Abstract
As a continuation of our earlier study on the in vitro antiprotozoal activity of 40 natural sesquiterpene lactones (STLs), we extended the set of tested compounds from our laboratories to 59. On the basis of this extended data set, further enriched by literature data for 10 compounds tested under the same conditions, our quantitative structure-activity relationship (QSAR) analyses for activity against T. brucei rhodesiense (etiologic agent of human African trypanosomiasis, or sleeping sickness) were continued, and the QSAR model thus obtained with 69 structures was used to predict the activity of a virtual library of 1,750 STL structures. As a major result from these calculations, furanoheliangolide-type compounds, a subclass of STLs hitherto untested against T. brucei rhodesiense, were predicted to have an exceptionally high level of in vitro activity. Four representative compounds of this type, goyazensolide, 4,5-dihydro-2′,3′-epoxy-15-deoxygoyazensolide, budlein A, and 4,15-isoatriplicolide tiglate, were therefore tested. They displayed 50% inhibitory concentrations (IC50s) of 0.07, 0.20, 0.07, and 0.015 μM, respectively, so that the in silico prediction was experimentally confirmed. 4,15-Isoatriplicolide tiglate is the most potent STL against T. b. rhodesiense found. Furanoheliangolide STLs were thus identified as interesting leads against this parasite which deserve more detailed investigations.
INTRODUCTION
Infections with trypanosomatid parasites of the genera Trypanosoma and Leishmania are neglected diseases for which only few drug therapies exist, and these therapies are often complicated by severe adverse effects/high toxicity. Furthermore, problems associated with availability and applicability of the treatment, especially in cases of deadly Trypanosoma brucei infections (human African trypanosomiasis [HAT], or sleeping sickness), make the development of new drugs against these diseases an urgent task (1). Natural products have in many instances been found to provide interesting leads against protozoan infections (2–4).
In previous studies, it was shown by our group (5–8) and others (9–11) that sesquiterpene lactones (STLs) possess considerable activity especially against Trypanosoma brucei rhodesiense (etiologic agent of East African HAT). We previously presented quantitative structure-activity relationship (QSAR) models for a set of 40 STLs with respect to their antitrypanosomal and cytotoxic activities, and it was found that the underlying structure-activity relationships, although very similar, are not identical (7), indicating that a structural rationale for the selective activity of some compounds must exist. As a continuation of these studies, we tested further STLs and thus extended the set of data available for QSAR to 59 compounds from our laboratory. The activity data of 12 of these compounds against T. b. rhodesiense, as well as against other protozoan pathogens, including Trypanosoma cruzi, Leishmania donovani, and Plasmodium falciparum, and their cytotoxic activity against mammalian cells (L6 rat skeletal myoblasts) are reported for the first time. Besides these new data from our laboratories, we also included in the present QSAR analyses the activity data for 10 further STLs that were previously determined at the Swiss TPH laboratory under the same conditions and published with other coauthors (9–11), so that a set of 69 compounds could be analyzed.
QSAR analysis generally aims at a quantitative understanding of the structural properties responsible for bioactivity; i.e., the resulting models should have an explanatory character. At the same time, and possibly even more importantly, QSAR models may also be used for predictive purposes within certain bounds, i.e., they should allow activity predictions for compounds not previously tested and thus help rationalize compound selection for further testing. In the present study, we have extended and refined the previous QSAR model for anti-T. b. rhodesiense activity of STLs (7) and used it to predict the activities of a virtual library of 1,750 natural STLs in order to find representatives of hitherto-untested types of STLs that might show high antitrypanosomal activity. The predictions made on the basis of the present QSAR analysis were experimentally confirmed for four STLs of the furanoheliangolide type.
MATERIALS AND METHODS
Test compounds.
For compounds 1 to 40, see our previous report (7). Compound 41 was isolated from Arnica angustifolia (Asteraceae) (12), compounds 42 and 44 from Inula peacockiana (Asteraceae) (13, 14), compound 43 from Ambrosia artemisiifolia (13, 14), compounds 49 and 63 from Artemisia glabela (Asteraceae) (13, 14), compounds 50, 51, and 64 from Eupatorium perfoliatum (15), compound 57 from Arctium lappa (Asteraceae) (13, 14), compounds 58 to 61 from Inula montbretiana (Asteraceae) (8), compound 62 from Liriodendron tulipifera (Magnoliaceae) (13, 14), compound 65 from Chamaemelum nobile (Asteraceae) (13, 14), compounds 70, 71, and 72 from Lychnophora diamantinana (16), Viguiera robusta (17), and Eremanthus goyazensis (18) (Asteraceae), respectively, and compound 73 from Helianthus tuberosus (Asterceae) (13, 14). Compounds obtained from commercial sources were compounds 45 and 55 (PhytoLab GmbH & Co KG, Vestenbergsgreuth, Germany) and 52 (Karl Roth GmbH & Co., Karlsruhe, Germany).
The purity of isolated compounds was assessed by 1H nuclear magnetic resonance (1H-NMR), high-pressure liquid chromatography (HPLC), and/or thin-layer chromatography (TLC) analyses and found to be >80% in all cases.
The data for activity against T. b. rhodesiense of compounds 46 to 48, 53, 54, 56 (9, 10), and 66 to 69 (11), determined under the same experimental conditions at the Swiss Tropical and Public Health Institute (STPH) laboratory, were taken from the literature. The chemicals used as positive controls in the bioassays were obtained from commercial sources except for melarsoprol, which was a gift from WHO; their purity was specified by the suppliers to be >95% in all cases.
In vitro assays and 50% inhibitory concentration (IC50) determination.
The biological data reported in this study (Table 1) were determined essentially as reported in our previous communication (7) using bloodstream forms of T. brucei rhodesiense (STIB900 strain), intracellular amastigotes of T. cruzi (Tulahuen C4 strain, cultivated in L6 rat skeletal myoblasts), axenic amastigotes of L. donovani (MHOM-ET-67/L8 strain), and intraerythrocytic forms of P. falciparum strains K1 and NF54 (7). Cytotoxicity determinations were carried out with L6 rat skeletal myoblasts.
TABLE 1.
In vitro antiprotozoal and cytotoxic activity of STLs determined for this study and not previously published
| Compound | IC50 (μM)a |
||||
|---|---|---|---|---|---|
| T. brucei rhodesiense | T. cruzi | L. donovani | P. falciparum | Cytotoxicity (L6 myoblasts) | |
| 41 | 3.56 ± 1.69 | 13.7 ± 0.9 | 9.33 ± 1.78 | 2.91 ± 0.27b | 10.7 ± 4.0 |
| 42 | 24.4 ± 2.9 | 78.3 ± 16.4 | 22.9 ± 0.71 | 22.2 ± 1.9b | 53.3 ± 12.7 |
| 43 | 24.6 ± 1.4 | 106.4 ± 27.3 | 50.8 ± 0.6 | 25.4 ± 1.6b | 58.9 ± 8.6 |
| 44 | 13.3 ± 5.8 | 40.1 ± 12.8 | 7.67 ± 0.36 | 18.0 ± 1.5b | 22.5 ± 5.6 |
| 45 | 1.98 ± 0.29 | 17.6 ± 2.8 | 2.13 ± 0.36 | 12.1 ± 1.8b | 7.20 ± 0.46 |
| 49 | 2.52 ± 0.42 | 18.9 ± 2.0 | 2.49 ± 0.13 | 7.38 ± 1.04c | 6.18 ± 0.28 |
| 52 | 234.5d | 254.7 ± 67.2 | >400e | 196.0 ± 7.3 | >400e |
| 55 | 2.03 ± 0.26 | 27.7 ± 6.4 | 4.17 ± 0.61 | 6.29 ± 0.09b | 15.5 ± 0.4 |
| 57 | 4.31 ± 2.29 | 13.8 ± 2.2 | 6.39 ± 0.62 | 7.71 ± 0.22b | 5.30 ± 1.40 |
| 62 | 0.227 ± 0.002 | 10.8 ± 2.8 | 4.72 ± 0.71 | 8.26 ± 0.62b | 4.70 ± 0.90 |
| 63 | 2.46 ± 0.27 | 22.1 ± 0.7 | 21.9 ± 1.6 | 4.53 ± 0.69c | 7.64 ± 0.35 |
| 65 | 2.08 ± 0.09 | 11.3 ± 3.3 | 2.69 ± 1.08 | 3.12 ± 0.52c | 4.91 ± 0.53 |
| 70f | 0.073 ± 0.002 | 1.08 ± 0.10 | 0.448 ± 0.021 | 0.291 ± 0.158b | 0.494 ± 0.086 |
| 71f | 0.072 ± 0.0006 | 1.75 ± 0.01 | 5.14 ± 0.21 | 1.24 ± 0.33c | 0.384 ± 0.020 |
| 72f | 0.202 ± 0.005 | 2.13 ± 0.30 | 0.518 ± 0.053 | 2.03 ± 0.53c | 4.31 ± 0.17 |
| 73f | 0.015 ± 0.003 | 3.74 ± 1.34 | NA | 1.03 ± 0.24c | 1.15 ± 0.53 |
| Positive controls | 0.006 ± 0.002g | 1.68 ± 0.31h | 0.411 ± 0.068i | 0.241 ± 0.025b,j; 0.008 ± 0.002c,j | 0.016 ± 0.004k |
Each entry is the mean of two independent measurements ± the deviation of the minimum and maximum values. For the positive controls, data are means of 3 to 8 independent measurements ± standard deviations. The data for anti-T. b. rhodesiense activity were used together with those of compounds 1 to 40, 46 to 48, 50, 51, 53, 54, 56, 58 to 61, 64, and 66 to 69 in the current QSAR analysis. A full list with the pIC50 data of all compounds against T. b. rhodesiense is presented in Table SA1 in the supplemental material. NA, data for the axenic amastigote model were not available (inactive at 3 μg/ml against intracellular amastigotes).
Strain K1.
strain NF54.
Only one definite measurement of IC50 was obtained; the second measurement yielded an IC50 of >400 μM.
Highest concentration tested; IC50 was not determined.
Compound selected for testing after QSAR-based prediction of anti-T. b. rhodesiense activity.
Melarsoprol.
Benznidazole.
Miltefosine.
Chloroquine.
Podophyllotoxin.
Computational methods.
Three-dimensional (3D) models of all compounds were generated with MOE (versions 2009.8 to 2011.10; Chemical Computing Group) using the MMFF94x force field. A stochastic conformational search was performed for each compound (default settings of MOE) and the resulting conformers with the lowest force field energy was minimized using the AM1 hamiltonian (MOPAC module of MOE). The AM1-minimized geometries were used for the QSAR study.
A total of 123 QSAR descriptors were calculated for each of the 69 structures using the QSAR module of MOE. A full list of descriptors considered is reported as supplemental information (see Table SA2 in the supplemental material). For the QSAR analyses, the data for antitrypanosomal IC50 against T. b. rhodesiense, expressed on the molar scale, were converted to negative decadic logarithms (pIC50; for data for all compounds, see Table SA1 in the supplemental material).
The compounds were divided into a training set (n = 46) and a test set (n = 23). In order to ensure an optimal coverage of chemical space for both subsets, this division was based on a structural diversity calculation using MOE's “diverse subset” function, which calculates for each molecule in a database its similarity to each other entry with respect to a predefined similarity scheme and then ranks the structures in the order of their difference from all other structures. Structural diversity was calculated based on the 3D 3-point pharmacophore molecular fingerprint scheme (piDAPH3) as implemented in MOE, and the Tanimoto coefficient was used as a similarity measure. The 46 top-ranking (i.e., most diverse) molecules thus obtained constituted the training set, and the 23 remaining structures yielded the test set of similar diversity (see Table SA1 in the supplemental material; both sets also cover the range of biological activity in a similar manner, as shown in Fig. SA1 in the supplemental material).
For QSAR modeling, genetic algorithm-driven variable selection combined with multiple linear regression (GA-MLR) as implemented in the MOE script GA.svl (available for MOE users at the MOE svl exchange website [http://www.chemcomp.com/Support-SVL_Exchange.htm]) was used. Fixed-length GA calculations with 4, 5, and 6 variables were performed on a population of 100 equations and the full descriptor matrix. The optimization criterion was minimization of the lack of fit. The optimized final populations were cross validated using the leave-one-out scheme. Equations were then ranked by their cross-validated coefficient of determination (Q2). The best combination obtained with respect to the number of variables and Q2 is represented by equation 1 (see Results). Besides internal leave-one-out cross validation and test set predictions, a set of three independent Y-scrambling tests was performed on equation 1 in order to exclude chance correlation. The Y data were manually reassigned in a random manner (“scrambled”) to the compounds of the training set, and the regression was repeated. None of the regression equations obtained yielded a significant correlation between calculated and experimental data (R2 = 0.06, 0.04, and 0.14), nor was any of these scrambled models able to predict the activities of the test set with a significant coefficient of determination (P2 = 0.26, 0.18, and 0.04). Equation 1 was then used to predict the activity data of the test set and of the database of 1,750 STL structures.
This in-house database in its full form contains optimized 3D models (lowest-energy conformers after a stochastic conformational search with the MMFF94x force field) of all compounds included in the treatment of 13C-NMR data of STL used by Budesinsky and Saman (19). In some cases where aspects of 3D geometry were unknown, several geometric isomers were included for one compound, so that the total number of entries in the database is 2,218, exceeding the actual number of compounds (2,071). These structures were filtered with MOE's descriptor lip_violation, which counts for each molecule the number of violations of Lipniski's rule of 5 (20). Compounds yielding violation counts of >0 were eliminated from the database, yielding a total of 1,750 molecular structures for which activity predictions were made. Of these, 71 were predicted to show an IC50 of ≤0.1 μM. These comprised 31 germacranolides (including 15 furanoheliangolides), 6 eudesmanolides, 14 guaianolides, 10 pseudoguaianolides, and 10 seco-STLs of various subtypes (elemanolides, seco-guaianolides, and seco-pseudoguaianolides).
RESULTS
Antiprotozoal activity of STLs.
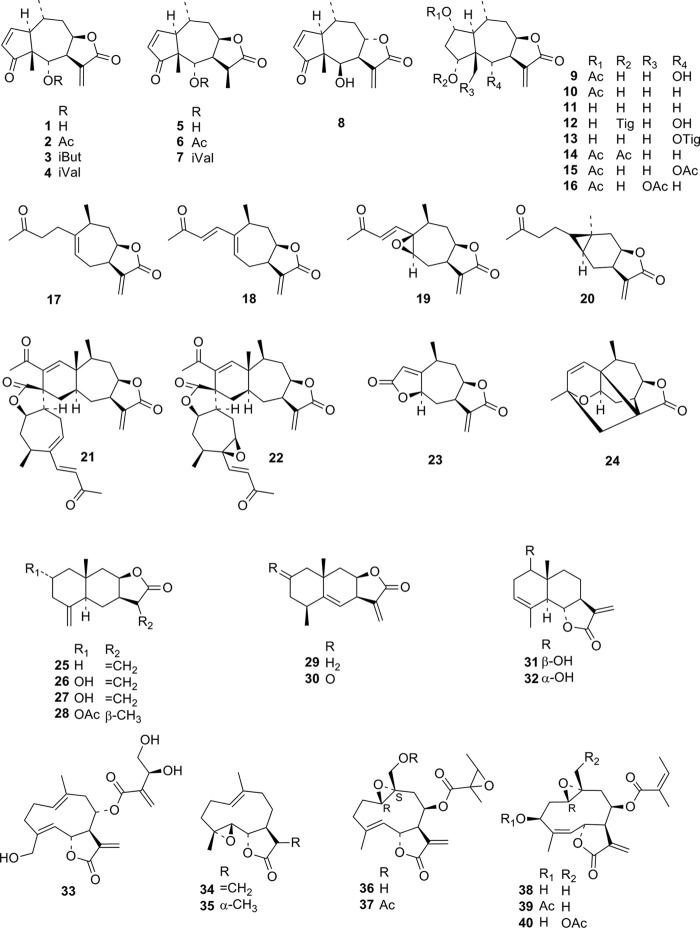
The bioactivity data of 47 sesquiterpene lactones (compounds 1 to 40, 50, 51, 58 to 61, and 64) against T. brucei rhodesiense, T. cruzi, L. donovani, and P. falciparum as well as cytotoxicity against L6 rat skeletal myoblasts were published previously (7, 8, 15). Their activity data were included in the present QSAR analysis but are reported only as supplemental information (see Table SA1 in the supplemental material). For the present investigation, the in vitro activity against the mentioned target organisms/cells of 12 further STLs (compounds 41 to 45, 49, 52, 55, 57, 62, 63 and 65; for structures, see Fig. 1; for biological data, see Table 1) was tested. The pseudoguaianolides helenalin 1 and helenalin acetate 2 remained the most active compounds against T. b. rhodesiense in our series, with IC50s of 0.05 and 0.06 μM (5, 7). Three further pseudoguaianolides included in the present series, namely, arnifolin, 2,3-dihydroaromaticin, and peruvin (compounds 41 to 43), displayed only a low level of activity. All three have a saturated ketone structure in the cyclopentane ring. Their low activity is thus in accordance with our previous structure-activity relationship studies, where the importance of a cyclopentenone structure for activity was already pointed out (7).
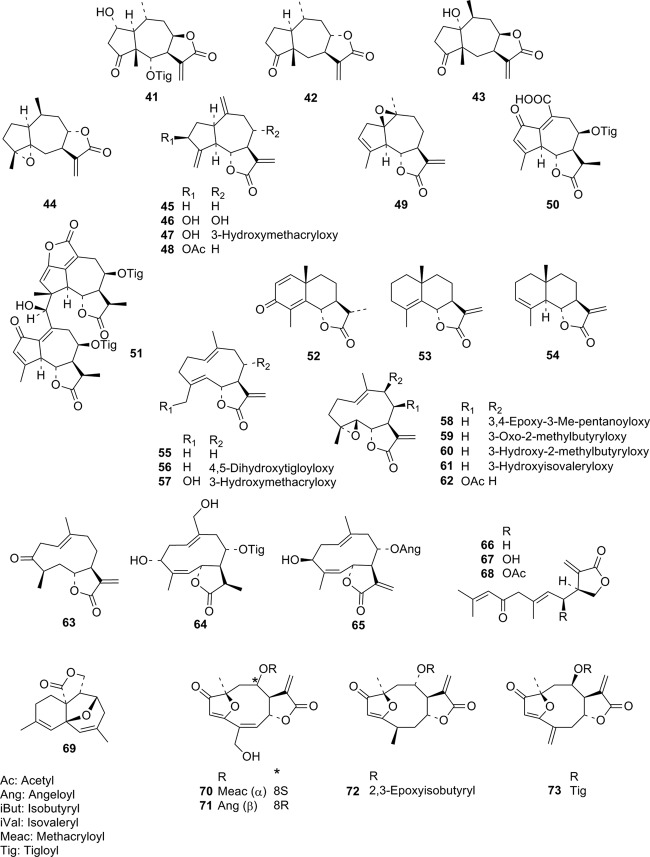
FIG 1.
Structures of STLs used in the present QSAR analysis. Compounds 1 to 40 were included in our earlier QSAR study. They are shown here for a complete overview.
The highest antitrypanosomal activity within the newly tested series of STLs was displayed by the germacranolide lipiferolide (= 8β-hydroxyparthenolide acetate [compound 62]), a constituent of the tulip tree, Liriodendron tulipifera (Magnoliaceae), which showed an IC50 of 0.23 μM against T. b. rhodesiense. It is noteworthy that this compound, recently also reported independently by others to exhibit activity against Plasmodium falciparum (21), was significantly more active against T. b. rhodesiense than four structurally related 9β-hydroxyparthenolide esters (compounds 58 to 61) recently isolated from Inula montbretiana (Asteraceae) (8) and also outmatched unsubstituted parthenolide (compound 34; IC50 = 0.39 [7]).
QSAR analysis for activity against T. brucei rhodesiense.
An attempt to test our previous quantitative structure-activity relationship (QSAR) model for compounds 1 to 40 (7) for external predictivity by calculating the activity data of 19 newly tested compounds (41 to 45, 49 to 52, 55, 57 to 65) failed to yield a significant correlation between experimental and predicted data (correlation coefficient [R] = −0.05; root mean square error of predictions [RMSEP] = 1.35 log units in pIC50). In order to arrive at a more predictive QSAR model to be used for external predictions of a large set of compounds, an extended QSAR analysis was performed after the newly determined in vitro activity data were combined with those of our earlier studies (7, 8, 15) and data for 10 further STLs determined at the STPH laboratory under identical conditions in cooperation with other coauthors (compounds 46 to 48, 53, 54, and 56 [9, 10] and 66 to 69 [11]). Thereby, a much broader chemical diversity of the data set than that in our previous study was obtained. For complete data on activity against T. b. rhodesiense of all 69 STL, see Table SA1 in the supplemental material.
Thus, a coherent data set for 69 STLs was available for the present QSAR study. Three-dimensional molecular models for all compounds were generated and 123 molecular descriptors (for the full list, see Table SA2 in the supplemental material) were calculated from these 3D structures (see Materials and Methods). The set of descriptors used in our previous QSAR study (7) was extended with a series of 78 vsurf descriptors implemented in newer releases of MOE, which represent hydrophobic/hydrophilic surface properties calculated from molecular interaction fields as originally formulated by Cruciani et al. (22).
The data set was separated into a training set (n = 46 compounds) and a test set (n = 23). In order to ensure a maximum chemical diversity of the training and test set, this division was performed after diversity ranking based on molecular fingerprints as implemented in MOE (for the selected fingerprint scheme and other details, see Materials and Methods). The 46 most diverse compounds were chosen as the training set, leaving 23 molecules for a test set with similar chemical diversity. Subsequently, QSAR analysis was performed with the training set by applying a genetic algorithm/multiple linear regression (GA/MLR) approach (see Materials and Methods). For the present study, the GA was used to select a predefined number of descriptors (4, 5, or 6) from the whole matrix, that yield an optimal correlation (coefficient of determination [R2]) of the calculated and experimental data for the training set. The result of a GA/MLR calculation is a family or population, of predefined size, of QSAR equations ranked by their R2 values. This population of optimized regression models is then submitted to leave-one-out cross validation (LOO-CV), yielding a cross validated coefficient of determination (Q2 value) for each equation. In the present study, a significant increase in Q2 was observed when the number of variables was increased from 4 to 5 (Q2max rose from 0.567 to 0.658), whereas a further increase in the number of descriptors led to only a marginal increase, i.e., Q2max = 0.667. Activity predictions for the test set molecules were then performed with the 10 best 5-variable equations. The best combination of internal and external predictivity was found for the following QSAR equation:
| (1) |
where Tbr is T. b. rhodesiense, n = 46, R2 = 0.75, RMSE = 0.42, Q2 = 0.65, and RMSEP = 0.50. Data were standardized to unit variance.
In this equation, the descriptor ENONCS refers to the accessible surface area of reactive carbon atoms in conjugated enone systems and thus represents a measure of the potential of an STL to react with nucleophilic groups of biological target molecules. ASAP4 is the fractional accessible surface area (23) attributable to atoms with a partial charge between +0.15 and +0.2 e (electrons). This charge interval is populated by hydrogen atoms attached to the double-bond carbons in α,β-unsaturated ketone groups such as H-2 and H-3 in helenalin (compound 1). This descriptor, already found to be influential in our previous antitrypanosomal QSAR (7), is also related to the presence of reactive structure elements. FASA− is the ratio of accessible surface area contributed by atoms with negative partial charge in relation to the total accessible surface area. The positive regression coefficient indicates an enhancing effect of negatively charged surface on activity. Since in this series of molecules, negative partial charge is mostly caused by oxygen, this could be interpreted in terms of the propensity to accept H bonds. In any case, this is in agreement with our previous observation that descriptors of positive surface area had a detrimental effect (7). The descriptors vsurf_D6 and vsurf_D8 refer to properties of molecular interaction fields with a hydrophobic DRY probe (22). They correspond to the tendency to generate attractive hydrophobic interactions. In this context, the descriptor D6 corresponds to the volume of regions with an interaction energy of −1.2 kcal/mol with the DRY probe, whereas D8 is calculated at −1.6 kcal/mol. The two descriptors show some colinearity (R = 0.76), as can be expected from the fact that they reflect very similar molecular properties (for a matrix of pairwise correlation between all variables in the model, see Table SA3 in the supplemental material). However, it was found that neither of them alone led to a significant improvement in comparison with a 3-variable model consisting only of the descriptors already discussed but that both have to be present in the equation to yield a very significant increase of the coefficients of determination. Their t test P values (both ≪10−4 [see Table SA4 in the supplemental material]) show that they both make highly significant contributions to the QSAR equation's overall performance. Although this combination of two descriptors is somewhat difficult to interpret, the positive and negative signs of their regression coefficients and the fact that they are both necessary for the model might indicate that it is more favorable for activity if a molecule has a relatively large region causing hydrophobic interactions of medium strength (vsurf_D6) than a large area forming strong hydrophobic interactions (vsurf_D8), i.e., that the lipophilicity should not be too high.
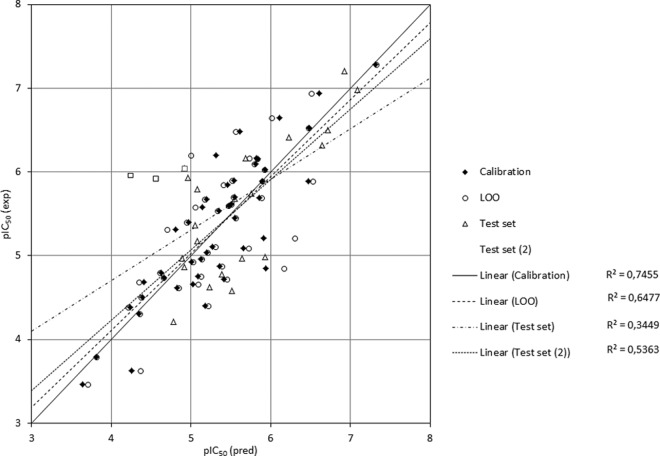
A plot of the experimental versus calculated pIC50 values for the training and test sets is shown in Fig. 2 (the underlying data are reported in Table SA2 in the supplemental material). As can be seen, the model performs satisfactorily to predict the activity of most test set molecules. The predictions are quite reasonable, especially in the region of molecules with high activity (i.e., a pIC50 of >6). Overall, however, the coefficient of determination, P2, for the predictions was only 0.35, which was due to three out of 23 compounds (no. 7, 32, and 56) whose predictions were of rather poor quality (i.e., more than 1 log unit too low). The model's failure to predict the activity of compounds 32 and 56 correctly can readily be attributed to the fact that their structural properties are not well represented in the training set. Thus, the predicted activity of compound 32 is much too low, since this compound differs only in stereochemistry from the much less active compound 31 (1-α-OH versus 1-β-OH), a difference which is not accounted for by the descriptors of the QSAR model. Compound 56 contains an ester side chain (dihydroxytigloyl) which does not occur in any other compound of the set but may be responsible for its increased activity in comparison with some similar compounds. In case of compound 7, the poor prediction is not as straightforwardly explained, since its congeners in the training set (compounds 5 and 6 with respect to the STL core and compound 4 with respect to the ester side chain) represent its structure quite well. Their calculated activity values are, however, generally somewhat too low, so that several factors leading to an underestimation of activity may sum up in this case.
FIG 2.
Correlation plot of experimental versus predicted pIC50 values for anti-T. brucei rhodesiense activity of 69 STLs as calculated by QSAR equation 1. Diamonds and circles, model calibration and leave-one-out (LOO) predictions, respectively, for the training set (46 compounds); triangles, predictions for the external test set (23 compounds); squares, three test set compounds (compounds 7, 32, and 56) with particularly poor predictions. Omission of these outliers yielded a much better correlation coefficient for the remaining 20 compounds [test set 2].
After omission of these outliers, P2 rose to 0.62 and was thus compatible with the Q2 value of the internal cross validation predictions.
It therefore appeared reasonable to attempt predictions for yet-untested STLs in order to find new candidates with high predicted activity for testing.
Activity predictions for a virtual library of 1,750 STL structures.
Our in-house database of STLs, originally created for the purpose of structure elucidation, contains optimized 3D molecular structures for a collection of almost 2,100 STLs published by Budesinsky and Saman along with their 13C NMR data (19). For the present study, structures violating Lipinski's rule of 5 (20) were eliminated from the database, so that 1,750 structures remained for further consideration. Using the QSAR model defined by equation 1 above, activity predictions were made for these “druglike” STL structures.
On the whole, 71 compounds were predicted to have a pIC50 value against T. b. rhodesiense equal to or larger than 7.0 (i.e., IC50s of ≤0.1 μM). Quite notably, among these 71 structures, 15 were found to belong to the furanoheliangolide subgroup of germacranolide-type STLs, of which, to the best of our knowledge, no representatives had hitherto been tested for activity against T. b. rhodesiense.
On this background, four furanoheliangolides (compounds 70 to 73 [Fig. 1]) available in sufficient purity and quantity were selected for in vitro testing against this parasite.
Experimental confirmation of activity predictions for furanoheliangolide-type STLs.
The furanoheliangolide goyazensolide (compound 70), with a predicted pIC50 of 7.62 (i.e., IC50pred = 0.02 μM) was chosen for testing along with its structural congener budlein A (compound 71), which was predicted to be less potent (pIC50pred = 6.60; IC50pred = 0.25 μM) but still active enough to make it interesting to test. Additionally, a structural analogue of compound 70 in which the potentially reactive α,β,γ,δ-unsaturated ketone system is reduced to an α,β-unsaturated system, i.e., 4,5-dihydro-2′,3′-epoxy-15-deoxygoyazensolide (compound 72), was also chosen. Its activity was predicted to be considerably lower (pIC50pred = 6.17, i.e., IC50pred = 0.68 μM) due to the hydrogenation of the conjugated γ,δ-double bond between C-4 and C-5. Furthermore, a structural analog with an exocyclic orientation of the γ,δ double bond (i.e., Δ4,15 instead of Δ4,5 as in compounds 70 and 71), namely, 4,15-isoatripicolide tiglate 73 (pIC50pred = 7.04; i.e., IC50pred = 0.09 μM), was also tested.
In the T. b. rhodesiense bioassay, compounds 70, 71, and 73 were highly active, with IC50s of 0.073 and 0.072 μM, respectively (70 and 71; pIC50 = 7.14) and of 0.015 μM (73; pIC50 = 7.82). Comparison with the predicted values shows that the QSAR model slightly overestimated the activity of compound 70 and underestimated that of compounds 71 and 73. However, all three had correctly been predicted to show very high activities comparable to those of compounds in the upper range of the training set, so that the validity of the QSAR model was also confirmed experimentally with these truly external predictions. At the same time, the 4,5-dihydrogoyazensolide derivative compound 72 was considerably less active than compounds 70 and 71, as predicted. Although its measured IC50 of 0.20 μM was somewhat lower than the predicted one, this prediction once more confirms that the chosen QSAR model allows external predictions with reasonable accuracy.
Comparison of compound 70 with compound 71 shows that the nature of the acyl moiety as well as the stereochemistry at C-8 does not have any significant influence on anti-T. b. rhodesiense activity, whereas changes in the α,β,γ,δ-unsaturated carbonyl structure as observed in compounds 72 and 73 had a higher impact. In case of compound 72, the lower activity is readily explained by the loss of one potentially reactive site at C-5. Better accessibility of the reactive exocyclic methylene group C-15 for Michael addition to a potential trypanosomal target can be held responsible for the increased activity of compound 73 in comparison with the activated methine carbon (C-5) in compounds 70 and 71.
Compound 73 is the STL with the highest in vitro activity against T. b. rhodesiense identified until now. It also displays a much higher selectivity index (SI = 77) than its congeners 70 and 71 (SI = 7 and 10, respectively) as well as the most active pseudoguaianolides reported so far (e.g., helenalin 1 and helenalin acetate 2, with SIs of 19 and 13, respectively).
DISCUSSION
Sesquiterpene lactones have been known for their broad diversity of chemical structure as well as biological activities for a long time. The structure-activity relationships of their various pharmacological and toxicological effects have been reviewed extensively (24, 25). Most of these activities have been attributed to the potential of many STLs to form covalent bonds with biological nucleophiles, especially cysteine residues of proteins. The results of our QSAR analyses for antiprotozoal activity clearly point in the same direction, since descriptors corresponding to the presence and size of potential Michael acceptor structures play an important role in both our previous QSAR model (7) and the present investigation, in which the data set was extended to a wider variety of different STL structures. It is quite noteworthy that several QSAR studies on the cytotoxic activity of STLs against mammalian cells have obtained similar results (23, 26). As we pointed out earlier, the mechanisms underlying toxicity of STLs toward T. b. rhodesiense and mammalian cells are probably very similar. We demonstrated that high selectivity toward T. b. rhodesiense among these compounds is directly correlated with high anti-T. b. rhodesiense potency rather than with low mammalian cytotoxicity (7). It is interesting that we very recently reported in a study on the inhibitory activity of STLs against the proto-oncogene c-Myb that compounds of the helenalin and furanoheliangolide types, representing the most active and selective STLs against T. b. rhodesiense as well as against c-Myb-dependent gene expression in vertebrate cells, also share a high degree of similarity in 3D molecular structure (13). A biophore pattern very similar to that postulated for the c-Myb inhibitory activity (13) could also result for the activity against T. b. rhodesiense. However, in vitro antitrypanosomal activity of these compounds against African trypanosomes is about 10 to 100 times stronger than their anti-c-Myb activity. (It should be noted that the discovery of high activity of furanoheliangolides against T. b. rhodesiense, guided by predictions made with the QSAR model presented here, was made chronologically before that of the anti-c-Myb-activity of these compounds. The former finding actually prompted us to also subject some of these compounds to the c-Myb inhibition assay.) In any case, the similar pattern of activity of STL in these unrelated biological systems gives rise to the hypothesis that the molecular mechanism underlying these activities should be closely related. Neither the molecular target in trypanosomes nor the exact binding site on c-Myb or a cooperating protein being known at present, it is still tempting to speculate that these targets share some similarity with respect to their structure and susceptibility to the binding of particular STLs.
An important issue in QSAR modeling is the predictive power of such models. The present QSAR analysis, based on 3D molecular descriptors, has proven its external predictivity, since it led to the discovery of hitherto-unknown high in vitro activity of furanoheliangolide-type STLs against T. brucei rhodesiense. Within the limited number of representatives of this group tested so far, 4,15-isoatriplicolide tiglate (compound 73) was found to be the most active compound, with an impressive IC50 of only 15 nM and a selectivity index of 77. Our previous observation that parasite selectivity among STLs is a function of high antitrypanosomal activity rather than of low cytotoxicity (7) is once more confirmed by this finding.
Given the considerable structural variation within the furanoheliangolide group, it will be of high interest to investigate further compounds of this type, which, as a result of this study, can be considered interesting leads against T. brucei rhodesiense. It was recently demonstrated by Zimmermann et al. that the STL cynaropicrin (compound 47; IC50 = 0.28 μM) is active against T. b. rhodesiense in an in vivo mouse model (10). It remains to be shown whether the much higher level of in vitro activity of the furanoheliangolides found in our study also leads to increased in vivo potency.
Supplementary Material
ACKNOWLEDGMENTS
This work was performed as part of the activities of the Research Network Natural Products against Neglected Diseases (ResNetNPND; http://www.uni-muenster.de/ResNetNPND/index.html).
Support from the Chemical Computing Group (CCG), Montreal, Canada, in the form of a software license grant is gratefully acknowledged. F.B.D.C. is grateful to the Brazilian funding agencies FAPESP, CAPES, and CNPq.
Footnotes
Published ahead of print 28 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01263-13.
REFERENCES
- 1.Crompton DWT, Peters P. (ed). 2010. First WHO report on neglected tropical diseases: working to overcome the global impact of neglected tropical diseases. Publication WHO/HTM/NTD/2010.1 WHO Press, Geneva, Switzerland [Google Scholar]
- 2.Schmidt TJ, Khalid SA, Romanha AJ, Alves TMA, Biavatti MW, Brun R, Da Costa FB, de Castro SL, Ferreira VF, de Lacerda MVG, Lago JHG, Leon LL, Lopes NP, das Neves Amorim RC, Niehues M, Ogungbe IV, Pohlit AM, Scotti MT, Setzer WN, de NC Soeiro M, Steindel M, Tempone AG. 2012. The potential of secondary metabolites from plants as drugs or leads against protozoan neglected diseases—part I. Curr. Med. Chem. 19:2128–2175. 10.2174/092986712800229023 [DOI] [PubMed] [Google Scholar]
- 3.Schmidt TJ, Khalid SA, Romanha AJ, Alves TMA, Biavatti MW, Brun R, Da Costa FB, de Castro SL, Ferreira VF, de Lacerda MVG, Lago JHG, Leon LL, Lopes NP, das Neves Amorim RC, Niehues M, Ogungbe IV, Pohlit AM, Scotti MT, Setzer WN, de NC Soeiro M, Steindel M, Tempone AG. 2012. The potential of secondary metabolites from plants as drugs or leads against protozoan neglected diseases—part II. Curr. Med. Chem. 19:2176–2228. 10.2174/092986712800229087 [DOI] [PubMed] [Google Scholar]
- 4.Hoet S, Opperdoes F, Brun R, Quetin-Leclercq J. 2004. Natural products active against African trypanosomes: a step towards new drugs. Nat. Prod. Rep. 21:353–364. 10.1039/b311021b [DOI] [PubMed] [Google Scholar]
- 5.Schmidt TJ, Willuhn G, Brun R, Khalid SA. 2002. Antitrypanosomal activity of helenalin and some related sesquiterpene lactones. Planta Med. 68:750–751. 10.1055/s-2002-33799 [DOI] [PubMed] [Google Scholar]
- 6.Nour AMM, Khalid SA, Kaiser M, Brun R, Abdallah WE, Schmidt TJ. 2009. The antiprotozoal activity of sixteen Asteraceae species native to Sudan and bioactivity-guided isolation of xanthanolides from Xanthium brasilicum Vell. Planta Med. 75:1363–1368. 10.1055/s-0029-1185676 [DOI] [PubMed] [Google Scholar]
- 7.Schmidt TJ, Nour AMM, Khalid SA, Kaiser M, Brun R. 2009. Quantitative structure-antiprotozoal activity relationships of sesquiterpene lactones. Molecules 14:2062–2076. 10.3390/molecules14062062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gökbulut A, Kaiser M, Brun R, Sarer E, Schmidt TJ. 2012. 9β-Hydroxyparthenolide esters from Inula montbretiana DC. and their antiprotozoal activity. Planta Med. 78:225–229. 10.1055/s-0031-1280371 [DOI] [PubMed] [Google Scholar]
- 9.Julianti T, Hata Y, Zimmermann S, Kaiser M, Hamburger M, Adams M. 2011. Antitrypanosomal sesquiterpene lactones from Saussurea costus. Fitoterapia 82:955–959. 10.1016/j.fitote.2011.05.010 [DOI] [PubMed] [Google Scholar]
- 10.Zimmermann S, Kaiser M, Brun R, Hamburger M, Adams M. 2012. Cynaropicrin: the first plant natural product with in vivo activity against Trypanosoma brucei. Planta Med. 78:553–556. 10.1055/s-0031-1298241 [DOI] [PubMed] [Google Scholar]
- 11.Karioti A, Skaltsa H, Kaiser M, Tasdemir D. 2009. Trypanocidal, leishmanicidal and cytotoxic effects of anthecotulide-type linear sesquiterpene lactones from Anthemis auriculata. Phytomedicine 16:783–787. 10.1016/j.phymed.2008.12.008 [DOI] [PubMed] [Google Scholar]
- 12.Schmidt TJ, Willuhn G, Steigel A, Wendisch D. 1995. Sesquiterpene lactones and inositol esters from Arnica angustifolia. Planta Med. 61:544–550. 10.1055/s-2006-959368 [DOI] [PubMed] [Google Scholar]
- 13.Schomburg C, Schuehly W, Da Costa FB, Klempnauer KH, Schmidt TJ. 2013. Natural sesquiterpene lactones as inhibitors of Myb-dependent gene expression: structure-activity relationships. Eur. J. Med. Chem. 63:313–320. 10.1016/j.ejmech.2013.02.018 [DOI] [PubMed] [Google Scholar]
- 14.Schomburg C. 2013. Naturstoffe als Inhibitoren c-Myb-abhängiger Transkriptionsprozesse. Dissertation; University of Münster, Münster, Germany [Google Scholar]
- 15.Maas M, Hensel A, Da Costa FB, Brun R, Kaiser M, Schmidt TJ. 2011. An unusual dimeric guaianolide with antiprotozoal activity and further sesquiterpene lactones from Eupatorium perfoliatum. Phytochemistry 72:635–644. 10.1016/j.phytochem.2011.01.025 [DOI] [PubMed] [Google Scholar]
- 16.Da Costa FB, Dias Da, Lopes JLC, Vichnewski W. 1993. Flavonoids and heliangolides from Lychnophora diamantinana. Phytochemistry 34:261–263. 10.1016/S0031-9422(00)90815-X [DOI] [Google Scholar]
- 17.Arakawa NS, Schorr K, Ambrosio SR, Merfort I, Da Costa FB. 2008. Further sesquiterpene lactones from Viguiera robusta and the potential anti-inflammatory activity of a heliangolide: inhibition of human neutrophil elastase release. Z. Naturforsch. 63c:533–538 [DOI] [PubMed] [Google Scholar]
- 18.Vichnewski W, Sarti SJ, Gilbert B, Herz W. 1976. Goyazensolide, a schistosomicidal heliangolide from Eremanthus goyazensis. Phytochemistry 15:191–193. 10.1016/S0031-9422(00)89082-2 [DOI] [Google Scholar]
- 19.Budesinsky M, Saman D. 1995. Carbon-13 NMR spectra of sesquiterpene lactones. Ann. Rep. NMR Spectr. 30:231–475. 10.1016/S0066-4103(08)60027-7 [DOI] [Google Scholar]
- 20.Lipinski CA. 2000. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 44:235–249. 10.1016/S1056-8719(00)00107-6 [DOI] [PubMed] [Google Scholar]
- 21.Graziose R, Rathinasabapathy T, Lategan C, Poulev A, Smith PJ, Grace M, Lila MA, Raskin I. 2011. Antiplasmodial activity of aporphine alkaloids and sesquiterpene lactones from Liriodendron tulipifera L. J. Ethnopharmacol. 133:26–30. 10.1016/j.jep.2010.08.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cruciani G, Crivori P, Carrupt PA, Testa B. 2000. Molecular fields in quantitative structure—permeation relationships: the VolSurf approach. J. Mol. Struct. (Theochem) 503:17–30. 10.1016/S0166-1280(99)00360-7 [DOI] [Google Scholar]
- 23.Schmidt TJ, Heilmann J. 2002. Quantitative structure-cytotoxicity relationships of sesquiterpene lactones derived from partial charge (Q)-based fractional accessible surface area descriptors (Q_frASAs). Quant. Struct. Act. Relat. (QSAR) 21:276–287. [DOI] [Google Scholar]
- 24.Schmidt TJ. 1999. Toxic activities of sesquiterpene lactones—structural and biochemical aspects. Curr. Org. Chem. 3:577–605 [Google Scholar]
- 25.Schmidt TJ. 2006. Structure-activity relationships of sesquiterpene lactones, p 309–392 In Atta-ur-Rahman (ed), Studies in natural products chemistry, vol. 33 Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 26.Schmidt TJ. 1999. Quantitative structure-cytotoxicity relationships within a series of helenanolide type sesquiterpene lactones. (Helenanolide type sesquiterpene lactones, IV.) Pharm. Pharmacol. Lett. 9:9–13 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.