a nitrogen substitution at the C-18 or C-17 carbons resulted in MICs of 20 ng/ml, whereas a C-16 substitution or the 18-methylfuran modifications were inactive (Fig. 1m to p).
Together, these data suggest a narrow structure-activity relationship for the EV-086 class of molecules. The distance between carbon atoms C-1 and C-9 cannot be changed. The C-9–C-10 carbons must not be in the trans configuration, whereas the cis and saturated forms are active. A minor degree of freedom was found for the furan part of the molecule, whereas the diyne moiety did not accept changes.
Chemical-genetic profiling of yeast identified lipid metabolism and organelle organization and biogenesis as targets of EV-086.
Chemical-genetic profiling of S. cerevisiae knockout mutants was carried out to identify the target pathway of EV-086 (Fig. 3a). Briefly, 4,917 homozygous diploid S. cerevisiae strains, each with one defined open reading frame (ORF) deleted (30), were individually cultured and replica plated on agar plates containing a permissive (0.5 μg/ml), semipermissive (2 μg/ml), or restrictive (8 μg/ml) concentration of EV-086. Plates were incubated and colony growth recorded. Strains hypersensitive to EV-086 were verified in a secondary drop spot assay (Fig. 3b and c). This revealed 44 genes that rendered S. cerevisiae hypersensitive to EV-086 when deleted. The S. cerevisiae GO-Slim vocabulary was used to map the identified genes to higher-level biological processes (Table 2) (19, 20). The 44 identified genes mapped to 24 out of 39 available biological processes. A significant overrepresentation was determined for lipid metabolic process (P < 0.004) and organelle organization and biogenesis (P < 0.04).
FIG 3.
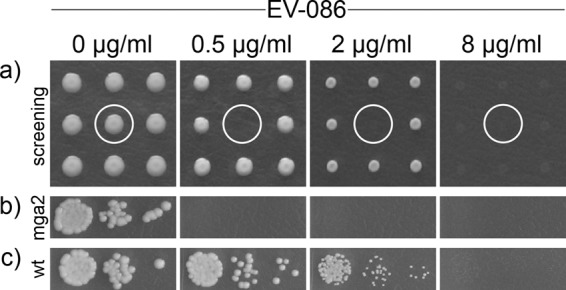
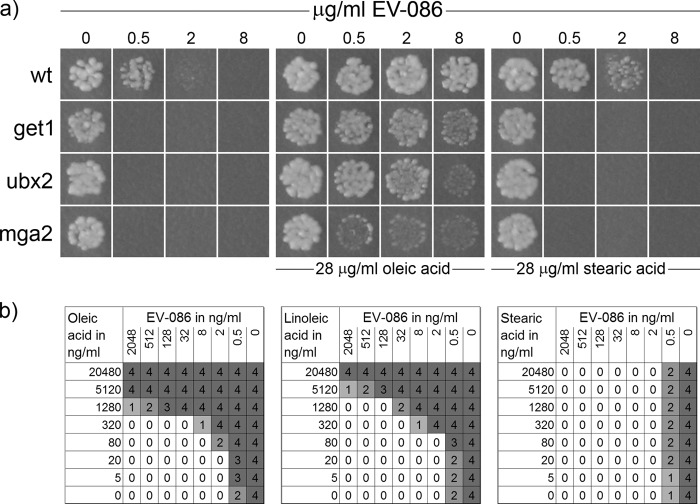
Chemical-genetic profiling of the S. cerevisiae knockout mutant collection. The concentrations of EV-086 in the YPD agar test plates are indicated, with 0.5 μg/ml as permissive, 2 μg/ml as semipermissive, and 8 μg/ml as restrictive. (a) Example frame of the primary screening, showing 9 different knockout strains. The white circle highlights a hypersensitive deletion strain (mga2 deletion). (b and c) Hypersensitive deletion strain mga2 (b) and the wild-type strain (BY4743) (c) in secondary drop spot assays. The frames in panels b and c show the 3-step 10-fold serial dilution of cell suspensions spotted on agar plates. Growth of the hypersensitive deletion strain (b) was inhibited at 0.5 μg/ml EV-086, whereas the wild-type strain (c) showed about 50% growth at 2 μg/ml.
TABLE 2.
Deleted genes and associated biological processes of the S. cerevisiae knockout mutants hypersensitive to EV-086
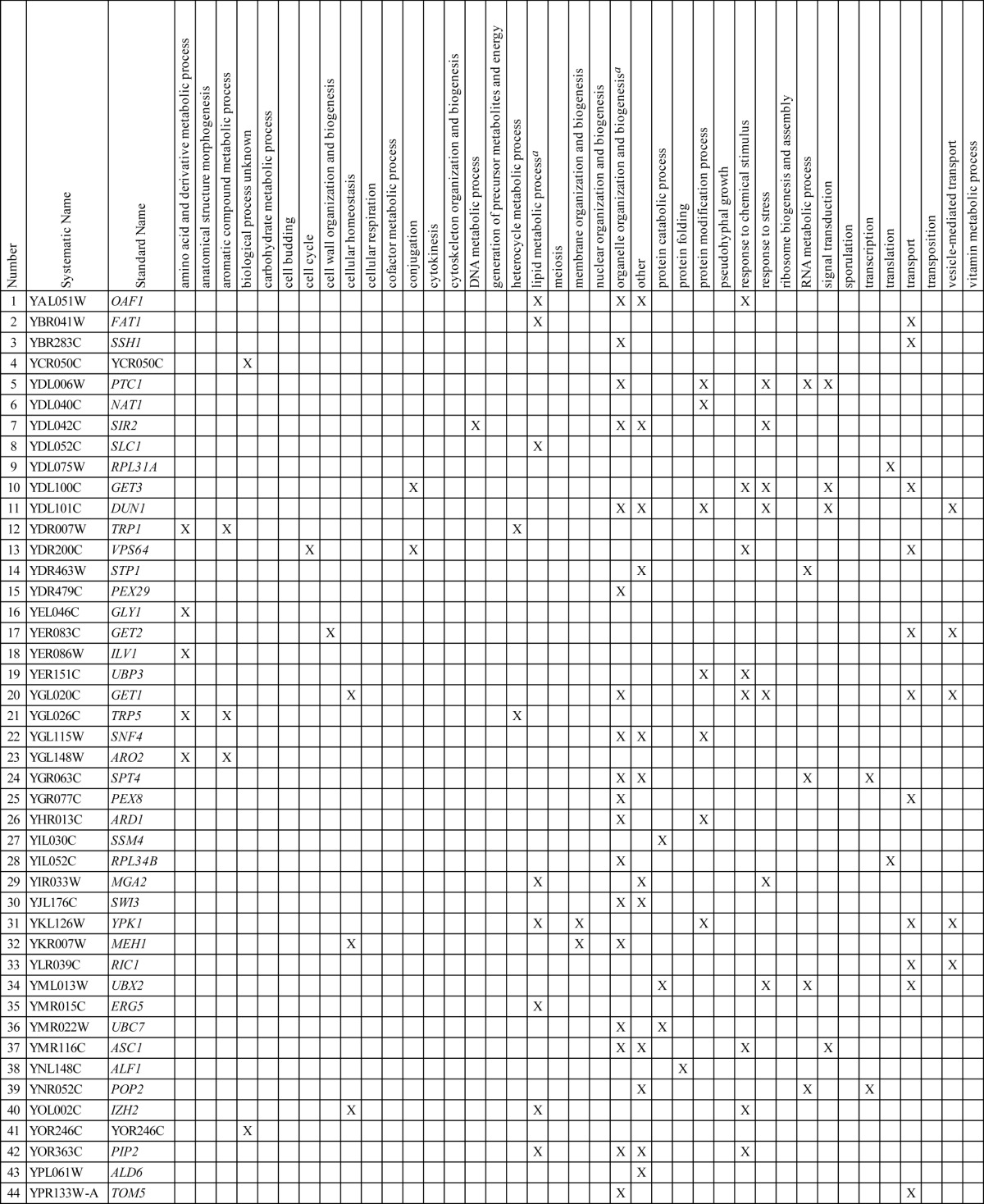
Significantly overrepresented biological process (P < 0.05). Genes in the category “biological process unknown” were significantly underrepresented.
Assembly of a putative pathway as the target of EV-086.
Based on the identified genes, associated biological processes, and data in the literature, we assembled putative pathways as targets of EV-086. Most promising was a pathway controlling delta-9 fatty acid desaturation. In this pathway, the genes GET1, GET2, GET3, SSM4, UBX2, UBC7, and MGA2, which were identified in the chemical-genetic profiling, are included (Fig. 4).
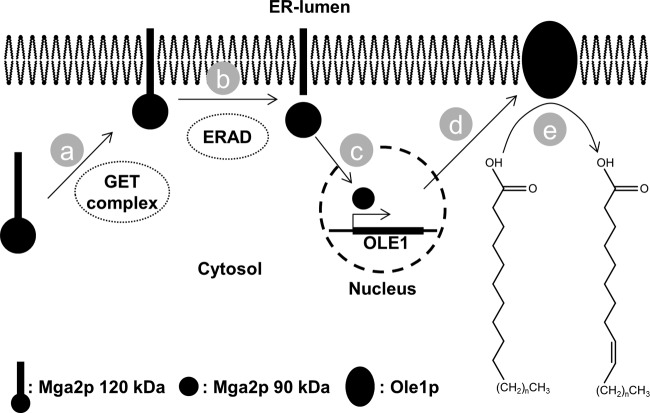
FIG 4.
Putative signaling pathway controlling delta-9 fatty acid desaturase activity. (a) The GET complex directs the putative tail-anchored protein Mga2 to the ER membrane. (b) Membrane-bound Mga2p is proteolytically cleaved in an ERAD-dependent process to release the soluble 90-kDa form from its 120-kDa membrane-bound form. (c) The soluble 90-kDa form of Mga2p is shuttled to the nucleus, where it activates transcription of the OLE1 gene. (d) Transcribed OLE1 is translated into Ole1p, which is targeted to the ER membrane. (e) Ole1p catalyzes delta-9 fatty acid desaturation. n = number of CH2 units; 5 for palmitic and palmitoleic acid; 7 for stearic and oleic acid.
As reported in the literature, the GET complex, consisting of Get1p, Get2p, and Get3p, mediates insertion of tail-anchored proteins into the endoplasmic reticulum (ER) membrane (31). Furthermore, the transcription factor Mga2p has been reported to localize to the ER membrane (32). Sequence analysis predicts a C-terminal transmembrane domain in Mga2p (33), and multiple genetic interactions between MGA2 and GET1, GET2, or GET3 have been reported (34–37). We therefore hypothesized that the GET complex also mediates insertion of Mga2p into the ER membrane. It has further been reported that the 120-kDa ER-associated form of Mga2p is proteolytically processed to a soluble 90-kDa form in a Cdc48p-dependent ER-associated protein degradation (ERAD) process (32). Ubx2p is a bridging factor that links Cdc48p to Ssm4p, Ssm4p is a membrane-associated ubiquitin protein ligase, and Ubc7p is a ubiquitin-conjugating enzyme (38, 39). We therefore hypothesized that Ubx2p, Ssm4p, and Ubc7p are involved in an ERAD-dependent proteolysis of Mga2p. Indeed, both UBX2 and SSM4 genetically interact with MGA2 (24, 44), and Spt23p, a functional homologue of Mga2p in S. cerevisiae, has been identified as a Ubx2p substrate (40). Finally, it has been well described that the soluble 90-kDa Mga2p shuttles to the nucleus, where it induces transcription of the OLE1 gene, which encodes the delta-9 fatty acid desaturase Ole1p, an essential gene in S. cerevisiae (41). Ole1p subsequently converts saturated fatty acyl-coenzyme A (CoA) substrates to monounsaturated fatty acid species by an oxygen-dependent mechanism (32).
In summary, the identified genes from the chemical-genetic profiling in the context of the data in the literature allowed us to assemble a putative pathway as the possible target of EV-086. From this work, we propose delta-9 fatty acid desaturation as the target of EV-086 and an involvement of the GET complex and ERAD in Mga2p-dependent transcriptional activation of OLE1.
EV-086 targets delta-9 fatty acid desaturation.
In order to test if EV-086 targets delta-9 fatty acid desaturation, we investigated whether delta-9 unsaturated fatty acids antagonized the growth-inhibitory effect of EV-086 in S. cerevisiae and C. albicans (Fig. 5).
FIG 5.
Antagonism between EV-086 and delta-9 unsaturated fatty acids. (a) Wild-type (wt) S. cerevisiae BY4743 and get1, ubx2, and mga2 deletion strains (BY4743 background) were prepared as suspensions and spotted onto YPD agar plates containing different concentrations of EV-086. Oleic acid and stearic acid were added to agar plates at the indicated concentrations. (b) Serial dilutions of EV-086 were combined with oleic acid, linoleic acid, or stearic acid in a checkerboard setup and inoculated with C. albicans ATCC 24433. Growth was recorded by visual inspection according to the following scale: 0, optically clear; 1, slightly hazy; 2, prominent decrease (50%) in visible growth; 3, slight reduction in visible growth; 4, no reduction in visible growth.
S. cerevisiae was grown in the presence of EV-086 supplemented with oleic acid or stearic acid. Indeed, oleic acid but not stearic acid antagonized EV-086 in wild-type S. cerevisiae. In the presence of 28 μg/ml oleic acid, the inhibitory effect of up to 8 μg/ml EV-086 was antagonized. Further, oleic acid complemented the EV-086-dependent growth defect of get1, ubx2, and mga2 deletion mutants, representing the GET complex, ERAD function, and OLE1 transcription, respectively. The growth defects in these mutants, however, were not fully complemented, most likely due to other functions of these genes (Fig. 5a). We further demonstrated an antagonism between EV-086 and oleic acid for C. albicans and T. mentagrophytes (Fig. 5b and data not shown). The oleic acid antagonism was also confirmed for the analogues EV-086–9-saturated, –18-oxazole, and –17-oxazole in C. albicans (Fig. 1 and data not shown).
Together, these data showed that EV-086 targets delta-9 fatty acid desaturation and that the target is conserved at least in S. cerevisiae, C. albicans, and T. mentagrophytes.
EV-086 induces a transcriptional upregulation of OLE1.
Based on our proposed pathway, we wanted to investigate at which level EV-086 targets delta-9 fatty acid desaturation: either at the Mga2p-dependent transcriptional control of OLE1 or posttranscriptionally. We therefore determined the expression of the OLE1 gene in EV-086-treated S. cerevisiae cells by using quantitative PCR.
S. cerevisiae BY4743 was cultured with EV-086 at concentrations of 1 and 10 μg/ml. Culture samples were taken at 10, 60, 120, and 240 min, and the relative transcription of OLE1 was determined as the fold change compared to the untreated control at 10 min (Fig. 6). A 10- to 20-fold increase in expression was observed between 60 and 240 min of incubation with 10 μg/ml EV-086 (P < 0.01), whereas the expression of OLE1 in the untreated control remained unchanged during the incubation period. At 1 μg/ml EV-086, the increase was around 5-fold (P > 0.05). Since oleic acid antagonized the growth inhibition of EV-086, the expression of OLE1 was further examined in the presence of 10 μg/ml EV-086 plus 10 μg/ml oleic acid. Under these conditions, the expression of OLE1 was reduced to the level of oleic acid alone. In analogy, the expression of OLE1 was also determined in C. albicans ATCC 24433. After 240 min, the expression increased about 5-fold compared to the untreated control (data not shown).
FIG 6.
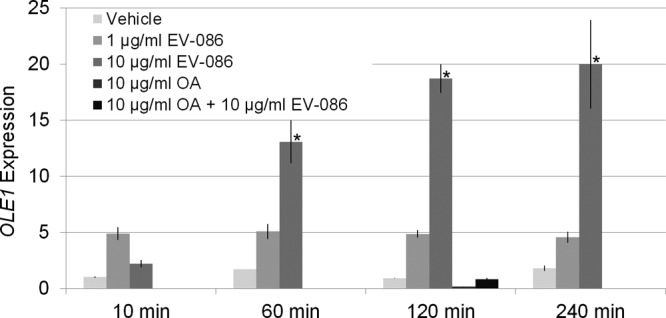
Expression of OLE1 and dependence on EV-086. Diploid S. cerevisiae BY4743 cells were inoculated in SC medium at a density of 5 × 106 cells/ml. Cultures were supplemented with ethanol (vehicle), 1 μg/ml EV-086, 10 μg/ml EV-086, 10 μg/ml oleic acid (OA), or 10 μg/ml EV-086 plus 10 μg/ml OA. Culture samples were taken at 10, 60, 120, and 240 min after compound addition. The expression of OLE1 was determined by quantitative PCR and normalized to the level of alpha-tubulin gene expression (TUB1) at 10 min without EV-086 or OA. OLE1 expression levels for 10 μg/ml OA and 10 μg/ml EV-086 plus 10 μg/ml OA were only analyzed at 120 min. *, P < 0.01 by one-way ANOVA with Dunnett's post hoc test, compared to vehicle at 10 min. Data are means ± standard errors (n = 3).
These findings suggest that EV-086 does not target delta-9 fatty acid desaturation at the Mga2p-dependent transcriptional control of OLE1 level but rather posttranscriptionally. This mechanism of inhibition is conserved between S. cerevisiae and C. albicans, and homologues of both MGA2 and OLE1 have been identified in C. albicans (42, 43).
EV-086 increases the ratio of saturated to unsaturated fatty acids.
Inhibition of delta-9 fatty acid desaturation is expected to change the saturation profile of free fatty acids and phospholipid fatty acyl chains. In order to verify EV-086's inhibitory activity against delta-9 fatty acid desaturation, the saturation degrees of free fatty acids and phosphatidylethanolamine (PE) fatty acyl chains were determined in S. cerevisiae.
Haploid wild-type S. cerevisiae BY4742 was cultured in SC medium with 10 μg/ml EV-086, and culture samples were taken after 240 min of incubation. Lipids were extracted, and relative levels of free C16 and C18 fatty acids were analyzed by mass spectrometry (Fig. 7a). For both C16 and C18 free fatty acids, EV-086 increased the ratio of saturated to unsaturated fatty acid by about 2.5- to 3-fold compared to the untreated control: the ratios of 16:0/16:1 fatty acids were 0.5 in untreated cells and 1.7 in drug-treated cells, and the ratios of 18:0/18:1 fatty acids were 1.8 in untreated cells and 4.6 in drug-treated cells (P < 0.001). Accordingly, the saturation profile of PE fatty acyl chains was analyzed (Fig. 7b). Similarly, EV-086 increased the ratio of saturated to unsaturated fatty acid acyl chains by 3- to 6-fold for PE(32:1/32:2), PE(34:1/34:2), and PE(36:1/36:2). Further, PE was shifted toward species with elevated levels of shorter fatty acyl chain substitutions.
FIG 7.
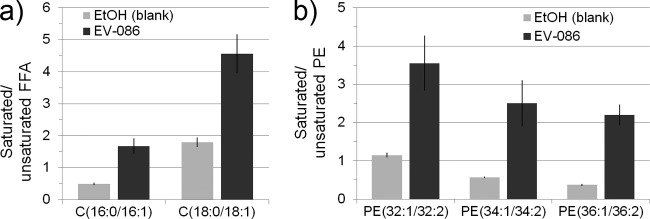
Ratio of saturated to unsaturated free fatty acids and PE fatty acyl chains of EV-086-treated S. cerevisiae cells. (a) Ratios of saturated to unsaturated free fatty acids. Fatty acid nomenclature: 16:0, palmitic acid; 16:1, palmitoleic acid; 18:0, stearic acid; 18:1, oleic acid. (b) Ratios of major PE fatty acyl chains. The ratios between saturated and unsaturated fatty acyl chains of PE species with identical sum of carbon atoms is shown. PE nomenclature: sum of carbon atoms in acyl chains:number of double bonds in acyl chains. P < 0.001 (a) and P < 0.0003 (b) by one-way ANOVA with Dunnett's post hoc test, compared to vehicle. Data are means ± standard deviations (n = 3).
These profound increases in the ratio of free saturated to unsaturated fatty acids and PE fatty acyl chains corroborate delta-9 fatty acid desaturation as the target of EV-086. Further, the shift toward PE species with elevated levels of shorter fatty acyl chains likely reflects an adaptive response to the inhibition of delta-9 fatty acid desaturation.
Radiolabeled EV-086 is incorporated into phospholipids in S. cerevisiae.
Exogenous fatty acids added to the growth medium are rapidly incorporated into phospholipids in S. cerevisiae (44). The structural conservation between EV-086 and common fatty acids suggests that EV-086 is metabolized similarly. We thus investigated whether exogenous EV-086 is incorporated into phospholipids.
Haploid S. cerevisiae BY4742 was inoculated in SC medium at a density of 5 × 106 cells/ml. [14C]EV-086 or [14C]palmitic acid was added at a concentration of 5 μg/ml, and culture samples were taken at 0, 30, 60, 120, and 240 min. Lipids were extracted from the cell pellet and the cleared supernatant. The relative incorporation of [14C]EV-086 and [14C]palmitic acid into the cellular lipid fraction was quantified by scintillation counting (Fig. 8a). [14C]EV-086 accumulated in the lipid fraction in a time-dependent manner, reaching 60% after 240 min. In comparison, [14C]palmitic acid reached about 90% after 240 min. Separation of the lipid fractions by TLC indicated that both [14C]EV-086 and [14C]palmitic acid were incorporated into phospholipids, including phosphatidylcholines, phosphatidylinositols, and phosphatidylethanolamines (Fig. 8b).
FIG 8.
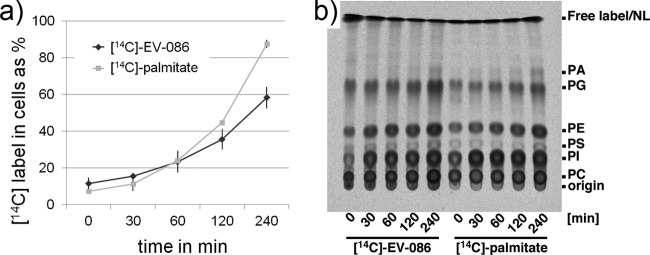
Uptake and incorporation of [14C]EV-086 into the lipid pool. (a) [14C]EV-086 and [14C]palmitate uptake, as the percentage of counts in the cellular lipid fraction versus the sum of counts in the cellular lipid fraction and the cleared culture medium. Error bars show standard deviations (n = 3). (b) TLC results for the phospholipid fraction. PC, phosphatidylcholines; PI, phosphatidylinositols; PS, phosphatidylserines; PE, phosphatidylethanolamines; PG, phosphatidylglycerols; PA, phosphatidic acids; NL, neutral lipids.
These results suggest that [14C]EV-086 is indeed recognized as a fatty acid and incorporated into the phospholipid fraction. The efficiency of uptake is 66%, compared to that of [14C]palmitic acid. This difference likely reflects not only uptake specificity but also the EV-086-mediated growth inhibition, which is not observed for palmitic acid.
EV-086 has in vivo efficacy in a guinea pig model of Trichophyton skin dermatophytosis.
EV-086 has proven broad-spectrum and potent antifungal activity in vitro. In order to investigate its potential in vivo, EV-086 was tested in a guinea pig model of Trichophyton skin dermatophytosis as previously described (27).
The activity of EV-086 against T. menagrophytes skin dermatophytosis in guinea pigs was assessed in five experimental groups: untreated control, vehicle control, 0.1% EV-086, 1% EV-086, and 1% terbinafine (Lamisil; Novartis) as the positive control. Square areas of shaved and abraded skin on the backs of the animals were infected with T. mentagrophytes ATCC 24953 on day 1. Treatment started on day 4 and lasted for 7 days, with daily applications of 0.2 ml of test product. On day 13, hair sample plugs were obtained and cultured for mycological evaluation, and local changes of the infected skin (redness, crusting, or lesions) were recorded for clinical assessment (Fig. 9).
FIG 9.
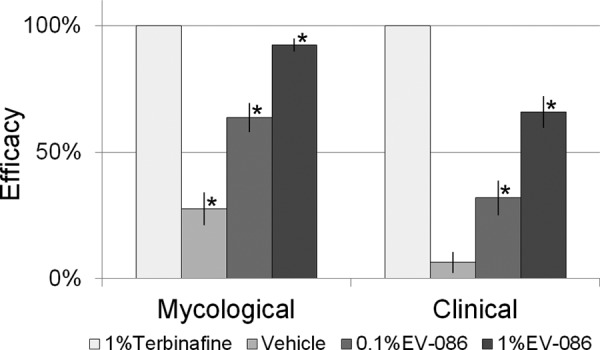
Efficacy of EV-086 in a guinea pig model of T. mentagrophytes skin dermatophytosis. The mycological and clinical efficacies of the test products are given as percentages in relation to the untreated control. *, P < 0.01 by one-way ANOVA with Dunnett's post hoc test, compared to the untreated control. Data are means ± standard errors (n ≥ 4).
At a concentration of 1% in Captex 350, EV-086 demonstrated a 92.5% mycological efficacy (reduction of fungal positive hair) and a 66% clinical efficacy compared to the untreated control (P < 0.01).
DISCUSSION
Here, we report the identification of the natural small molecule EV-086 from the plant A. bellidifolium. EV-086 is a diyne-furan fatty acid with potent and specific antifungal activity that targets delta-9 fatty acid desaturation. To our knowledge, EV-086 is a first-in-class fungal delta-9 fatty acid inhibitor. The triazolidinethiones ECC145 and ECC188 have so far been the only other published fungal delta-9 fatty acid desaturase inhibitors (45).
Chemical-genetic profiling of S. cerevisiae knockout mutants has proven to be an effective strategy for the identification of drug targets (46). Here, we screened the S. cerevisiae ORF deletion library and identified 44 genes that confer hypersensitivity to EV-086. Based on the identified genes and data in the literature, a putative pathway controlling delta-9 fatty acid desaturation was assembled as a possible target of EV-086. Whereas an involvement of the GET complex in the localization of Mga2p is hypothetical and needs further investigation, only recently has an involvement of Ubx2p in the ERAD-dependent proteolysis of Mga2p been reported (40). The roles of Ssm4p and Ubc7p in the ERAD process, however, need further investigation. Transcript profiling suggests that EV-086 targets delta-9 fatty acid desaturation not at the level of Mga2p-dependent OLE1 transcription but rather posttranscriptionally at the biosynthetic level of delta-9 fatty acid desaturation. The observed increase of OLE1 expression in response to EV-086 indicated a compensatory effect of the cell for the delta-9 unsaturated fatty acid auxotrophy. A decrease of OLE1 transcription would have been indicative of an inhibition of Mga2p or other transcriptional regulators. Ole1p would therefore be the most likely target of EV-086. However, purification and characterization of Ole1p in vitro has not been reported yet, and so far we have been unable to purify Ole1p for an enzymatic assay.
We further identified TRP1, TRP5 and ARO2 from the aromatic amino acid biosynthetic pathway, as well as other genes from amino acid biosynthetic pathways in the chemical-genetic profiling. Tryptophan did not antagonize the inhibitory effect of EV-086, as only a very slight decrease in sensitivity was observed for S. cerevisiae in the presence of tryptophan (unpublished results). We speculate that EV-086 alters the membrane composition via inhibition of delta-9 fatty acid desaturation, thereby affecting the function of various integral membrane proteins, including tryptophan transporters (47). In agreement with this is the finding that S. cerevisiae OLE1 mutants show a disintegration of the nuclear envelope, which irreversibly damages the cell (48). EV-086-dependent inhibition of delta-9 fatty acid desaturation thus likely affects different membrane-associated processes leading to multiple physiological failures and subsequent cell death. Furthermore, the heterodimeric transcription factors OAF1 and PIP2 (49), which activate genes involved in beta-oxidation of fatty acids, were identified in our screen. These mutants were more susceptible to EV-086, which might indicate that beta-oxidation is a mechanism for inactivation of EV-086 in yeast. Similarly, it has been reported that Oaf1p and Pip2p are involved in inactivation of the toxic fatty acids octanoic acid and decanoic acid (50), and OAF1 and PIP2 are upregulated in response to 6-nonadecynoic acid (45).
The analysis of EV-086 analogues demonstrated that no changes were tolerated at the carboxylic acid moiety and that the configuration of the C-9–C-10 carbon bond was critical for activity. Whereas a trans-9-decenoic configuration was not active (EV-086-trans), cis-9-decenoic acid (EV-086) or decanoic acid (EV-086–9-saturated) configurations were active. The cis-9-decenoic acid and decanoic acid configurations are conserved in common fatty acids, including palmitic acid, palmitoleic acid, stearic acid, oleic acid, or linoleic acid. Lipid analyses with labeled EV-086 suggested that EV-086 is integrated into phospholipids, likely as a phospholipid fatty acyl chain. The impact of this incorporation on the antifungal activity of EV-086 is so far unclear. However, it might be that EV-086 is targeted to membranes as an EV-086–phospholipid. Assuming that EV-086 inhibits Ole1p, EV-086–phospholipids might colocalize with Ole1p in the ER membrane. EV-086–phospholipids could then expose the EV-086 acyl chain to the hydrophobic side of the phospholipid bilayer, and inhibition of Ole1p might take place at the transmembrane portion of the protein.
Fatty acid biosynthesis as a target for antimicrobials has been controversial (51). Although platensimycin has demonstrated antibacterial activity in vivo by inhibiting FASII-dependent fatty acid biosynthesis, another study suggested that bacterial pathogens overcome drug-induced FASII pathway inhibition via complementation with serum fatty acids (52, 53). EV-086 has proven efficacy in a guinea pig model of T. mentagrophytes skin infection. This validates delta-9 fatty acid desaturation as a target for this indication and proves the efficacy of the compound in vivo. T. mentagrophytes therefore does not overcome EV-086-induced delta-9 fatty acid desaturase inhibition via complementation with skin unsaturated fatty acids. In contrast to the good efficacy observed in the guinea pig model of T. mentagrophytes skin infection, EV-086 was ineffective in rat and mouse models of systemic candidiasis (unpublished results). These findings could indicate that C. albicans overcomes EV-086-induced delta-9 fatty acid desaturase inhibition via complementation with serum unsaturated fatty acids. This in turn suggests that C. albicans can rely on serum fatty acids as a sole source of delta-9 unsaturated fatty acids and that C. albicans delta-9 fatty acid desaturation is dispensable for virulence in systemic infections. However, different studies have suggested that OLE1-mediated delta-9 fatty acid desaturation is an essential process for virulence (45, 54, 55). We therefore assume that the inactivity of EV-086 observed in the systemic candidiasis studies is related to the molecule and not the target. This, however, needs further investigation.
In conclusion, EV-086 represents a novel class of antifungal compounds that target delta-9 fatty acid desaturation, and it is a novel lead antifungal compound with opportunities in human fungal disease.
ACKNOWLEDGMENTS
We thank Thomas Tange and Jutta Heim for critical reading of the manuscript.
R.S. acknowledges support by the Swiss National Science Foundation (31003A_134742). M.G. served as a consultant for and received contracts from Evolva SA. R.S. received contracts from Evolva SA.
Footnotes
Published ahead of print 4 November 2013
REFERENCES
- 1.Anaissie EJ, McGinnis MR, Pfaller MA. 2009. Clinical mycology, 2nd ed. Elsevier, New York, NY [Google Scholar]
- 2.Gupta AK, Cooper EA. 2008. Update in antifungal therapy of dermatophytosis. Mycopathologia 166:353–367. 10.1007/s11046-008-9109-0 [DOI] [PubMed] [Google Scholar]
- 3.Kathiravan MK, Salake AB, Chothe AS, Dudhe PB, Watode RP, Mukta MS, Gadhwe S. 2012. The biology and chemistry of antifungal agents: a review. Bioorg. Med. Chem. 20:5678–5698. 10.1016/j.bmc.2012.04.045 [DOI] [PubMed] [Google Scholar]
- 4.Ostrosky-Zeichner L, Casadevall A, Galgiani JN, Odds FC, Rex JH. 2010. An insight into the antifungal pipeline: selected new molecules and beyond. Nat. Rev. Drug Discov. 9:719–727. 10.1038/nrd3074 [DOI] [PubMed] [Google Scholar]
- 5.Lass-Florl C. 2009. The changing face of epidemiology of invasive fungal disease in Europe. Mycoses 52:197–205. 10.1111/j.1439-0507.2009.01691.x [DOI] [PubMed] [Google Scholar]
- 6.Pfaller MA, Diekema DJ. 2010. Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 36:1–53. 10.3109/10408410903241444 [DOI] [PubMed] [Google Scholar]
- 7.Odds FC, Brown AJ, Gow NA. 2003. Antifungal agents: mechanisms of action. Trends Microbiol. 11:272–279. 10.1016/S0966-842X(03)00117-3 [DOI] [PubMed] [Google Scholar]
- 8.McAlpine JB, Pazoles C, Stafford A. 1999. Phytera's strategy for the discovery of novel anti-infective agents from plant cell cultures, p 159–166 In Bohlin L, Bruhn JG. (ed), Bioassay methods in natural product research and drug development, 43rd ed. Springer, Houten, Netherlands [Google Scholar]
- 9.Stafford A, Pazoles CJ, Siegel S, Yeh L-A. 1998. Plant cell culture: a vehicle for drug discovery, p 53–64 In Harvey AL. (ed), Advances in drug discovery techniques. John Wiley & Sons, New York, NY [Google Scholar]
- 10.Yang SW, Ubillas R, McAlpine J, Stafford A, Ecker DM, Talbot MK, Rogers B. 2001. Three new phenolic compounds from a manipulated plant cell culture, Mirabilis jalapa. J. Nat. Prod. 64:313–317. 10.1021/np0004092 [DOI] [PubMed] [Google Scholar]
- 11.Gillum AM, Tsay EY, Kirsch DR. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182 [DOI] [PubMed] [Google Scholar]
- 12.Gamborg OL, Miller RA, Ojima K. 1968. Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50:151–158. 10.1016/0014-4827(68)90403-5 [DOI] [PubMed] [Google Scholar]
- 13.CLSI 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard, 3rd ed. CLSI document M27-A3 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 14.CLSI 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard, 2nd ed. CLSI document M38-A2 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 15.Amberg DC, Burke D, Strathern JN. 2005. Methods in yeast genetics. CSHL Press, Cold Spring Harbor, NY [Google Scholar]
- 16.CLSI 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed. CLSI document M07-A8 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 17.Berridge MV, Herst PM, Tan AS. 2005. Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. Biotechnol. Annu. Rev. 11:127–152. 10.1016/S1387-2656(05)11004-7 [DOI] [PubMed] [Google Scholar]
- 18.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132 [DOI] [PubMed] [Google Scholar]
- 19.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25:25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.SGD Project 2009. Saccharomyces Genome Database. go_slim_mapping.tab. Stanford University, Stanford, CA: ftp://ftp.yeastgenome.org/yeast/ [Google Scholar]
- 21.Salkind NJ. 2007. Encyclopedia of measurement and statistics. SAGE Publications, Inc., Thousand Oaks, CA [Google Scholar]
- 22.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 23.Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386. 10.1385/1-59259-192-2:365 [DOI] [PubMed] [Google Scholar]
- 24.Schatz PJ, Pillus L, Grisafi P, Solomon F, Botstein D. 1986. Two functional alpha-tubulin genes of the yeast Saccharomyces cerevisiae encode divergent proteins. Mol. Cell. Biol. 6:3711–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stukey JE, McDonough VM, Martin CE. 1990. The OLE1 gene of Saccharomyces cerevisiae encodes the delta 9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J. Biol. Chem. 265:20144–20149 [PubMed] [Google Scholar]
- 26.Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A. 2009. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 106:2136–2141. 10.1073/pnas.0811700106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghannoum MA, Hossain MA, Long L, Mohamed S, Reyes G, Mukherjee PK. 2004. Evaluation of antifungal efficacy in an optimized animal model of Trichophyton mentagrophytes-dermatophytosis. J. Chemother. 16:139–144 [DOI] [PubMed] [Google Scholar]
- 28.Hefter GT, Tomkins RPT. 2003. The experimental determination of solubilities. John Wiley & Sons, Hoboken, NJ [Google Scholar]
- 29.Tetko IV, Gasteiger J, Todeschini R, Mauri A, Livingstone D, Ertl P, Palyulin VA, Radchenko EV, Zefirov NS, Makarenko AS, Tanchuk VY, Prokopenko VV. 2005. Virtual computational chemistry laboratory: design and description. J. Comput. Aided Mol. Des 19:453–463. 10.1007/s10822-005-8694-y [DOI] [PubMed] [Google Scholar]
- 30.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El BM, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906 [DOI] [PubMed] [Google Scholar]
- 31.Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. 2008. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134:634–645. 10.1016/j.cell.2008.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin CE, Oh CS, Jiang Y. 2007. Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim. Biophys. Acta 1771:271–285. 10.1016/S1388-1981(06)00189-2 [DOI] [PubMed] [Google Scholar]
- 33.UniProt Consortium 2008. The universal protein resource (UniProt). Nucleic Acids Res. 36:D190–D196. 10.1093/nar/gkm895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Lin ZY, Liang W, Marback M, Paw J, San Luis BJ, Shuteriqi E, Tong AH, DNvan Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pal C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras AC, Morris QD, Kim PM, Kaiser CA, Myers CL, Andrews BJ, Boone C. 2010. The genetic landscape of a cell. Science 327:425–431. 10.1126/science.1180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, Schuldiner M. 2009. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323:1693–1697. 10.1126/science.1167983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124:1069–1081. 10.1016/j.cell.2005.12.036 [DOI] [PubMed] [Google Scholar]
- 37.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, Weissman JS, Krogan NJ. 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123:507–519. 10.1016/j.cell.2005.08.031 [DOI] [PubMed] [Google Scholar]
- 38.Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. 2005. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat. Cell Biol. 7:993–998. 10.1038/ncb1298 [DOI] [PubMed] [Google Scholar]
- 39.Schuberth C, Buchberger A. 2005. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat. Cell Biol. 7:999–1006. 10.1038/ncb1299 [DOI] [PubMed] [Google Scholar]
- 40.Kolawa NJ, Sweredoski MJ, Graham RLJ, Oania R, Hess S, Deshaies RJ. 2013. Perturbations to the ubiquitin conjugate proteome in yeast Δubx mutants identify Ubx2 as a regulator of membrane lipid composition. Mol. Cell. Proteomics 12:2791–2803. 10.1074/mcp.M113.030163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stukey JE, McDonough VM, Martin CE. 1989. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J. Biol. Chem. 264:16537–16544 [PubMed] [Google Scholar]
- 42.Krishnamurthy S, Plaine A, Albert J, Prasad T, Prasad R, Ernst JF. 2004. Dosage-dependent functions of fatty acid desaturase Ole1p in growth and morphogenesis of Candida albicans. Microbiology 150:1991–2003. 10.1099/mic.0.27029-0 [DOI] [PubMed] [Google Scholar]
- 43.Oh CS, Martin CE. 2006. Candida albicans Spt23p controls the expression of the Ole1p Delta9 fatty acid desaturase and regulates unsaturated fatty acid biosynthesis. J. Biol. Chem. 281:7030–7039. 10.1074/jbc.M510746200 [DOI] [PubMed] [Google Scholar]
- 44.Bossie MA, Martin CE. 1989. Nutritional regulation of yeast delta-9 fatty acid desaturase activity. J. Bacteriol. 171:6409–6413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu D, Sillaots S, Davison J, Hu W, Jiang B, Kauffman S, Martel N, Ocampo P, Oh C, Trosok S, Veillette K, Wang H, Yang M, Zhang L, Becker J, Martin CE, Roemer T. 2009. Chemical genetic profiling and characterization of small-molecule compounds that affect the biosynthesis of unsaturated fatty acids in Candida albicans. J. Biol. Chem. 284:19754–19764. 10.1074/jbc.M109.019877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roemer T, Davies J, Giaever G, Nislow C. 2012. Bugs, drugs and chemical genomics. Nat. Chem. Biol. 8:46–56. 10.1038/nchembio.744 [DOI] [PubMed] [Google Scholar]
- 47.Santos AX, Riezman H. 2012. Yeast as a model system for studying lipid homeostasis and function. FEBS Lett. 586:2858–2867. 10.1016/j.febslet.2012.07.033 [DOI] [PubMed] [Google Scholar]
- 48.Zhang S, Skalsky Y, Garfinkel DJ. 1999. MGA2 or SPT23 is required for transcription of the delta9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics 151:473–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baumgartner U, Hamilton B, Piskacek M, Ruis H, Rottensteiner H. 1999. Functional analysis of the Zn(2)Cys(6) transcription factors Oaf1p and Pip2p. Different roles in fatty acid induction of beta-oxidation in Saccharomyces cerevisiae. J. Biol. Chem. 274:22208–22216 [DOI] [PubMed] [Google Scholar]
- 50.Xu T, Tripathi SK, Feng Q, Lorenz MC, Wright MA, Jacob MR, Mask MM, Baerson SR, Li XC, Clark AM, Agarwal AK. 2012. A potent plant-derived antifungal acetylenic acid mediates its activity by interfering with fatty acid homeostasis. Antimicrob. Agents Chemother. 56:2894–2907. 10.1128/AAC.05663-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fulmer T. 2009. Not so FAS. SciBX 2(11):1–2. 10.1038/scibx.2009.430 [DOI] [Google Scholar]
- 52.Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C. 2009. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature 458:83–86. 10.1038/nature07772 [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, Ha S, Dorso K, Motyl M, Jayasuriya H, Ondeyka J, Herath K, Zhang C, Hernandez L, Allocco J, Basilio A, Tormo JR, Genilloud O, Vicente F, Pelaez F, Colwell L, Lee SH, Michael B, Felcetto T, Gill C, Silver LL, Hermes JD, Bartizal K, Barrett J, Schmatz D, Becker JW, Cully D, Singh SB. 2006. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 441:358–361. 10.1038/nature04784 [DOI] [PubMed] [Google Scholar]
- 54.Fradin C, Kretschmar M, Nichterlein T, Gaillardin C, d'Enfert C, Hube B. 2003. Stage-specific gene expression of Candida albicans in human blood. Mol. Microbiol. 47:1523–1543. 10.1046/j.1365-2958.2003.03396.x [DOI] [PubMed] [Google Scholar]
- 55.Nguyen LN, Gacser A, Nosanchuk JD. 2011. The stearoyl-coenzyme A desaturase 1 is essential for virulence and membrane stress in Candida parapsilosis through unsaturated fatty acid production. Infect. Immun. 79:136–145. 10.1128/IAI.00753-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.National Research Council 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC [Google Scholar]