Abstract
Poland's first Enterobacteriaceae isolate producing the New Delhi metallo-β-lactamase (NDM) was identified in August 2011. Escherichia coli sequence type ST410 NDM-1 was cultured from a critically ill patient who had been transferred directly from the Congo. The blaNDM-1 gene was carried by conjugative IncFII-type plasmid pMC-NDM (87,619 bp), which showed structural similarity to plasmid pGUE-NDM, which was identified earlier in France in an E. coli ST131 isolate of Indian origin.
TEXT
Multidrug-resistant, carbapenemase-producing members of the family Enterobacteriaceae (CPE) are a public health problem of the highest global concern today (1, 2). They express diverse carbapenem-hydrolyzing enzymes, including metallo-β-lactamases (MBLs) (1, 2). Of a few MBL types observed in CPE so far, the NDM (New Delhi MBL) enzymes were described first in 2009 (3), and this commenced an avalanche of reports on their spread (4). NDM-producing organisms are endemic on the Indian subcontinent and likely also in some Middle Eastern and Balkan countries, from where numerous transfers have been recorded worldwide (4, 5). The blaNDM genes have a chimeric structure (6) and are linked with the bleomycin resistance gene bleMBL (7). The blaNDM-bleMBL operon (plus some other genes) was found in transposon Tn125 formed by two ISAba125 insertion elements and was probably preassembled in an Acinetobacter before transmission to Enterobacteriaceae (8). The presence of Tn125-like elements in diverse plasmids and a variety of Gram-negative species and clones (9–19) indicates that they play a key role in the spread of blaNDM genes. However, in contrast to Acinetobacter baumannii, in Enterobacteriaceae, the Tn125 structure has usually been altered into a variety of truncated variants (1, 4). The aim of this study was to analyze the first NDM case identified in Poland, which was probably linked to travel to equatorial Africa.
A 53-year-old male Polish citizen traveled to the Congo in 2011. He was admitted to an intensive care unit (ICU) because of sudden cardiac arrest on 24 July. After 24 h, the critically ill patient was transferred to Poland by air and directly admitted to the ICU of a Warsaw hospital (26 July). A week later, a sample of his urine yielded Escherichia coli isolate 5428/11, which was identified by Vitek 2 (bioMérieux, Marcy l'Etoile, France) as resistant to carbapenems. The MBL-positive phenotype of the isolate was determined by the EDTA double-disk test (20). MICs of antimicrobials were evaluated with Etest (bioMérieux) and interpreted according to EUCAST (http://www.eucast.org/). E. coli 5428/11 was susceptible only to tigecycline and colistin of all of the drugs tested (Table 1). The patient was treated with colistin for 12 days and isolated for his entire stay. However, despite these efforts, he died of multiorgan failure on 20 August without infection symptoms. There was no transmission of the MBL-producing E. coli strain to other ICU patients.
TABLE 1.
Susceptibilities of the E. coli 5428/11 isolate producing NDM-1, CTX-M-15, TEM-1, and OXA-1 and its E. coli A15 Rifr transconjugant producing NDM-1 and TEM-1
| Isolate | MIC (μg/ml)a |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMX | AMC | PIP | TZP | CAZ | CTX | FEP | ATM | ERT | IPM | MEM | AMK | GEN | CIP | TET | TGC | CST | |
| E. coli 5428/11 | >256 | >256 | >256 | >256 | >256 | >256 | 32 | 32 | >32b | >32b | >32b | >256 | >256 | >32 | >256 | 0.19 | 0.38 |
| R+ [5428/11]c | >256 | >256 | >256 | 24 | >256 | 32 | 6 | 0.064 | 2 | 2 | 4 | >256 | >256 | 0.016 | 2 | 0.19 | 0.38 |
| E. coli A15 Rifr | 4 | 4 | 1.5 | 0.5 | 0.125 | 0.032 | ≤0.016 | 0.047 | 0.008 | 0.19 | 0.032 | 1.5 | 0.5 | 0.032 | 2 | 0.25 | 0.38 |
Abbreviations: AMK, amikacin; AMC; amoxicillin-clavulanate; AMX, amoxicillin; ATM, aztreonam; CAZ, ceftazidime; CIP, ciprofloxacin; CST, colistin; CTX, cefotaxime; ERT, ertapenem; FEP, cefepime; GEN, gentamicin; IPM, imipenem; MEM, meropenem; PIP, piperacillin; TET, tetracycline; TGC, tigecycline; TZP, piperacillin-tazobactam.
With carbapenems, multiple scattered colonies were observed inside the growth inhibition zones around Etest strips.
R+ [5428/11], transconjugant of E. coli 5428/11 obtained by using E. coli A15 Rifr as the recipient.
PCR screening of E. coli 5428/11 for blaVIM-, blaIMP-, and blaNDM-like MBL genes was positive with blaNDM-specific primers (9), and sequencing of the amplicon revealed the presence of NDM-1 (3). Isoelectric focusing (21) showed that the isolate also produced β-lactamases with pIs of 8.9, 7.4, and 5.4 that were identified by PCR and sequencing (22) as CTX-M-15, OXA-1, and TEM-1, respectively (PCR screening for blaCMY-2- and blaOXA-48-like genes was negative). The presence of multiple β-lactamases in NDM producers, including those found in E. coli 5428/11, is the characteristic they have in common (3, 12, 23–25).
E. coli 5428/11 was subjected to multilocus sequence typing (26); the database available at http://mlst.ucc.ie/mlst/dbs/Ecoli was used to assign its sequence type (ST). The isolate belonged to ST410, clonal complex ST23, and by PCR assay (27) it was classified in E. coli phylogroup A. ST410 isolates have been recovered worldwide from human, meat, and animal samples with various resistance traits, such as extended-spectrum β-lactamases (ESBLs) of the CTX-M type (28–32). ST410 acquired the KPC-2 carbapenemase in Greece (33), and it has also been identified with NDM-1, as reported in Norway (patient from India) (34), the United Kingdom (9), Switzerland (patient from Serbia), France, and the United States (12).
E. coli 5428/11 (rifampin MIC, 12 μg/ml) was mated with E. coli A15 Rifr (21) by using 100 μg/ml rifampin and 0.5 μg/ml imipenem as selective agents. Transconjugants were obtained with high efficiency (∼2 × 10−3 per donor cell). The PCR-based replicon typing (PBRT) of plasmids (35) showed replicons FIA, FIB, and FII in the clinical isolate and only FII in the transconjugant. Plasmid profiling was carried out by pulsed-field gel electrophoresis (PFGE) of total DNA cut with nuclease S1 (TaKaRa, Otsu, Japan) (36). Following PFGE, the DNA was hybridized separately with seven probes by using the ECL Random-Prime Labeling and Detection system (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). The probes were the FIA, FIB, and FII PBRT amplicons, and PCR products specific for all of the β-lactamase genes were detected. Two plasmids were found in E. coli 5428/11, one of ∼90 kb with replicon FII and one of ∼110 kb with replicons FIA and FIB; only the first one was present in the transconjugant. Replicon sequence typing (37) showed that the ∼90-kb plasmid was of the F2:A−:B− type, whereas the ∼110 kb molecule was of the F−:A1:B1 type. Of the β-lactamase genes, blaNDM-1 hybridized only with the F2:A−:B− plasmid, whereas blaTEM-1 was located on both. The blaCTX-M-15 and blaOXA-1 genes did not hybridize with any of the plasmids but only with the E. coli 5428/11's highest-molecular-weight DNA band. The following I-Ceu I analysis (38) demonstrated cohybridization of the blaCTX-M-15 and rRNA gene probes with an ∼800-kb DNA fragment, indicating the chromosomal location of blaCTX-M-15 (results not shown). This has been another report of blaNDM-1 on an IncFII-type plasmid, following those from India, France (isolate of Indian origin), Switzerland (Serbian origin) and Turkey (12, 39). One of these plasmids, pGUE-NDM from an E. coli ST131 strain imported to France from India (40), has been well characterized, including its entire sequence (15, 41).
The ∼90-kb F2:A−:B− plasmid, named pMC-NDM, was purified from the transconjugant with the Qiagen Plasmid Midi kit (Qiagen, Hilden, Germany) and sequenced with the 454 GS Junior instrument (Roche Diagnostics, Indianapolis, IN) by Genomed (Warsaw, Poland). Sequence reads were assembled into 11 contigs by the CLC Genomics Workbench software (version 4.7.2; CLC Bio, Aarhus, Denmark). The contigs were closed by Sanger sequencing of PCR products. The sequence was analyzed with the DNA Lasergene (DNASTAR, Madison, WI), BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and RAST (42) tools.
pMC-NDM is a molecule of 87,619 bp with a GC content of 51%. It is similar to 87,022-bp pGUE-NDM (GenBank accession no. JQ364967) (15), with the high-homology part covering almost the entire backbone and a relatively large fragment of multidrug resistance regions in both plasmids (Fig. 1). This part comprises 87.0 and 90.6% of the pMC-NDM and pGUE-NDM structures, respectively. The IncFII replicon region from the repA2 gene to oriR is the same in both plasmids. However, 21 single nucleotide polymorphisms (SNPs) were found within an ∼500-bp-long fragment downstream of oriR, including repA4 (43), and in fact, pMC-NDM is more similar in this fragment to some other IncFII-type plasmids than to pGUE-NDM (e.g., Rsc13; GenBank accession no. J01783) (44). The conjugative transfer loci tra-trb differ only by two SNPs and the number of CAACAGCCG tandem repeats in the traD gene (11 and 10 repeats in pMC-NDM and pGUE-NDM, respectively). Of the two fragments associated with stable inheritance in pGUE-NDM, the region located downstream of repA4 and carrying the toxin-antitoxin type II system pemI/K is missing from pMC-NDM (see below). However, the extensive backbone fragment upstream of tra-trb with the toxin-antitoxin type I system hok/mok and the parB and parM partition genes is present in both plasmids. The differences between pMC-NDM and pGUE-NDM in this region included 27 point changes and different numbers of TACTGC repeats (six and seven repeats, respectively) and a group II intron inserted into pGUE-NDM but not into pMC-NDM.
FIG 1.
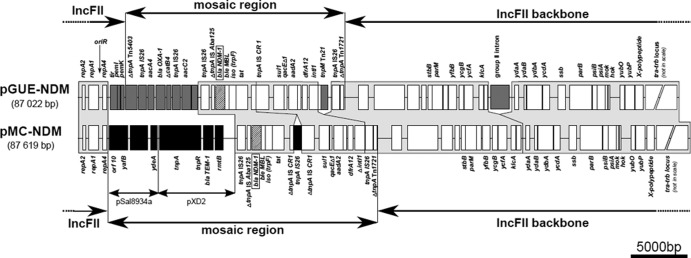
Comparison of IncFII-type plasmids pMC-NDM and pGUE-NDM carrying blaNDM-1 genes. Boxes indicate ORFs identified by sequence analysis. All regions are drawn to scale, except for the tra-trb locus. Gene designations refer to ORFs with putative functions that were assigned on the basis of homology searches performed by Bonnin et al. (15) or in this study. Parts of both plasmids showing similar structures and high sequence homology are indicated by frames shaded light gray. Dark gray and black boxes indicate ORFs found only in pGUE-NDM and pMC-NDM, respectively. The blaNDM-1 genes are represented by hatched boxes. The IncFII backbone, mosaic regions, and the pMC-NDM fragments that are highly homologous to plasmids pSal8934a and pXD2 are indicated by horizontal arrows.
pMC-NDM and pGUE-NDM share a unique arrangement of mobile elements that in both plasmids constitutes a large part of their mosaic regions. It is located directly upstream of the common stable inheritance region and contains remnants of three transposons, namely, Tn1721-, Tn21-, and Tn125-like elements, and two IS26 copies located upstream of ΔTn125 and ΔTn1721, respectively. Major differences between pMC-NDM and pGUE-NDM concern the Tn21-like element, containing a class 1 integron with the dfrA12-orfF-aadA2 cassette array and the ISCR1 element (45) behind the 3′-CS segment. In pMC-NDM, this element is truncated further by an ∼1.1-kb deletion covering tnpM and a fragment of the integrase gene intI1. pMC-NDM also has one more IS26 element inserted within ISCR1 by transposition, as indicated by the presence of 8-bp direct repeats at its boundaries. In addition, the ΔTn125-adjacent IS26 element has two SNPs when the two plasmids are compared. The remnant of Tn125 (8) containing the blaNDM-1-bleMBL operon is the same in pMC-NDM and pGUE-NDM.
The remaining parts of the mosaic regions in pMC-NDM and pGUE-NDM are different. Instead of an ∼9-kb fragment with the genes aacA4, blaOXA-1, ΔcatB4, and aacC2 in pGUE-NDM, pMC-NDM carries an ∼11.4-kb-long segment consisting of two parts. The first of these (∼6.9 kb) is located upstream of IS26-ΔTn125 and encompasses the Tn3 transposon with blaTEM-1 and the 16S rRNA methylase gene rmtB, conferring resistance to aminoglycosides (46). The same arrangement of Tn3 and rmtB was found in some other plasmids, including IncFII-type plasmid pXD2 (GenBank accession no. JN315966) (47). The second part (∼4.5 kb) is located between Tn3 and the replicon region and is almost identical (one SNP) to a fragment of IncI-type plasmids pSal8934a (GenBank accession no. JF274993) and Plm (GenBank accession no. JQ901381). This part contains several open reading frames (ORFs), including the gene yafB, which codes for a putative conjugation repressor domain, similar to FinO in IncFII plasmids (48), as has been annotated for numerous IncI-like plasmids, e.g., pSal8934a and ColIb-P9 (GenBank accession no. AB021078). A sequence of ∼280 bp upstream of yafB (and orf10) in pSal8934a is highly homologous to the region between the oriR and pemI/K genes in IncFII plasmids covering a fragment of repA4. Therefore, it might have been a site of recombination between IncFII- and IncI-type molecules during pMC-NDM evolution, resulting in the acquisition of the IncI-backbone fragment and deletion of the pemI/K toxin-antitoxin genes. Similar events between IncFII- and IncI-like plasmids have been observed before (49, 50).
A comparison of pMC-NDM and pGUE-NDM revealed differences but also clear similarities between the two IncFII-like plasmids with blaNDM-1, carried by unrelated E. coli isolates of different geographic and epidemiological origins. The key common element is essentially the same composition of the truncated Tn1721-, Tn21-, and Tn125-like transposons and their identical location with respect to the backbone. The described architecture of this region must have resulted from a series of DNA rearrangements that are rather unlikely to have occurred independently several times. Therefore, pMC-NDM and pGUE-NDM may have an ancestor in common and, with time, may have differentiated from each other by multiple events.
This work documents the first NDM-positive CPE in Poland, which was possibly imported from Africa, as suggested by the clinical data. Earlier studies showed the presence of NDM CPE in Cameroon (51), Kenya (24), Morocco (25, 52), and South Africa (53), while North Africa has also been described as a region where A. baumannii with NDMs has spread (54, 55). Comparative analysis of the pMC-NDM and pGUE-NDM plasmids indicates the probable evolution of NDM-1 genetic determinants proceeding on a large geographic scale.
Nucleotide sequence accession number.
The complete sequence of plasmid pMC-NDM has been submitted to the EMBL database and assigned accession number HG003695.
ACKNOWLEDGMENTS
We are very grateful to M. Herda and K. Bojarska for their excellent technical support and to I. Kern-Zdanowicz for very helpful discussions.
This work was partially financed by the grant Narodowy Program Ochrony Antybiotyków (NPOA) from the Polish Ministry of Health and the grant SPUB MIKROBANK from the Polish Ministry of Science and Higher Education.
Footnotes
Published ahead of print 18 November 2013
REFERENCES
- 1.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 18:263–272. 10.1016/j.molmed.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 2.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25:682–707. 10.1128/CMR.05035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol. 19:588–595. 10.1016/j.tim.2011.09.005 [DOI] [PubMed] [Google Scholar]
- 5.Struelens MJ, Monnet DL, Magiorakos AP, Santos O'Connor F, Giesecke J. 2010. New Delhi metallo-beta-lactamase 1-producing Enterobacteriaceae: emergence and response in Europe. Euro Surveill. 15:19716 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19716 [DOI] [PubMed] [Google Scholar]
- 6.Toleman MA, Spencer J, Jones L, Walsh TR. 2012. blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56:2773–2776. 10.1128/AAC.06297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dortet L, Nordmann P, Poirel L. 2012. Association of the emerging carbapenemase NDM-1 with a bleomycin resistance protein in Enterobacteriaceae and Acinetobacter baumannii. Antimicrob. Agents Chemother. 56:1693–1697. 10.1128/AAC.05583-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, Nordmann P. 2012. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56:1087–1089. 10.1128/AAC.05620-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, Livermore DM, Woodford N. 2011. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J. Antimicrob. Chemother. 66:2002–2005. 10.1093/jac/dkr226 [DOI] [PubMed] [Google Scholar]
- 10.Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I, Tong AH, Bao JY, Lok S, Lo JY. 2011. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One 6:e17989. 10.1371/journal.pone.0017989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poirel L, Bonnin RA, Nordmann P. 2011. Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob. Agents Chemother. 55:4224–4229. 10.1128/AAC.00165-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 55:5403–5407. 10.1128/AAC.00585-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekizuka T, Matsui M, Yamane K, Takeuchi F, Ohnishi M, Hishinuma A, Arakawa Y, Kuroda M. 2011. Complete sequencing of the blaNDM-1-positive IncA/C plasmid from Escherichia coli ST38 isolate suggests a possible origin from plant pathogens. PLoS One 6:e25334. 10.1371/journal.pone.0025334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11:355–362. 10.1016/S1473-3099(11)70059-7 [DOI] [PubMed] [Google Scholar]
- 15.Bonnin RA, Poirel L, Carattoli A, Nordmann P. 2012. Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS One 7:e34752. 10.1371/journal.pone.0034752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giske CG, Froding I, Hasan CM, Turlej-Rogacka A, Toleman M, Livermore D, Woodford N, Walsh TR. 2012. Diverse sequence types of Klebsiella pneumoniae contribute to the dissemination of blaNDM-1 in India, Sweden, and the United Kingdom. Antimicrob. Agents Chemother. 56:2735–2738. 10.1128/AAC.06142-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, Hao Q. 2012. Yang X, Yang X, Xiao X, Luan C, Yang Y, Cui Y, Yang R, Gao GF, Song Y, Zhu B: Novel plasmid and its variant harboring both a blaNDM-1 gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob. Agents Chemother. 56:1698–1702. 10.1128/AAC.06199-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villa L, Poirel L, Nordmann P, Carta C, Carattoli A. 2012. Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J. Antimicrob. Chemother. 67:1645–1650. 10.1093/jac/dks114 [DOI] [PubMed] [Google Scholar]
- 19.Dolejska M, Villa L, Poirel L, Nordmann P, Carattoli A. 2013. Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J. Antimicrob. Chemother. 68:34–39. 10.1093/jac/dks357 [DOI] [PubMed] [Google Scholar]
- 20.Lee K, Lim YS, Yong D, Yum JH, Chong Y. 2003. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-β-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 41:4623–4629. 10.1128/JCM.41.10.4623-4629.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauernfeind A, Grimm H, Schweighart S. 1990. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 18:294–298. 10.1007/BF01647010 [DOI] [PubMed] [Google Scholar]
- 22.Empel J, Baraniak A, Literacka E, Mrowka A, Fiett J, Sadowy E, Hryniewicz W, Gniadkowski M. 2008. Molecular survey of β-lactamases conferring resistance to newer β-lactams in Enterobacteriaceae isolates from Polish hospitals. Antimicrob. Agents Chemother. 52:2449–2454. 10.1128/AAC.00043-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. 2011. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006-2007. Antimicrob. Agents Chemother. 55:1274–1278. 10.1128/AAC.01497-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirel L, Revathi G, Bernabeu S, Nordmann P. 2011. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob. Agents Chemother. 55:934–936. 10.1128/AAC.01247-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirel L, Benouda A, Hays C, Nordmann P. 2011. Emergence of NDM-1-producing Klebsiella pneumoniae in Morocco. J. Antimicrob. Chemother. 66:2781–2783. 10.1093/jac/dkr384 [DOI] [PubMed] [Google Scholar]
- 26.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558. 10.1128/AEM.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.López-Cerero L, Egea P, Serrano L, Navarro D, Mora A, Blanco J, Doi Y, Paterson DL, Rodriguez-Bano J, Pascual A. 2011. Characterisation of clinical and food animal Escherichia coli isolates producing CTX-M-15 extended-spectrum β-lactamase belonging to ST410 phylogroup A. Int. J. Antimicrob. Agents 37:365-367. 10.1016/j.ijantimicag.2011.01.001 [DOI] [PubMed] [Google Scholar]
- 29.Peirano G, Asensi MD, Pitondo-Silva A, Pitout JD. 2011. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli from Rio de Janeiro, Brazil. Clin. Microbiol. Infect. 17:1039–1043. 10.1111/j.1469-0691.2010.03440.x [DOI] [PubMed] [Google Scholar]
- 30.Correia S, Pacheco R, Radhouani H, Diniz JC, Ponce P, Jones-Dias D, Canica M, Igrejas G, Poeta P. 2012. High prevalence of ESBL-producing Escherichia coli isolates among hemodialysis patients in Portugal: appearance of ST410 with the blaCTX-M-14 gene. Diagn. Microbiol. Infect. Dis. 74:423–425. 10.1016/j.diagmicrobio.2012.08.017 [DOI] [PubMed] [Google Scholar]
- 31.Huber H, Zweifel C, Wittenbrink MM, Stephan R. 2013. ESBL-producing uropathogenic Escherichia coli isolated from dogs and cats in Switzerland. Vet. Microbiol. 162:992–996. 10.1016/j.vetmic.2012.10.029 [DOI] [PubMed] [Google Scholar]
- 32.Izdebski R, Baraniak A, Fiett J, Adler A, Kazma M, Salomon J, Lawrence C, Rossini A, Salvia A, Vidal Samso J, Fierro J, Paul M, Lerman Y, Malhotra-Kumar S, Lammens C, Goossens H, Hryniewicz W, Brun-Buisson C, Carmeli Y, Gniadkowski M. 2013. Clonal structure, extended-spectrum β-lactamases, and acquired AmpC-type cephalosporinases of Escherichia coli populations colonizing patients in rehabilitation centers in four countries. Antimicrob. Agents Chemother. 57:309–316. 10.1128/AAC.01656-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mavroidi A, Miriagou V, Malli E, Stefos A, Dalekos GN, Tzouvelekis LS, Petinaki E. 2012. Emergence of Escherichia coli sequence type 410 (ST410) with KPC-2 β-lactamase. Int. J. Antimicrob. Agents 39:247–250. 10.1016/j.ijantimicag.2011.11.003 [DOI] [PubMed] [Google Scholar]
- 34.Samuelsen O, Thilesen CM, Heggelund L, Vada AN, Kummel A, Sundsfjord A. 2011. Identification of NDM-1-producing Enterobacteriaceae in Norway. J. Antimicrob. Chemother. 66:670–672. 10.1093/jac/dkq483 [DOI] [PubMed] [Google Scholar]
- 35.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- 36.Samuelsen O, Naseer U, Tofteland S, Skutlaberg DH, Onken A, Hjetland R, Sundsfjord A, Giske CG. 2009. Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. J. Antimicrob. Chemother. 63:654–658. 10.1093/jac/dkp018 [DOI] [PubMed] [Google Scholar]
- 37.Villa L, Garcia-Fernandez A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65:2518–2529. 10.1093/jac/dkq347 [DOI] [PubMed] [Google Scholar]
- 38.Liu SL, Hessel A, Sanderson KE. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:6874–6878. 10.1073/pnas.90.14.6874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poirel L, Ozdamar M, Ocampo-Sosa AA, Turkoglu S, Ozer UG, Nordmann P. 2012. NDM-1-producing Klebsiella pneumoniae now in Turkey. Antimicrob. Agents Chemother. 56:2784–2785. 10.1128/AAC.00150-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poirel L, Hombrouck-Alet C, Freneaux C, Bernabeu S, Nordmann P. 2010. Global spread of New Delhi metallo-β-lactamase 1. Lancet Infect. Dis. 10:832. 10.1016/S1473-3099(10)70279-6 [DOI] [PubMed] [Google Scholar]
- 41.Potron A, Poirel L, Nordmann P. 2011. Plasmid-mediated transfer of the blaNDM-1 gene in Gram-negative rods. FEMS Microbiol. Lett. 324:111–116. 10.1111/j.1574-6968.2011.02392.x [DOI] [PubMed] [Google Scholar]
- 42.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang T, Min YN, Liu W, Womble DD, Rownd RH. 1993. Insertion and deletion mutations in the repA4 region of the IncFII plasmid NR1 cause unstable inheritance. J. Bacteriol. 175:5350–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryder TB, Davidson DB, Rosen JI, Ohtsubo E, Ohtsubo H. 1982. Analysis of plasmid genome evolution based on nucleotide-sequence comparison of two related plasmids of Escherichia coli. Gene 17:299–310. 10.1016/0378-1119(82)90146-9 [DOI] [PubMed] [Google Scholar]
- 45.Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296–316. 10.1128/MMBR.00048-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wachino J, Arakawa Y. 2012. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist. Updat. 15:133–148. 10.1016/j.drup.2012.05.001 [DOI] [PubMed] [Google Scholar]
- 47.Li DX, Zhang SM, Hu GZ, Wang Y, Liu HB, Wu CM, Shang YH, Chen YX, Du XD. 2012. Tn3-associated rmtB together with qnrS1, aac(6′)-Ib-cr and blaCTX-M-15 are co-located on an F49:A-:B- plasmid in an Escherichia coli ST10 strain in China. J. Antimicrob. Chemother. 67:236–238. 10.1093/jac/dkr428 [DOI] [PubMed] [Google Scholar]
- 48.Wong JJ, Lu J, Glover JN. 2012. Relaxosome function and conjugation regulation in F-like plasmids—a structural biology perspective. Mol. Microbiol. 85:602–617. 10.1111/j.1365-2958.2012.08131.x [DOI] [PubMed] [Google Scholar]
- 49.Kato A, Mizobuchi K. 1994. Evolution of the replication regions of IncI alpha and IncFII plasmids by exchanging their replication control systems. DNA Res. 1:201–212. 10.1093/dnares/1.5.201 [DOI] [PubMed] [Google Scholar]
- 50.Osborn A. M., da Silva Tatley F. M., Steyn L. M., Pickup R. W., Saunders JR. 2000. Mosaic plasmids and mosaic replicons: evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology 146(Pt 9):2267–2275 http://mic.sgmjournals.org/content/146/9/2267.long [DOI] [PubMed] [Google Scholar]
- 51.Dortet L, Poirel L, Anguel N, Nordmann P. 2012. New Delhi metallo-β-lactamase 4-producing Escherichia coli in Cameroon. Emerg. Infect. Dis. 18:1540–1542. 10.3201/eid1809.120011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barguigua A, El Otmani F, Lakbakbi El Yaagoubi F, Talmi M, Zerouali K, Timinouni M. 2013. First report of a Klebsiella pneumoniae strain coproducing NDM-1, VIM-1 and OXA-48 carbapenemases isolated in Morocco. APMIS 121:675–677. 10.1111/apm.12034 [DOI] [PubMed] [Google Scholar]
- 53.Brink AJ, Coetzee J, Clay CG, Sithole S, Richards GA, Poirel L, Nordmann P. 2012. Emergence of New Delhi metallo-β-lactamase (NDM-1) and Klebsiella pneumoniae carbapenemase (KPC-2) in South Africa. J. Clin. Microbiol. 50:525–527. 10.1128/JCM.05956-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaase M, Nordmann P, Wichelhaus TA, Gatermann SG, Bonnin RA, Poirel L. 2011. NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J. Antimicrob. Chemother. 66:1260–1262. 10.1093/jac/dkr135 [DOI] [PubMed] [Google Scholar]
- 55.Boulanger A, Naas T, Fortineau N, Figueiredo S, Nordmann P. 2012. NDM-1-producing Acinetobacter baumannii from Algeria. Antimicrob. Agents Chemother. 56:2214–2215. 10.1128/AAC.05653-11 [DOI] [PMC free article] [PubMed] [Google Scholar]


