Abstract
Curcumin (CUR) shows antifungal activity against a range of pathogenic fungi, including Candida albicans. The reported mechanisms of action of CUR include reactive oxygen species (ROS) generation, defects in the ergosterol biosynthesis pathway, decrease in hyphal development, and modulation of multidrug efflux pumps. Reportedly, each of these pathways is independently linked to the cell wall machinery in C. albicans, but surprisingly, CUR has not been previously implicated in cell wall damage. In the present study, we performed transcriptional profiling to identify the yet-unidentified targets of CUR in C. albicans. We found that, among 348 CUR-affected genes, 51 were upregulated and 297 were downregulated. Interestingly, most of the cell wall integrity pathway genes were downregulated. The possibility of the cell wall playing a critical role in the mechanism of CUR required further validation; therefore, we performed specific experiments to establish if there was any link between the two. The fractional inhibitory concentration index values of 0.24 to 0.37 show that CUR interacts synergistically with cell wall-perturbing (CWP) agents (caspofungin, calcofluor white, Congo red, and SDS). Furthermore, we could observe cell wall damage and membrane permeabilization by CUR alone, as well as synergistically with CWP agents. We also found hypersusceptibility in calcineurin and mitogen-activated protein (MAP) kinase pathway mutants against CUR, which confirmed that CUR also targets cell wall biosynthesis in C. albicans. Together, these data provide strong evidence that CUR disrupts cell wall integrity in C. albicans. This new information on the mechanistic action of CUR could be employed in improving treatment strategies and in combinatorial drug therapy.
INTRODUCTION
Candidiasis, caused by Candida albicans, is the most common opportunistic fungal infection and a serious medical problem that causes significant morbidity and mortality, particularly in AIDS patients, transplant recipients, and other immunocompromised people (1). In spite of the continuous expansion of the arsenal of antifungal drugs, the available drugs cannot meet the ever-increasing requirement to combat Candida infections in patients. Infections caused by pathogenic C. albicans are commonly treated with either azoles or some nonazole antifungal agents, but they have pronounced side effects (2, 3). In the clinical setting, excessive use of these antifungals causes liver damage, altered estrogen levels, allergic reactions, and, above all, development of drug resistance among patients (4). Hence, there has been an urgent need to develop effective drugs for combating these fungal diseases.
Researchers in recent years have turned their attention toward utilizing natural products as the building blocks for the development of effective next-generation drugs. Curcumin (CUR) (a mixture of curcuminoids) is extracted from the rhizome of Curcuma longa (a small perennial herb native to India) and is known to have therapeutic potential due to its anti-inflammatory, anticarcinogenic, and anti-infectious properties (5, 6). It has long been used as a common household medicine and as a spice in Southeast Asia, and previously, we reported its antifungal activity against C. albicans (7). Antifungal activity of CUR has also also been reported against Cryptococcus neoformans, Sporothrix schenckii, Paracoccidioides brasiliensis, and Aspergillus spp. (5). Reports show that the antifungal activity of CUR occurs via multiple pathways in C. albicans. For example, CUR leads to cell death via reactive oxygen species (ROS)-induced apoptosis in C. albicans (8). This ROS-induced cell killing was also observed when CUR was used synergistically, along with azoles and polyenes (7). In another study, CUR was shown to target ERG3 of C. albicans, leading to altered ergosterol biosynthesis, which in turn is critical to drug resistance (9). Additionally, the activities of multidrug efflux pumps, namely, CaCdr1p, CaCdr2p, CaMdr1p, and ScPdr5p, have been shown to be modulated by CUR (10). Overall, CUR targets a majority of the pathways known to be directly involved in acquisition of drug resistance.
Studies show that there is a linkage between the cell wall and induction of drug resistance in C. albicans. There is strong evidence that protein kinase C (PKC) signaling, which comprises Hsp90 and calcineurin, governs the drug resistance phenomenon of C. albicans (11). Also, resistance to cell wall-targeting drugs like echinocandins (nonazole drugs) has been shown to develop through calcineurin-mediated signaling (3) and through 1,3-glucan biosynthesis inhibition (12) in C. albicans.
Considering the above-mentioned facts, one may question whether CUR has any link with cell wall pathways of C. albicans. In the present study, we used a microarray approach to identify global changes that occur upon CUR treatment in C. albicans. Interestingly, apart from the other known target pathways of CUR, many genes related to cell wall biogenesis were also found to be downregulated. This result was in line with our earlier presumption that CUR may target the cell wall of C. albicans. Therefore, we have extended this study to discuss the mode of action of CUR in relation to cell wall disruption. Interestingly, CUR alone and in combination with cell wall-perturbing (CWP) agents (like caspofungin [CAS], calcofluor white [CFW], Congo red [CR], and SDS) at its synergistic concentration modulates the cell wall architecture of C. albicans. Using mutant variants of calcineurin and the mitogen-activated protein (MAP) kinase pathway, we were able to show that CUR targets cell wall integrity and promotes cell death by disrupting the cell wall of C. albicans.
MATERIALS AND METHODS
Chemicals used.
Most medium components were obtained from HiMedia (Mumbai, India), while Bacto agar was obtained from Difco (BD Biosciences, NJ, USA) and RPMI 1640 medium from Gibco BRL (Gaithersburg, MD, USA). CFW (4-methyl-7-diethylamino coumarin), CUR (a commercial grade mixture of curcuminoids), CR, SDS, sodium chloride, and other chemicals used in the experiments were purchased from Sigma Chemical Co. (St. Louis, MO, USA). propidium iodide (PI) was purchased from Molecular Probes (Eugene, OR, USA).
Candida strains, media, and growth conditions.
All the C. albicans strains/mutants used in this study are listed in Table S1 in the supplemental material. All the strains were maintained on yeast extract-peptone-dextrose (YEPD) broth and agar plates (HiMedia, Mumbai, India) at 30°C.
Cell culture and CUR exposure for microarray experiments.
Candida cells were grown at 30°C in YEPD medium to an optical density at 600 nm (OD600) of 0.1 and were then split into two cultures. For CUR treatment, cells at an OD of 0.1 were allowed to reach an OD of 0.4, and then 251 μM (92.45 μg/ml) CUR (MIC50) was added to one of the cultures and the second culture was subjected to an equivalent mock treatment (water). After 14 h of CUR treatment, the cells were harvested and used for RNA isolation (13). Three independent cultures were prepared for biological repeats.
RNA isolation, cDNA synthesis, labeling, and hybridization.
RNA isolation was done by following the TRIzol (Sigma) method according to the manufacturer's specifications, except that 300 μl acid-washed 0.4- to 0.6-mm glass beads (Sigma, St. Louis, MO, USA) was used during cell lysis (14). Ten milligrams of purified total RNA was used to synthesize cDNA by using the protocol described previously (www.transcriptome.ens.fr/sgdb/protocols/labeling.yeast.php). We used a previously described direct-labeling method to label cDNA with Cy3 and Cy5 dyes (15). The labeled cDNA was mixed with hybridization buffer and applied immediately to microarray slides. It was covered with 22- by 60-mm coverslips, sealed in Corning hybridization chambers, and allowed to incubate overnight at 42°C. The slides were washed and scanned. Each experimental condition was independently repeated three times, including a dye swap. C. albicans microarrays (batch C050G) containing a cDNA probe deposited in duplicate for 98% of Candida open reading frames (ORFs) (assembly 19) were obtained from Eurogentec (Seraing, Belgium) (13).
Scanning and cDNA microarray data analysis.
Slides were scanned using a Scan Array Express microarray scanner from PerkinElmer. The raw data obtained were quantified using the adaptive-circle method and normalized using locally weighted scatterplot smoothing (LOWESS). The value obtained by this method was background corrected, and the ratio of treated versus untreated intensities was converted into log2 ratios, which were averaged for replicates of the same set of experiments. We used the www.candida.bri.nrc website to obtain the annotations for the ORFs. To address the biological description and relevance of the genes presented in Table S2 in the supplemental material, we looked for gene functional categories in the Candida Genome Database (CGD) and gene ontology (GO) term finder tools (16).
RT-PCR.
For validation of the microarray results, reverse transcriptase (RT) PCR was done as described in the RevertAid H Minus kit (MBI, Fermentas). The amplified products were subjected to gel electrophoresis and quantitated using Quantity One software with the Bio-Rad gel documentation system. The densities of bands (for genes of interest) were measured and normalized to that of the endogenous actin gene (ACT1). We have taken three biological replicates and measured the standard deviation of the intensity per unit area to validate the RT-PCR. The primers used in this experiment are listed in Table S3 in the supplemental material.
Determination of MICs.
The MICs of CWP agents (CAS, CFW, CR, and SDS) against C. albicans strain SC5314 were determined by broth microdilution using 2-fold serial dilutions in RPMI 1640 medium, as described in method M27-A3 from the Clinical and Laboratory Standards Institute (CLSI) (formerly NCCLS) (17).
Checkerboard assay.
The interaction of CUR with CWP agents was evaluated by the checkerboard method recommended by the CLSI (17) and expressed as the sum of the fractional inhibitory concentration index (FICI) for each agent. The checkerboard assay was performed in triplicate as described previously (8). The FICI of each agent was calculated as the MIC of the agent in combination divided by the MIC of the agent alone (18). The FICI was interpreted as synergistic when it was ≤0.5 and antagonistic when it was ≥4.0, and any value in between was interpreted as indifferent (12).
Time-kill assay.
C. albicans cells (∼106) were inoculated in RPMI 1640 medium containing CUR, CFW, CR, and SDS alone and in combination. The concentrations of drugs used here are the synergistic concentrations. At predetermined time points (0, 1, 4, 8, 12, 16, 20, and 24 h at 30°C incubation; agitation, 200 rpm), a 100-μl aliquot was removed, serially diluted (10-fold) in saline, and plated on YPD agar plates. Colony counts were determined after incubation at 30°C for 24 h in triplicate (19).
Growth percentage assessment.
The interaction of CUR with different CWP agents was evaluated by the growth curve via three independent experiments. After inoculation of 104 cells in RPMI 1640 medium and synergistic concentrations of the above-mentioned drugs (alone and in combination with CUR) in the microdilution plates, they were agitated for 15 s and incubated at 37°C for 24 h, and the OD600 was recorded. The changes in OD over time were used to generate a growth percentage at each drug concentration and for the drug-free growth control.
Spot assay.
The susceptibilities of C. albicans strain SC5314 to CUR, CFW, CR, and SDS in solid medium were measured using the spot assay method as described previously, and differences were monitored after growth for 48 h at 30°C (20). Spot assays were also performed with calcineurin, MAP kinase, and HSP90 pathway mutants (see Table S1 in the supplemental material).
Confocal microscopy.
Candida cells (∼106) were suspended in RPMI 1640 medium and incubated with 1.5 μM PI plus CUR alone at their MIC80 and PI plus CUR plus CWP agents at the synergy concentration at 30°C with constant shaking (200 rpm) for 4 h and visualized by confocal microscopy (Olympus Fluoview FV1000) at a wavelength of >560 nm (21). The MIC80 of CUR was used, but not the MIC50, as in the transcriptome-profiling experiment, to visualize the dye uptake.
Electron microscopy.
Cell damage was observed in CUR-treated cells (∼106) at the MIC100 and in cells treated with CUR plus CWP agents at synergistic concentrations using scanning (SEM [Zeiss EV040]) and transmission electron (TEM [JEOL 2100F]) microscopy. The MIC100 of CUR was used to determine if a combination of CUR and CWP at synergistic concentrations can have the same impact on the cell wall. The sample preparation and analysis methods were similar to those described previously (21). The images were acquired at ×30,000 magnification.
RESULTS
Transcriptional response of C. albicans to CUR.
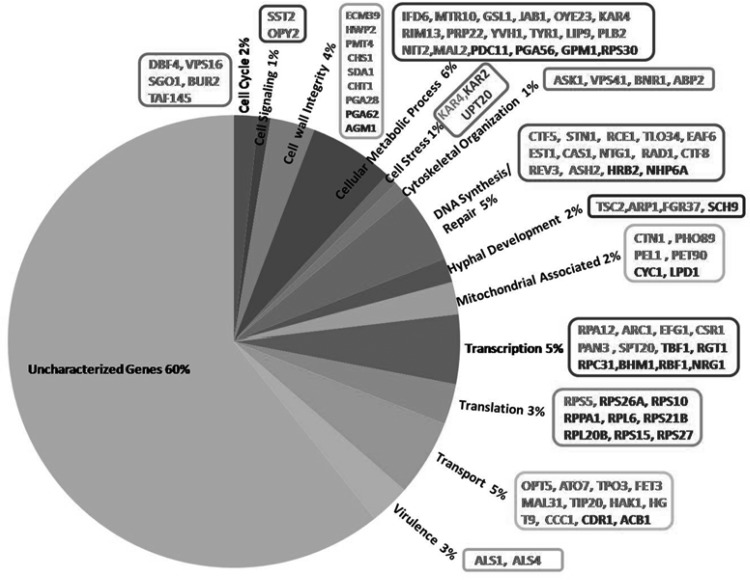
To obtain a detailed overview of the mechanistic action of CUR, transcriptome profiling of C. albicans was performed at the MIC50 (251 μM; 92.45 μg/ml) of CUR only. We used the MIC50 for the array experiments to avoid the growth defects/inhibition that would have occurred at higher MIC values, and we found that the MIC50 was just sufficient to trigger the transcriptional response. Microarray data analysis showed that 348 genes were differentially expressed in CUR-treated C. albicans cells (SC5314) versus the C. albicans wild-type strain not treated with CUR. Among them, 51 genes were significantly upregulated and 297 genes were significantly downregulated. The cutoff value as a log2 ratio is shown in Table S2 in the supplemental material. The transcriptome data showed that CUR was effective against C. albicans via multiple pathways, including the cell cycle, cell signaling, cell wall integrity, cellular metabolic processes, stress, cytoskeletal organization, DNA synthesis/repair, hyphal development, mitochondria, transcription, translation, transport, virulence, and several uncharacterized genes (Fig. 1). Further, to validate the microarray data, we performed RT-PCR in response to CUR under similar growth conditions. We randomly picked a few genes, both up- and downregulated, for RT-PCR analysis and found that the RT-PCR data corroborated the microarray results (see Fig. S1 in the supplemental material).
FIG 1.
Microarray results showing percent distributions of various categories of genes that were differentially expressed in CUR-treated C. albicans. The gene names in boldface indicate upregulation in response to CUR treatment, while those in gray type indicate downregulation.
CUR targets the genes of the cell wall integrity pathway.
Among the categories of genes that were found to be downregulated upon CUR treatment, we found prominent changes in the genes that maintain cell wall integrity in C. albicans (see Table S2 in the supplemental material). The cell wall integrity pathway is known to play a significant role in combating various drug treatments (22), but surprisingly, no study has shown any link between CUR and the cell wall. We found that upon CUR exposure, 11 genes of this class were downregulated. Specifically, PGA28, ECM39, GFA1, HWP2, PMT4, HYR3, HDA1, CHS1, CHT1, orf19.5271, and orf19.376 were significantly downregulated upon CUR treatment (Fig. 1). This result suggests that the cell wall integrity might be compromised in C. albicans when exposed to CUR. We also analyzed the enrichment for GO terms, where the cell wall category was found to be enriched (see Table S4 in the supplemental material). Therefore, in the later part of this study, we specifically focus on discussing the link between CUR and cell wall integrity.
Antifungal activities of CUR and CWP agents.
To elaborate on the microarray results that suggest influences of CUR on cell wall stress, we first determined the antifungal activities (in terms of the MIC100) of CUR and CWP agents against C. albicans. The MIC was defined as the lowest concentration of drug exhibiting 100% inhibition of growth compared with that of the drug-free control. The MIC100s were as follows: CUR, 695 μM (256 μg/ml); CAS, 0.39 μM (0.42 μg/ml); CFW, 62.5 μM (57.5 μg/ml); CR, 15.7 μM (11 μg/ml); and SDS, 250 μM (72 μg/ml).
CUR is synergistic with CWP agents.
The in vitro antifungal effects of CUR in combination with CWP agents (CFW, CR, and SDS) were tested against C. albicans. The results showed that combinations of noncandidacidal concentrations of CUR and CWP agents were highly active against C. albicans, suggesting their synergistic action. Combinations of noncandidacidal concentrations of 86.8 μM (31.97 μg/ml) for CUR plus 0.048 μM (0.051 μg/ml) for CAS, 173.7 μM (63.98 μg/ml) for CUR plus 1.95 μM (1.79 μg/ml) for CFW, 173.7 μM (63.98 μg/ml) for CUR plus 1.90 μM (1.36 μg/ml) for CR, and 86.8 μM (31.97 μg/ml) for CUR plus 31.25 μM (9 μg/ml) for SDS showed almost complete killing of C. albicans. FICI values of CUR with CWP agents were determined as described in Materials and Methods. As shown from the FICI values, CUR was synergistic with all tested CWP agents (Table 1). In combination with CUR, the MIC100 of CWP agents was reduced 8-fold for CAS, 32-fold for CFW, and 8-fold for CR and SDS, and the MIC100 of CUR was reduced from 4- to 8-fold with CWP agents.
TABLE 1.
FICI values of CWP agents with CUR against C. albicans
| Concn of CUR alone (μg/ml) | Concn of CWP agent alone (μg/ml) | Concn of CUR (μg/ml) and CWP agent (μg/ml) in combination | FICI | Effect |
|---|---|---|---|---|
| 256 | 0.42 (CAS) | 31.97 (CUR) + 0.051 (CAS) | 0.24 | Synergy |
| 256 | 57.5 (CFW) | 63.98 (CUR) + 1.79 (CFW) | 0.28 | Synergy |
| 256 | 21.85 (CR) | 63.98 (CUR) + 1.36 (CR) | 0.37 | Synergy |
| 256 | 72 (SDS) | 31.97 (CUR) + 9 (SDS) | 0.25 | Synergy |
Time-kill curves, growth percentage assessment, and spot assay reconfirm the synergism of CUR with CWP agents.
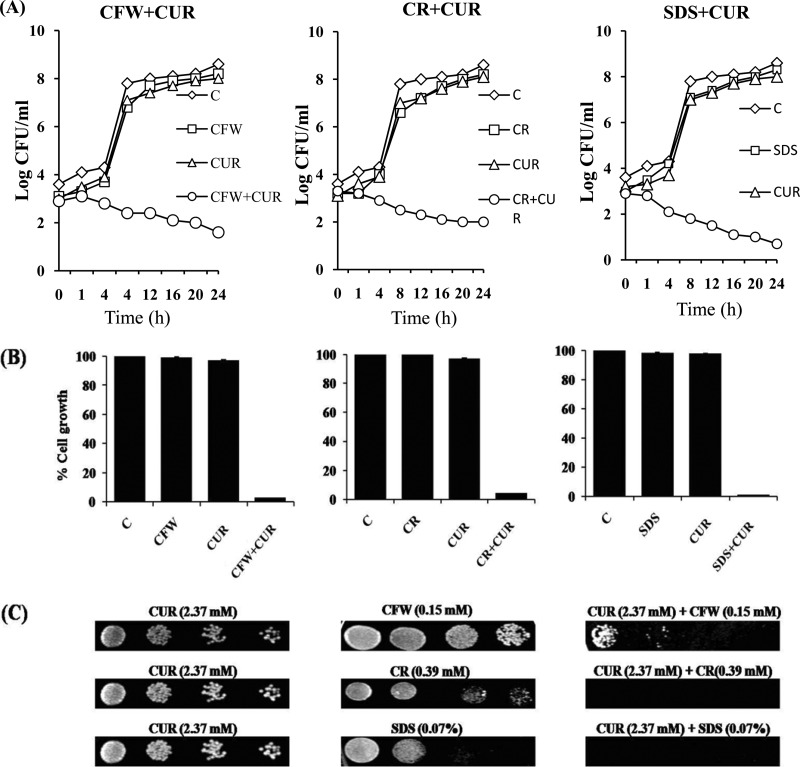
To validate the synergy result, we performed a time-kill assay, a growth percentage assessment, and a spot assay that further confirmed the synergistic interaction of CUR and CWP agents (Fig. 2). Time-kill curves showed that CUR and either of the tested CWP agents at the indicated concentrations alone did not affect the growth of C. albicans (Fig. 2A). However, a combination of CUR with CWP agents significantly affected the CFU count (Fig. 2A), as well as the cell growth percentage, of C. albicans after 24 h (Fig. 2B). A solid-medium spot assay also showed synergy of CUR with CWP agents in C. albicans (Fig. 2C). The data from these experiments confirmed the high susceptibility of the C. albicans strain tested with CUR in combination with CWP agents, indicating synergistic action.
FIG 2.
(A) Time-kill curve of CUR in combination with CWP agents: 173.7 μM CUR (63.98 μg/ml) plus 1.95 μM (1.79 μg/ml) CFW, 173.7 μM (63.98 μg/ml) CUR plus 1.90 μM (1.36 μg/ml) CR, and 86.8 μM (31.97 μg/ml) CUR plus 31.25 μM (9 μg/ml) SDS. (B) Percent growth of C. albicans at the same synergy concentrations of CUR with CWP agents as for panel A. (C) Solid-medium spotting assay. The wild-type (SC5314) strain was tested by spotting decreasing numbers of cells on YPD agar plates with CUR and CWP agents alone and in combination.
CUR induced membrane permeabilization in C. albicans.
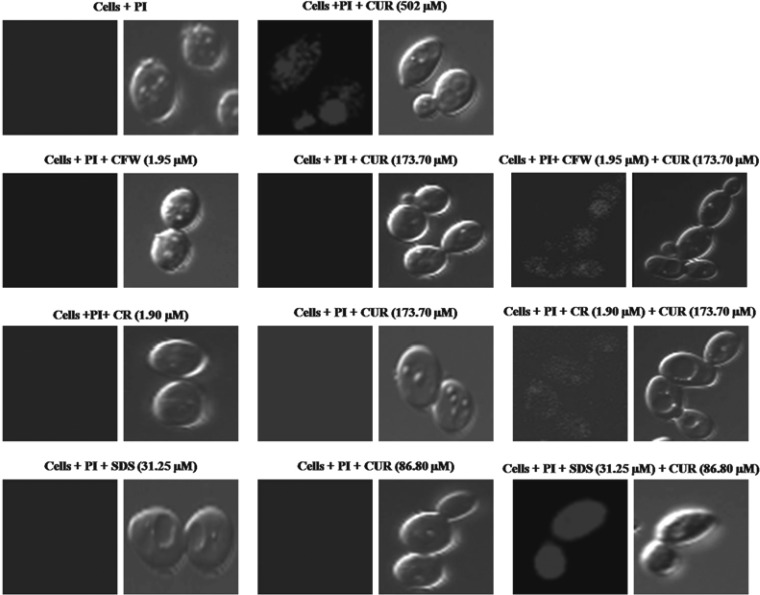
Alteration in membrane permeability indicates a change in the physical state of the membrane or toward compromised cell wall integrity. To determine whether CUR treatment induces membrane permeabilization in C. albicans, we measured the uptake of membrane-impermeant dye, PI. Incubation of CUR (MIC80) with Candida cells resulted in PI uptake, as monitored by confocal microscopy (Fig. 3). We also observed PI uptake in C. albicans cells when CUR was used in combination with CWP agents at synergistic concentrations (Fig. 3). This result showed that when used in combination, CUR acted synergistically with CWP agents to cause membrane permeabilization at concentrations that are very low compared to their individual MIC values. No PI uptake was observed when C. albicans cells were treated with PI alone or with a nonantifungal concentration (MIC50) of CUR or CWP agents (control).
FIG 3.
Confocal microscopy analysis of membrane permeabilization assay by PI uptake. C. albicans was incubated with CUR alone (251 μM [92.45 μg/ml] and 502 μM [184.9 μg/ml]) and at synergistic concentrations with CWP agents (1.95 μM [1.79 μg/ml] CFW plus 173.7 μM [63.98 μg/ml] CUR, 1.95 μM [1.36 μg/ml] CR plus 173.7 μM [63.98 μg/ml] CUR, and 31.25 μM [9 μg/ml] SDS plus 86.8 μM [31.97 μg/ml] CUR) for 4 h.
CUR treatment causes abrogation of cell surface morphology in C. albicans.
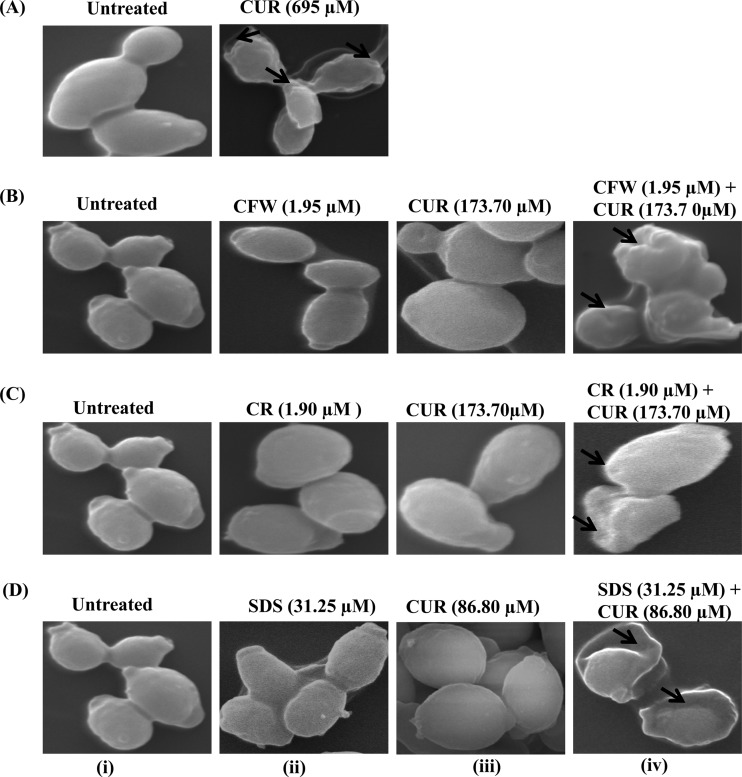
Higher membrane permeability could be due to altered cell surface morphology of C. albicans. To verify this fact, we looked for morphological changes in CUR-treated C. albicans cells by SEM (Fig. 4). Cells treated with CUR alone at its MIC80 for 24 h showed wrinkling and corrugation of the cell surface compared with the smooth surfaces of untreated cells (Fig. 4A). Further, we assessed the morphological changes in C. albicans cells when treated with CUR and CWP agents in combination (CUR plus CFW, CUR plus CR, and CUR plus SDS) at noncandidacidal synergistic concentrations (Fig. 4B, C, and D). We found that upon treatment with CUR plus CWP agents at synergistic concentrations, cells showed wrinkling and corrugation of the cell surface (Fig. 4B-iv, C-iv, and D-iv, arrows). In the control experiment, untreated cells (Fig. 4B-i, C-i, and D-i) or cells treated with synergistic nonfungicidal concentrations of CUR (Fig. 4B-iii, C-iii, and D-iii), CFW (Fig. 4B-ii), CR (Fig. 4C-ii), and SDS (Fig. 4D-ii) alone showed a smooth surface. These data showed that CUR in combination with CWP agents at synergistic concentrations could cause extensive damage to the cell surface morphology.
FIG 4.
Scanning electron micrographs showing corrugation of the cell surface, squeezing of the cell, and leakage of cytoplasmic content due to cell wall damage (arrows) in C. albicans after 24-h treatment with CUR and CWP agents at synergistic concentrations. (A) SEM of C. albicans treated or not with CUR (695 μM; 256 μg/ml) for 24 h showing corrugation of the cell wall and squeezing of cells. (B-i, C-i, and D-i) Controls; cells are intact and evenly shaped. (B-ii to -iv) Cells after treatment with 1.95 μM (1.79 μg/ml) CFW (B-ii), 173.7 μM (63.98 μg/ml) CUR (B-iii), and 1.95 μM (1.79 μg/ml) CFW plus 173.7 μM (63.98 μg/ml) CUR (B-iv). (C-ii to -iv) Cells after incubation with 1.90 μM (1.36 μg/ml) CR (C-ii), 173.7 μM (63.98 μg/ml) CUR (C-iii), and 1.90 μM (1.36 μg/ml) CR plus 173.7 μM (63.98 μg/ml) CUR (C-iv). (D-ii to -iv) Cells during incubation with 31.25 μM SDS (D-ii), 86.8 μM CUR (D-iii), and 31.25 μM (9 μg/ml) SDS plus 86.8 μM (31.97 μg/ml) CUR (C-iv).
CUR mediated cell wall damage in C. albicans.
To get detailed insight into the structural changes induced upon CUR treatment, we monitored the ultrastructure of C. albicans cells using TEM (see Fig. S2 in the supplemental material). Ultrathin sections of CUR-treated (MIC100) C. albicans cells showed damage and alteration to the cell wall (see Fig. S2A in the supplemental material). We also found alteration and breakage in the walls of Candida cells after treatment with CUR in combination with CWP agents at synergistic concentrations (see Fig. S2B-iv, C-iv, and D-iv in the supplemental material). At noncandidacidal concentrations of CUR and CWP agents, the cell surface is intact, just like untreated cells (see Fig. 2B-i, C-i, and D-i in the supplemental material). These results support the idea that CUR might also target the cell wall to kill C. albicans cells.
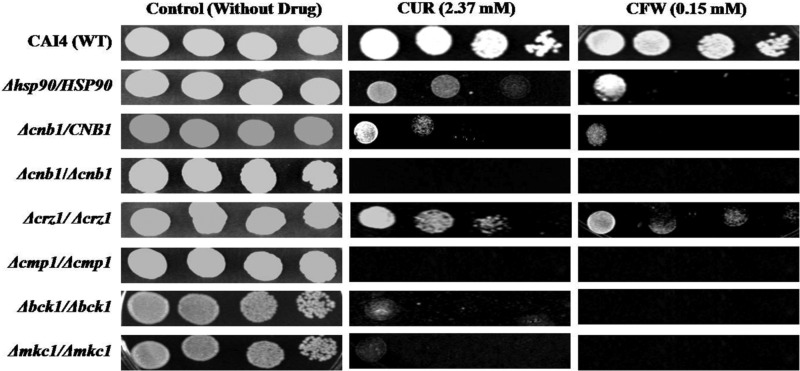
CUR confers hypersusceptibility on mutants of the calcineurin and MAP kinase pathways in C. albicans.
Our microarray results and drug susceptibility assays suggested that CUR might target critical genes of the cell wall integrity pathways. Reportedly, calcineurin and MAP kinase are two well-established pathways required for the maintenance of cell wall integrity in C. albicans and several other fungi (23–25). Therefore, the mutants of the C. albicans calcineurin signaling pathway (Δcmp1/Δcmp1, Δcnb1/CNB1, Δcnb1/Δcnb1, and Δcrz1/Δcrz1), MAP kinase pathway (Δbck1/Δbck1 and Δmkc1/Δmkc1), and stress pathway (hsp90Δ/HSP90) were examined for the effect of CUR (2.37 mM; nonfungicidal concentration) treatment. All the tested calcineurin and MAP kinase mutants exhibited hypersusceptiblity to CUR (Fig. 5). Moreover, the observed effect of CUR was comparable to the growth of these mutants in the presence of CFW (Fig. 5), a well-known agent that compromises cell wall integrity by binding to chitin (26). The data suggest that CUR disrupts cell wall integrity by targeting the MAP kinase pathway and calcineurin-mediated signaling.
FIG 5.
Calcineurin, MAP kinase, and stress mutants showed enhanced fungicidal activity of CUR in C. albicans. Cells were grown overnight in YPD at 30°C, 5-fold serially diluted, and spotted onto YPD medium containing CFW and CUR at the concentrations indicated. The plates were incubated at 30°C for 48 h.
DISCUSSION
Previously we demonstrated the antifungal activity of CUR against C. albicans and postulated that several mechanisms are responsible for cell death (7–10). Many of these mechanisms are also crucial for the development of drug tolerance in C. albicans (27). To increase our understanding of the possible mechanism of action of CUR at a molecular level, in the present study, we performed a microarray study of C. albicans response to CUR treatment. Our microarray analyses further showed that CUR employs multiple mechanisms to kill cells, viz., the cell cycle, signaling alteration, loss of cell wall integrity, metabolic shift, cell stress, DNA synthesis and repair, hyphal development, mitochondrial integrity, transcriptional and translational regulation, etc. (Fig. 1; see Table S2 in the supplemental material). Among these mechanisms, CUR shows a significant influence on the genes affecting cell wall integrity, because most of the genes of the pathway were downregulated.
Based on microarray results, we extended and verified the CUR-mediated cell wall damage hypothesis through other experiments (described in Materials and Methods). We first tested the synergistic effect of CUR with some known cell wall-perturbing agents (CFW, CR, and SDS). CUR synergized the effects of CWP agents, which suggests that the cell wall is a major target of CUR in the cell death mechanism of C. albicans. We also found loss of cell viability at synergistic concentrations of CUR and CWP agents by the time-kill, growth, and spot assays, showing that CUR and CWP agents share a target. Overall, these results validate the microarray outcome and confirm that CUR is involved in C. albicans cell wall damage.
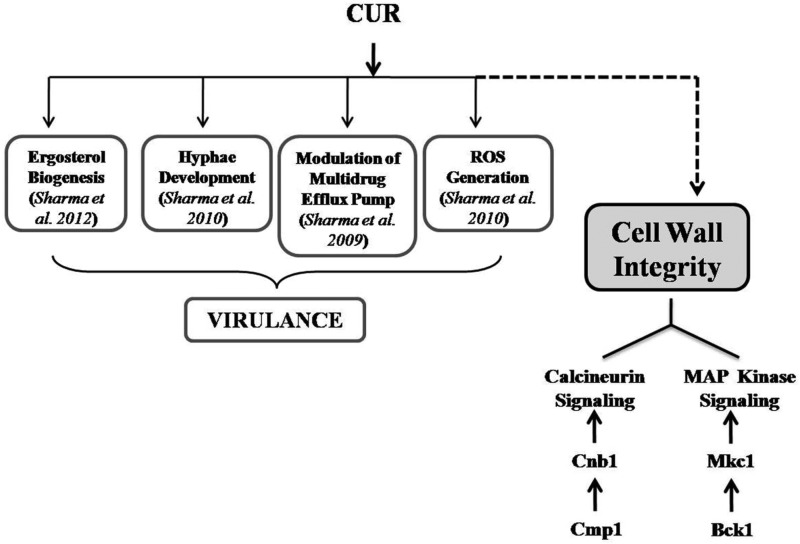
Alteration of membrane permeability is one of the major mechanisms underlying the drug tolerance phenomenon, and damage to the cell wall should affect overall cell permeability (28). We found that the cell membrane became permeable to a membrane-impermeant dye, PI (29), when cells were exposed to CUR along with CWP agents at synergistic concentrations. Further, we found extensively altered cell surface morphology and cell wall ultrastructure of C. albicans after CUR treatment, using SEM and TEM studies to gain further insights into the cell wall perturbation mechanisms of CUR. Fungal growth and development and the ability to survive under environmental stress conditions are dependent on the integrity of the cell wall (27). Reports show that calcineurin plays an important role in governing cell wall integrity, which might involve related pathways, such as the PKC and HSP90 pathways (2, 30–32). Previously, LaFayette et al. described the level of regulatory control circuitry that links PKC and calcineurin signaling and governs the cell wall integrity pathway in C. albicans (11). Our data showed that calcineurin and MAP kinase pathway mutants of C. albicans are hypersusceptible to CUR, suggesting that these genes render fungicidal activity of CUR ineffective and play a critical role in the mechanism of CUR-induced cell wall integrity defect of C. albicans. Phenotypic susceptibility assays in response to CUR in calcineurin mutants validated the microarray data. Evidently, both the cell wall integrity pathway and the calcineurin pathway are working against CUR stress, but whether these pathways are connected or work independently remains to be validated. Together, our data show that CUR in C. albicans causes direct damage to the cell wall and loosening of membrane permeability, which finally leads to cell death by leakage of the cytoplasmic content of the cells. Figure 6 summarizes the known mechanism of CUR-mediated cell death, along with our recent findings that affect cell wall integrity in C. albicans.
FIG 6.
Summary of altered mechanisms in C. albicans cells after CUR treatment. These mechanisms are crucial for the development of drug tolerance and virulence in C. albicans.
Although emerging cell wall-targeting drugs have shown great potential in treating Candida infections, many cases of development of resistance to these drugs are already appearing (7). Combinational drug therapy is an alternate and effective strategy to treat candidiasis at concentrations of drugs much lower than those used individually. In the present study, we were able to show that CUR synergizes the effects of CWP agents by targeting the cell wall in C. albicans. Thus, the doses of drugs like candins if given in combination with CUR could be significantly reduced in clinical settings. Further, this study highlights some important mechanisms of the anti-Candida effects of CUR, which include loss of cell viability, membrane permeability, and ultrastructural alteration of the cell wall. Together, our data provide mechanistic insights that demonstrate a link between the CUR susceptibility of C. albicans and the calcineurin signaling pathway and present an opportunity to employ this new information in improving treatment strategies.
Supplementary Material
ACKNOWLEDGMENTS
This work is financially supported by the University Grants Commission (UGC), New Delhi, India, under the D. S. Kothari Postdoctoral Fellowship (UGC-DSK-PDF) Scheme.
We are grateful to Genotypic Solutions, Bangalore, India, for assisting us in microarray data analysis. We acknowledge the assistance of Ashok Kumar Sahu, Ruchita Pal, and Gajender Saini of the Advanced Instrumentation Research Facility (AIRF) at Jawaharlal Nehru University, India, in confocal microscopy, SEM, and TEM experiments. We thank Joseph Heitman, Joachim Morschhauser, Dominique Sanglard, Malcolm Whiteway, and Leah Cowen for providing Candida mutant strains as generous gifts.
Footnotes
Published ahead of print 21 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01385-13.
REFERENCES
- 1.Low CY, Rotstein C. 2011. Emerging fungal infections in immunocompromised patients. F1000 Med. Rep. 3:14. 10.3410/M3-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh SD, Robbins N, Zaas AK, Schell WA, Perfect JR, Cowen LE. 2009. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 5:e1000532. 10.1371/journal.ppat.1000532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White TC, Marr KA, Bowden RA. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanglard D, Odds FC. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73–85. 10.1016/S1473-3099(02)00181-0 [DOI] [PubMed] [Google Scholar]
- 5.Martins CVB, Silva DL, Neres ATM, Magalhaes TFF, Watanabe GA, Modolo LV, Sabino AA, Fatima A, Resende MA. 2009. Curcumin as a promising antifungal of clinical interest. J. Antimicrob. Chemother. 63:337–339. 10.1093/jac/dkn488 [DOI] [PubMed] [Google Scholar]
- 6.Ravindran J, Prasad JS, Aggarwal BB. 2009. Curcumin and cancer cells: how many ways can curry kill tumor cells selectively? AAPS J. 11:495–510. 10.1208/s12248-009-9128-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma M, Manoharlal R, Negi AS, Prasad R. 2010. Synergistic anticandidal activity of pure polyphenolcurcumin I in combination with azoles and polyenes generates reactive oxygen species leading to apoptosis. FEMS Yeast Res. 10:570–578. 10.1111/j.1567-1364.2010.00637.x [DOI] [PubMed] [Google Scholar]
- 8.Sharma M, Manoharlal R, Puri N, Prasad R. 2010. Antifungal curcumin induces reactive oxygen species and triggers an early apoptosis but prevents hyphae development by targeting the global repressor TUP1 in Candida albicans. Biosci. Rep. 30:391–404. 10.1042/BSR20090151 [DOI] [PubMed] [Google Scholar]
- 9.Sharma M, Dhamgaye S, Singh A, Prasad R. 2012. Lipidome analysis reveals antifungal polyphenol curcumin affects membrane lipid homeostasis. Front. Biosci. (Elite ed) 4:1195–1209. 10.2741/451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma M, Manoharlal R, Shukla S, Puri N, Prasad T, Ambudkar SV, Prasad R. 2009. Curcumin modulates efflux mediated by yeast ABC multidrug transporters and is synergistic with antifungals. Antimicrob. Agents Chemother. 53:3256–3265. 10.1128/AAC.01497-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LaFayette SL, Collins C, Zaas AK, Schell WA, Betancourt-Quiroz M, Gunatilaka AA, Perfect JR, Cowen LE. 2010. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog. 6:e1001069. 10.1371/journal.ppat.1001069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odds FC, Brown AJ, Gow NA. 2003. Antifungal agents: mechanisms of action. Trends Microbiol. 11:272–279. 10.1016/S0966-842X(03)00117-3 [DOI] [PubMed] [Google Scholar]
- 13.Dhamgaye S, Devaux F, Manoharlal R, Vandeputte P, Shah AH, Singh A, Blugeon C, Sanglard D, Prasad R. 2012. In vitro effect of malachite green on Candida albicans involves multiple pathways and transcriptional regulators UPC2 and STP2. Antimicrob. Agents Chemother. 56:495–506. 10.1128/AAC.00574-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee D, Lelandais G, Shukla S, Mukhopadhyay G, Jacq C, Devaux F, Prasad R. 2008. Responses of pathogenic and nonpathogenic yeast species to steroids reveal the functioning and evolution of multidrug resistance transcriptional networks. Eukaryot. Cell 7:68–77. 10.1128/EC.00256-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hameed S, Dhamgaye S, Singh A, Goswami SK, Prasad R. 2011. Calcineurin signaling and membrane lipid homeostasis regulates iron mediated multidrug resistance mechanisms in Candida albicans. PLoS One 6:e18684. 10.1371/journal.pone.0018684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnaud MB, Costanzo MC, Skrzypek MS, Binkley G, Lane C, Miyasato SR, Sherlock G. 2005. The Candida Genome Database (CGD), a community resource for Candida albicans gene and protein information. Nucleic Acids Res. 33:D358–D363. 10.1093/nar/gki003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard, 3rd ed CLSI document M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 18.Guo N, Wu X, Yu L, Liu J, Meng R, Jin J. 2010. In vitro and in vivo interactions between fluconazole and allicin against clinical isolates of fluconazole-resistant Candida albicans determined by alternative methods. FEMS Immunol. Med. Microbiol. 58:193–201. 10.1111/j.1574-695X.2009.00620.x [DOI] [PubMed] [Google Scholar]
- 19.Quan H, Cao YY, Xu Z, Zhao JX, Gao PH, Qin XF, Jiang YY. 2006. Potent in vitro synergism of fluconazole and berberine chloride against clinical isolates of Candida albicans resistant to fluconazole. Antimicrob. Agents Chemother. 50:1096–1099. 10.1128/AAC.50.3.1096-1099.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukhopadhyay K, Kohli AK, Prasad R. 2002. Drug susceptibilities of yeast cells are affected by membrane lipid composition. Antimicrob. Agents Chemother. 46:3695–3705. 10.1128/AAC.46.12.3695-3705.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maurya IK, Pathak S, Sharma M, Sanwal H, Chaudhary P, Tupe S, Deshpande M, Chauhan VS, Prasad R. 2011. Antifungal activity of novel synthetic peptides by accumulation of reactive oxygen species (ROS) and disruption of cell wall against Candida albicans. Peptides 32:1732–1740. 10.1016/j.peptides.2011.06.003 [DOI] [PubMed] [Google Scholar]
- 22.Gillum AM, Tsay EY, Kirsch DR. 1984. Isolation of the Candida albicans genes for orotidine-59-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182. 10.1007/BF00328721 [DOI] [PubMed] [Google Scholar]
- 23.Kraus PR, Deborah SF, Cox GM, Heitman J. 2003. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol. Microbiol. 48:1377–1387. 10.1046/j.1365-2958.2003.03508.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cruz MC, Goldstein AL, Blankenship JR, Del Poeta M, Davis D, Cardenas ME, Perfect JR, McCusker JH, Heitman J. 2002. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 21:546–559. 10.1093/emboj/21.4.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanglard DF, Ischer O, Marchetti J, Entenza B, Bille J. 2003. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 48:959–976. 10.1046/j.1365-2958.2003.03495.x [DOI] [PubMed] [Google Scholar]
- 26.Roncero C, Duran A. 1985. Effect of calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J. Bacteriol. 163:1180–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prasad R, Kapoor K. 2005. Multidrug resistance in yeast Candida. Int. Rev. Cytol. 242:215–248. 10.1016/S0074-7696(04)42005-1 [DOI] [PubMed] [Google Scholar]
- 28.El-Nakeeb MA, Abou-Shleib HM, Khalil AM, Omar HG, El-Halfawy OM. 2011. Membrane permeability alteration of some bacterial clinical isolates by selected antihistaminics. Braz. J. Microbiol. 42:992–1000. 10.1590/S1517-83822011000300019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shapiro NM. 1988. Practical flow cytometry, 2nd ed. Alan R. Liss Press, New York, NY [Google Scholar]
- 30.Bader T, Bodendorfer B, Schroppel K, Morschhauser J. 2003. Calcineurin is essential for virulence in Candida albicans. Infect. Immun. 71:5344–5354. 10.1128/IAI.71.9.5344-5354.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyazaki T, Yamauchi S, Inamine T, Nagayoshi Y, Saijo T, Izumikawa K, Seki M, Kakeya H, Yamamoto Y, Yanagihara K, Miyazaki Y, Kohno S. 2010. Roles of calcineurin and Crz1 in antifungal susceptibility and virulence of Candida glabrata. Antimicrob. Agents Chemother. 54:1639–1643. 10.1128/AAC.01364-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blankenship JR, Wormley FL, Boyce MK, Schell WA, Filler SG. 2003. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryot. Cell 2:422–430. 10.1128/EC.2.3.422-430.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.