Abstract
Conjugative transposon Tn5253, an integrative conjugative element (ICE) of Streptococcus pneumoniae carrying the cat and tet(M) genes, was shown to be 64,528 bp in size and to contain 79 open reading frames, of which only 38 could be annotated. Two distinct genetic elements were found integrated into Tn5253: Tn5251 (18,033 bp), of the Tn916-Tn1545 family of ICEs, and Ωcat(pC194) (7,627 bp), which could not conjugate but was capable of intracellular mobility by excision, circularization, and integration by homologous recombination. The highest conjugation frequency of Tn5253 was observed when Streptococcus pyogenes was the donor (6.7 × 10−3 transconjugants/donor).
TEXT
Comparative analysis of sequenced Streptococcus pneumoniae genomes indicates that many genes clustered in integrative and conjugative elements (ICEs) are responsible for pneumococcal genome evolution (1). ICEs, which comprise conjugative transposons, are found integrated into bacterial genomes and can be excised to form covalently closed circular intermediates that can either integrate within the cell at a different genomic site or move to a new bacterial host upon conjugative transfer (2). One of the first ICEs to be identified was Tn5253, originally called Ω(cat-tet) BM6001, which is a conjugative transposon found in the chromosome of a serogroup 19 clinical isolate of S. pneumoniae (3, 4). Tn5253 was shown to be a genetic element conferring resistance to chloramphenicol and tetracycline that is capable of conjugal transfer and site-specific chromosomal integration in different Gram-positive bacteria belonging to the genera Streptococcus and Enterococcus (5–7). Tn5253 was described as a composite conjugative transposon since it carried a complete copy of another ICE, named Tn5251, integrated into its sequence (8, 9). Tn5251 is a conjugative transposon of the Tn916-Tn1545 family (10) that was shown to be a fully autonomous ICE capable of conjugal transfer to a variety of bacterial species, including S. pneumoniae, Streptococcus gordonii, Streptococcus pyogenes, Streptococcus agalactiae, Enterococcus faecalis, and Bacillus subtilis (11). Here we report the complete annotated sequence of Tn5253 and its phenotypic characterization, which allowed the identification of a new genetic element, Ωcat(pC194), integrated into the Tn5253 sequence and containing a linearized copy of cat-carrying plasmid pC194 (12).
The nucleotide sequence of Tn5253 (GenBank accession no. EU351020) was determined by direct sequencing of seven PCR fragments spanning the whole element and obtained from S. pneumoniae strain DP1322 (13). PCR primers (see Table S1 in the supplemental material) were designed on the basis of (i) available Tn5253 sequences (8, 14), (ii) Tn5253 chromosomal junction sequences (7), and (iii) the cat gene of staphylococcal plasmid pC194 (GenBank accession no. V01277), which is homologous to the cat gene of Tn5253 (15). The DNA sequence was confirmed on the other strand by using short PCR fragments as sequencing templates. PCR, DNA sequencing, and sequence analysis were performed as already described (11).
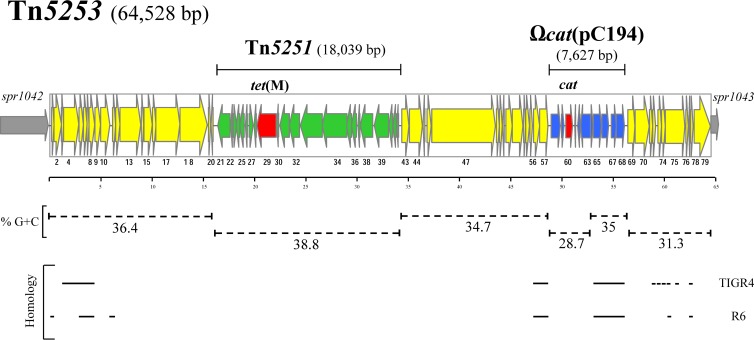
Tn5253 was found to be 64,528 bp in size and to contan 79 open reading frames (ORFs). Two distinct genetic elements were found integrated into Tn5253, (i) Tn5251, 18,033 bp long, belonging to the Tn916-Tn1545 family of tet(M)-carrying ICEs, and (ii) Ωcat(pC194), 7,627 bp long, an element carrying cat-containing plasmid pC194 (Fig. 1). Insertion of Tn5251 and Ωcat(pC194) identified three regions differing in GC content in Tn5253, (i) the left arm, containing ORFs 1 to 20, with a GC content of 36.4%; (ii), the central region, ORFs 43 to 57, with a GC content of 34.7%; and (iii) the right arm, ORFs 69 to 79, with a GC content of 31.3%. Tn5251 showed the highest GC content (38.8%), the integrated copy of pC194 showed the lowest GC content (28.7%), and the GC content of the rest of Ωcat(pC194) was 35%. (Fig. 1). A further mark of separation between the left arm and the rest of Tn5253 is the presence of orf20, coding for a truncated transposase of the IS110 family, right before the att site of Tn5251 (Table 1). Manual homology-based annotation (11) with functional prediction of the hypothetical gene product was possible for only 38 out of the 79 predicted ORFs, whereas 41 ORFs encoded hypothetical proteins that showed no homology to other characterized sequences (Table 1). Tn5253 showed a typical ICE modular organization (16) with an “intercellular mobility module” spanning orf13 to orf18 and a “recombination module” spanning orf74 to orf79.
FIG 1.
Structure of S. pneumoniae Tn5253. Sequence analysis of Tn5253 showed that it is 64,528 bp in size and contains 79 ORFs. Tn5253 contains two distinct integrated genetic elements, the 18,033-bp-long ICE Tn5251 carrying tet(M) and the 7,627-bp-long element Ωcat(pC194) containing cat. ORFs and their directions of transcription are represented by arrows, and the annotated ORFs are indicated only by their numbers. The regions corresponding to Tn5251 and Ωcat(pC194) are indicated by solid bars, and their ORFs are represented by thinner arrows. Tetracycline and chloramphenicol resistance genes are represented by red arrows. Chromosomal genes flanking the Tn5253 insertion site are represented by thin gray arrows. The different GC contents of the various regions are indicated by dotted bars. Homologies with pneumococcal chromosomal regions are marked by solid lines. The scale is in kilobases.
TABLE 1.
Annotated ORFs of Tn5253
| ORF (no. of amino acids)a,b | Annotation and comments; reference | Pfam domain(s)c (amino acids) [E value(s)] | Homologous protein ID/origin | No. of amino acids identical/total (%)-no. similar/total (%) | E valued |
|---|---|---|---|---|---|
| repA/orf2 (259) | Plasmid replication initiator protein A, N terminal; 20 | RepA_N (13-88) [4.1e-28] | |||
| orf4 (452) | DNA cytosine methyltransferase; 21 | ||||
| orf8 (195) | Protease, putative | Abi-CAAX protease self-immunity (103-188) [1.6e-13] | |||
| orf9 (196) | Abortive infection protein AbiEi; 22 | DUF4095 (29-146) [2.1e-23] | AAB52382.1/pNP40 L. lactis | 53/203 (26)-91/203 (44) | 6e-06 |
| orf10 (278) | Abortive infection protein AbiEii; 22 | DUF1814 (37-260) [2.1e-19] | AAB52383.1/pNP40 L. lactis | 81/279 (29)-136/279 (48) | 3e-18 |
| traG/orf13 (625) | TraG-like protein, involved in bacterial conjugal transfer; 23 | T4SS-DNA_transf (133-574) [1.1e-66] | |||
| orf15 (284) | Membrane protein, putative | TrbL (123-269) [0.00026] | YP_195781.1 (prgH)/pCF10 E. faecalis | 88/261 (33)-150/261 (57) | 5e-24 |
| orf16 (119) | Type IV secretion system protein | PrgI (4-95) [2.1e-22] | |||
| orf17 (785) | Type IV secretion protein, VirB4, ATPase | AAA_10 (449-736) [3.8e-44] | YP_195783.1 (prgJ)/pCF10 E. faecalis | 284/750 (37)-448/750 (59) | 5e-134 |
| orf18 (937) | Peptidoglycan hydrolase, putative | CHAP (805-926) [8.1e-30] | |||
| orf20 (64) | ISSth5, transposase, IS110 family, truncated | YP_139721.1/S. thermophilus | 43/64 (67)-47/64 (73) | 1e-13 | |
| orf43 (243) | ABC transporter: ATP-binding protein, putative | ABC_tran (27-173) [2.8e-30] | |||
| orf44 (438) | ABC transporter: permease (10 transmembrane helices predicted) | FtsX (55-176) [4.7e-07] | |||
| orf47 (2028) | Restriction-modification protein, putative | Methyltransf_26 (504-615) [1e-17], SNF2_N (1253-1600) [2.3e-14], Helicase_C (1698-1768) [2.2e-05] | |||
| orf52 (361) | DNA primase, putative | DUF3991 (113-198) [2e-13], Toprim_2 (203-329) [4.7e-10] | |||
| pezA/orf56 (158) | Antitoxin (TA system); 18 | ||||
| pezT/orf57 (252) | Toxin (TA system); 18 | ||||
| cat/orf60 (216) | CATe | CAT (1-205) [2.8e-95] | NP_040437.1/pC194 S. aureus | 216/216 (100) | |
| orf63 (281) | Plasmid replication protein | Rep_1 (54-250) [1e-48] | NP_040435.3/pC194 S. aureus | 194/195 (99)-195/195 (100) | 6e-111 |
| nplT/orf65 (245) | Truncated neopullulanase (α-amylase); 24 | Alpha-amylase (2-141) [5.5e-25] | |||
| pezA/orf67 (158) | Antitoxin (TA system); 18 | ||||
| pezT/orf68 (256) | Toxin (TA system); 18 | ||||
| umuD/orf69 (227) | UmuD MucA homolog; 25 | HTH_3 (7-61) [2.3e-17], Peptidase_S24 (143-205) [8e-12] | |||
| umuC/orf70 (471) | SOS response UmuC protein; 25 | IMS (17-207) [2.2e-17], IMS_HHH (223-255) [5.1e-06], IMS_C (268-382) [5.9e-12] | |||
| orf74 (121) | Bacterial mobilization protein, putative; 26 | MobC (60-107) [5.4e-15] | |||
| orf75 (609) | Relaxase; 27 | Relaxase (12-264) [1.6e-73] | |||
| orf76 (75) | Transcriptional regulator, putative; 28 | HTH_3 (5-60) [2.1e-15] | |||
| xis/orf78 (77) | DNA excisionase; 29 | AAA72427.1/Tn5276 L. lactis | 26/60(43)-43/60 (71) | 5e-06 | |
| int/orf79 (502) | DNA integrase; 30 | Phage_integrase (188-394) [2.3e-24] | NP_150133.1/bacteriophage MM1 S. pneumoniae | 121/413 (29)-193/413 (46) | 4e-26 |
The number of amino acids is shown in parentheses.
orf21 through orf42 correspond to orf1 to orf22 of Tn5251 and are annotated elsewhere (11). orf58 through orf68 belong to Ωcat(pC194).
The numbers in parentheses represent the part of the protein homologous to the Pfam domain.
Determined by compositional matrix adjustment.
CAT, chloramphenicol acetyltransferase.
Transconjugants of different bacterial species (Streptococcus agalactiae, Streptococcus gordonii, Streptococcus pyogenes, and Enterococcus faecalis) harboring Tn5253 were used as donors in mating experiments with S. pneumoniae FP11 (17) as the recipient (Table 2). The frequency of transfer of the element from S. pyogenes transconjugant FR40 is, to our knowledge, the highest obtained for an ICE (6.7 × 10−3 transconjugants per donor), while we could not transfer Tn5253 from the enterococcal donor FR49 (<1.8 × 10−8 transconjugants per donor). The frequency of Tn5253 transfer was also investigated in capsulated S. pneumoniae recipients with different genetic backgrounds. Conjugal transfer of Tn5253 from the standard rough donors to capsulated pneumococci occurred at frequencies ranging from 3.2 × 10−5 to 4.4 × 10−7 transconjugants per donor, while the frequency was 1.6 × 10−4 transconjugants per donor among the rough type 2 derivatives.
TABLE 2.
Conjugation frequencies of Tn5253
| Donor strain (species) | Pneumococcal recipient | Transfer frequencya | Representative transconjugant |
|---|---|---|---|
| FR67 (S. agalactiae) | FP11 (type 2 rough) | 1.1 × 10−6 | FR81 |
| FR43 (S. gordonii) | FP11 (type 2 rough) | 8.3 × 10−7 | FR61 |
| FR40 (S. pyogenes) | FP11 (type 2 rough) | 6.7 × 10−3 | FR59 |
| FR49 (E. faecalis) | FP11 (type 2 rough) | <1.8 × 10−8 | |
| FR50 (E. faecalis) | FP11 (type 2 rough) | <2.7 × 10−8 | |
| FR22 (S. pneumoniae) | FP11 (type 2 rough) | 1.6 × 10−4 | FR58 |
| FR58 (S. pneumoniae) | FP58 (type 2) | 2 × 10−5 | FR38 |
| FR58 (S. pneumoniae) | HB565 (type 3) | 4.4 × 10−7 | FR39 |
| FR22 (S. pneumoniae) | FP47 (type 4) | 3.2 × 10−5 | FR54 |
| FR58 (S. pneumoniae) | FR55 (type 6) | 1.3 × 10−5 | FR56 |
The transfer frequency is expressed as the number of transconjugant CFU per donor CFU, and each result is the mean of at least three mating experiments.
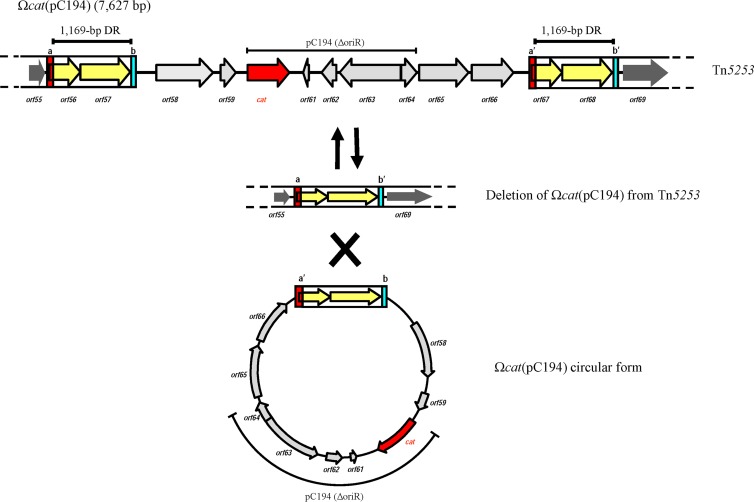
Ωcat(pC194) was found to be 7,627 bp long and to carry a copy of cat-containing plasmid pC194 harboring a 93-bp deletion involving the replication origin (Fig. 1 and 2). The element in its Tn5253-integrated form was flanked by two 1,169-bp direct repeats (DRs) corresponding to orf56-orf57 and orf67-orf68 (Fig. 2). The 1,169-bp DRs were part of two longer 1,357-bp imperfect DRs; 9 out of 13 mismatches (including two deletions) were located at the 5′ end, clustered in the first 146 nucleotides (designated a and a′ in Fig. 2), whereas 4 out of 13 are located at the 3′ end, clustered in the last 42 nucleotides (designated b and b′ in Fig. 2). PCR and sequencing analysis showed that the Ωcat(pC194) element excised from Tn5253 produced a circular intermediate and a deletion in Tn5253 (Fig. 2). The recombination event leading to the excision of Ωcat(pC194) from Tn5253 occurred within the 1,169-bp segment, leaving in Tn5253 a copy of the 1,169-bp segment flanked by imperfect DRs a and b′, whereas in the circular intermediate it was flanked by imperfect DRs a′ and b. The element contained 11 ORFs (orf58 to orf68), 5 out of 11 belonging to plasmid pC194, while 4 out of 11, orf65 to orf68, showed a high level of homology (>90%) to region spr0948 to spr0952 of R6 S. pneumoniae (Table 1). When investigating Ωcat(pC194) mobility, we found that under our experimental conditions, the element could not be transferred by conjugation (<4.1 × 107 transconjugants per donor), but it was capable of intracellular mobility. In fact, we found and sequenced a copy of Ωcat(pC194) integrated into the chromosomal spr0948-to-spr0952 region (GenBank accession no. GU808561). DNA sequence analysis of three different transconjugants containing Tn5253 indicated that ectopic integration of Ωcat(pC194) occurred by insertion duplication at different positions between spr0948 and spr0952 (data not shown), as expected for homologous recombination. This ectopic integration generated two long DRs that flank the element and are homologous to the long DRs that also flank Ωcat(pC194) when it is inserted into Tn5253. It is interesting that the long DRs at both sites each contain a complete copy of the genes coding for a toxin-antitoxin system (18), the presence of which may contribute to the stable maintenance of these genetic elements in the bacterial host.
FIG 2.
S. pneumoniae Ωcat(pC194) is a 7,627-bp-long element integrated into Tn5253 and contains a copy of plasmid pC194 lacking its replication origin. It is flanked by two 1,169-bp DRs (boxed yellow arrows) that are part of two longer 1,357-bp imperfect DRs. The 5′ and 3′ ends of the imperfect DRs are represented by red (a and a′) and blue (b and b′) boxes. Ωcat(pC194) is excised from Tn5253 and produces a circular form and a deletion in Tn5253. Excision of Ωcat(pC194) occurs by recombination between the flanking DRs and leaves in Tn5253 a single DR flanked by a and b′, while the DR of the circular form is flanked by a′ and b.
In conclusion, sequencing of the whole element allowed us to clearly define the composite nature of Tn5253, which was shown to contain two independent genetic elements, (i) fully functional conjugative transposon Tn5251 (11) and (ii) the newly recognized cat-containing element Ωcat(pC194). It is noteworthy that a large fraction (52%) of the Tn5253 ORFs code for putative proteins of unknown function, confirming that previously undescribed genes are preferentially carried by mobile genetic elements and reinforcing the notion that investigation of the mobilome is essential for understanding bacterial genomes (19).
Supplementary Material
ACKNOWLEDGMENTS
This work is dedicated to the memory of Walter Guild, who, by working on Tn5253, pioneered the field of research on integrative and conjugative elements. We also acknowledge the decisive contribution to the understanding of Tn5253 of the people from Walter Guild's lab: Nadja Shoemaker, Mike Smith, Scott Priebe, Judith Hageman, and Moses Vijayakumar.
The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement 241446 (project ANTIRESDEV).
Footnotes
Published ahead of print 2 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01764-13.
REFERENCES
- 1.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, MacGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parker J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434. 10.1126/science.1198545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullany P, Roberts AP, Wang A. 2002. Mechanism of integration and excision in conjugative transposons. Cell. Mol. Life Sci. 59:2017–2022. 10.1007/s000180200001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shoemaker NB, Smith MD, Guild WR. 1979. Organization and transfer of heterologous chloramphenicol and tetracycline resistance genes in pneumococcus. J. Bacteriol. 139:432–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vijayakumar MN, Priebe SD, Pozzi G, Hageman JM, Guild WR. 1986. Cloning and physical characterization of chromosomal conjugative elements in streptococci. J. Bacteriol. 166:972–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shoemaker NB, Smith MD, Guild WR. 1980. DNase-resistant transfer of chromosomal cat and tet insertions by filter mating in pneumococcus. Plasmid 3:80–87. 10.1016/S0147-619X(80)90036-0 [DOI] [PubMed] [Google Scholar]
- 6.Pozzi G, Musmanno RA, Renzoni EA, Oggioni MR, Cusi MG. 1988. Host-vector system for integration of recombinant DNA into chromosomes of transformable and nontransformable streptococci. J. Bacteriol. 170:1969–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vijayakumar MN, Ayalew S. 1993. Nucleotide sequence analysis of the termini and chromosomal locus involved in site-specific integration of the streptococcal conjugative transposon Tn5252. J. Bacteriol. 175:2713–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Provvedi R, Manganelli R, Pozzi G. 1996. Characterization of conjugative transposon Tn5251 of Streptococcus pneumoniae. FEMS Microbiol. Lett. 135:231–236. 10.1111/j.1574-6968.1996.tb07994.x [DOI] [PubMed] [Google Scholar]
- 9.Ayoubi P, Kiliç AO, Vijayakumar MN. 1991. Tn5253, the pneumococcal Ω(cat tet) BM6001 element, is a composite structure of two conjugative transposons, Tn5251 and Tn5252. J. Bacteriol. 173:1617–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rice LB. 1998. Tn916 family of conjugative transposons and determination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 42:1871–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santoro F, Oggioni MR, Pozzi G, Iannelli F. 2010. Nucleotide sequence and functional analysis of the tet(M)-carrying conjugative transposon Tn5251 of Streptococcus pneumoniae. FEMS Microbiol. Lett. 308:150–158. 10.1111/j.1574-6968.2010.02002.x [DOI] [PubMed] [Google Scholar]
- 12.Widdowson CA, Adrian PV, Klugman KP. 2000. Acquisition of chloramphenicol resistance by the linearization and integration of the entire staphylococcal plasmid pC194 into the chromosome of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:393–395. 10.1128/AAC.44.2.393-395.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith MD, Hazum S, Guild WR. 1981. Homology among tet determinants in conjugative elements of streptococci. J. Bacteriol. 148:232–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiliç AO, Vijayakumar MN, Al-khaldi SF. 1994. Identification and nucleotide sequence analysis of a transfer-related region in the streptococcal conjugative transposon Tn5252. J. Bacteriol. 176:5145–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pozzi G, Guild WR. 1988. Two genes for chloramphenicol resistance common to staphylococci and streptococci. Eur. J. Epidemiol. 4:20–24. 10.1007/BF00152687 [DOI] [PubMed] [Google Scholar]
- 16.Burrus V, Waldor MK. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155:376–386. 10.1016/j.resmic.2004.01.012 [DOI] [PubMed] [Google Scholar]
- 17.Iannelli F, Pozzi G. 2007. Protocol for conjugal transfer of genetic elements in Streptococcus pneumoniae, p 525–529 In Hakenbeck R, Chhatwal GS. (ed), Molecular biology of streptococci. Horizon Bioscience, Norfolk, United Kingdom [Google Scholar]
- 18.Khoo SK, Loll B, Chan WT, Shoeman RL, Ngoo L, Yeo CC, Meinhart A. 2007. Molecular and structural characterization of the PezAT chromosomal toxin-antitoxin system of the human pathogen Streptococcus pneumoniae. J. Biol. Chem. 282:19606–19618. 10.1074/jbc.M701703200 [DOI] [PubMed] [Google Scholar]
- 19.Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722–732. 10.1038/nrmicro1235 [DOI] [PubMed] [Google Scholar]
- 20.Díaz-López T, Lages-Gonzalo M, Serrano-López A, Alfonso C, Rivas G, Diaz-Orejas R, Giraldo R. 2003. Structural changes in RepA, a plasmid replication initiator, upon binding to origin DNA. J. Biol. Chem. 278:18606–18616. 10.1074/jbc.M212024200 [DOI] [PubMed] [Google Scholar]
- 21.Sampath J, Vijayakumar MN. 1998. Identification of a DNA cytosine methyltransferase gene in conjugative transposon Tn5252. Plasmid 39:63–76 [DOI] [PubMed] [Google Scholar]
- 22.Garvey P, Fitzgerald GF, Hill C. 1995. Cloning and DNA sequence analysis of two abortive infection phage resistance determinants from the lactococcal plasmid pNP40. Appl. Environ. Microbiol. 61:4321–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schröder G, Krause S, Traxler B, Yeo HJ, Lurz R, Waksman G, Lanka E. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184:2767–2789. 10.1128/JB.184.10.2767-2779.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HS, Kim MS, Cho HS, Kim JI, Kim TJ, Choi JH, Park C, Lee HS, Oh BH, Park KH. 2002. Cyclomaltodextrinase, neopullulanase, and maltogenic amylase are nearly indistinguishable from each other. J. Biol. Chem. 277:21891–21897. 10.1074/jbc.M201623200 [DOI] [PubMed] [Google Scholar]
- 25.Munoz-Najar U, Vijayakumar MN. 1999. An operon that confers UV resistance by evoking the SOS mutagenic response in streptococcal conjugative transposon Tn5252. J. Bacteriol. 181:2782–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang S, Meyer R. 1997. The relaxosome protein MobC promotes conjugal plasmid mobilization by extending DNA strand separation to the nick site at the origin of transfer. Mol. Microbiol. 25:509–516. 10.1046/j.1365-2958.1997.4861849.x [DOI] [PubMed] [Google Scholar]
- 27.Srinivas P, Kiliç AO, Vijayakumar MN. 1997. Site-specific nicking in vitro at oriT by the DNA relaxase of Tn5252. Plasmid 37:42–50. 10.1006/plas.1996.1268 [DOI] [PubMed] [Google Scholar]
- 28.Brennan RG, Matthews BW. 1989. The helix-turn-helix DNA binding motif. J. Biol. Chem. 4264:1903–1906 [PubMed] [Google Scholar]
- 29.Rauch PJG, De Vos WM. 1994. Identification and characterization of genes involved in excision of the Lactococcus lactis conjugative transposon Tn5276. J. Bacteriol. 176:2165–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gindreau E, Lopez R, Garcia P. 2000. MM1, a temperate bacteriophage of the type 23F Spanish/USA multiresistant epidemic clone of Streptococcus pneumoniae: structural analysis of the site-specific integration system. J. Virol. 74:7803–7813 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC112310/pdf/jv007803.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.