Abstract
Cadazolid is a new oxazolidinone-type antibiotic currently in clinical development for the treatment of Clostridium difficile-associated diarrhea. Here, we report investigations on the mode of action and the propensity for spontaneous resistance development in C. difficile strains. Macromolecular labeling experiments indicated that cadazolid acts as a potent inhibitor of protein synthesis, while inhibition of DNA synthesis was also observed, albeit only at substantially higher concentrations of the drug. Strong inhibition of protein synthesis was also obtained in strains resistant to linezolid, in agreement with low MICs against such strains. Inhibition of protein synthesis was confirmed in coupled transcription/translation assays using extracts from different C. difficile strains, including strains resistant to linezolid, while inhibitory effects in DNA topoisomerase assays were weak or not detectable under the assay conditions. Spontaneous resistance frequencies of cadazolid were low in all strains tested (generally <10−10 at 2× to 4× the MIC), and in multiple-passage experiments (up to 13 passages) MICs did not significantly increase. Furthermore, no cross-resistance was observed, as cadazolid retained potent activity against strains resistant or nonsusceptible to linezolid, fluoroquinolones, and the new antibiotic fidaxomicin. In conclusion, the data presented here indicate that cadazolid acts primarily by inhibition of protein synthesis, with weak inhibition of DNA synthesis as a potential second mode of action, and suggest a low potential for spontaneous resistance development.
INTRODUCTION
Clostridium difficile is a Gram-positive, anaerobic, toxin- and spore-forming bacterium that is the most common infectious cause of antibiotic-associated diarrhea and colitis. Clostridium difficile infection (CDI, or CDAD for C. difficile-associated diarrhea) is a major health care problem with significant morbidity and mortality, especially in elderly hospitalized patients (1). The frequency and severity of CDAD have increased in recent years, and new hypervirulent and epidemic strains of C. difficile that are characterized by acquired resistance to fluoroquinolones such as ciprofloxacin and moxifloxacin have been discovered (1–4). Vancomycin and metronidazole are the mainstay of antibiotic therapy of CDAD; however, treatment success in severe disease is limited and high recurrence rates have been reported (5, 6). A new macrocyclic antibiotic, fidaxomicin, has recently been shown to be effective in clinical studies, with lower recurrence rates than those observed with vancomycin (7–9). Cadazolid (formerly ACT-179811) is a new antibiotic currently in clinical development for the treatment of CDAD. Cadazolid showed potent in vitro activity against C. difficile (10, 11) and has an antibacterial spectrum largely limited to Gram-positive bacteria, while activity against Gram-negative bacteria is weak or not detectable (12). The chemical structure of cadazolid holds elements of both the oxazolidinone and the fluoroquinolone classes of antibacterials (Fig. 1). Oxazolidinones, such as linezolid (LZD), act by interfering with an early step in bacterial protein synthesis, whereas fluoroquinolone antibiotics inhibit the function of bacterial type II DNA topoisomerases (DNA gyrase and topoisomerase IV) and hereby interfere with DNA replication (13).
FIG 1.
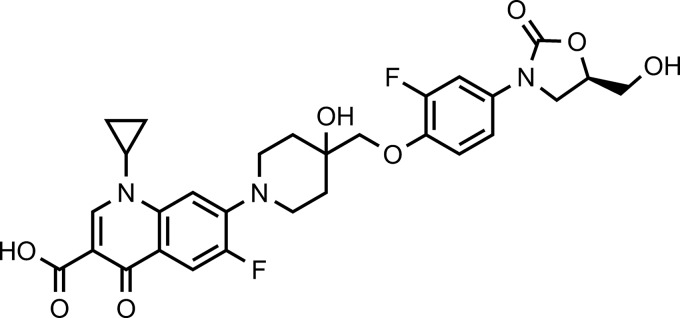
Chemical structure of cadazolid (1-cyclopropyl-6-fluoro-7-{4-[2-fluoro-4-((R)-5-hydroxymethyl-2-oxo-oxazolidin-3-yl)-phenoxymethyl]-4-hydroxy-piperidin-1-yl}-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid).
In this study, we investigated the mode of action of cadazolid in C. difficile by macromolecular labeling studies and in biochemical assays and we assessed the in vitro propensity for resistance development and the potential cross-resistance to other antibiotics. Linezolid and fluoroquinolone(s) were included as comparators due to structural similarities to cadazolid, while vancomycin and fidaxomicin (lipiarmycin A3), approved antibiotics for treatment of CDAD, were included in experiments addressing resistance development.
Part of this work was previously presented as a poster at the 52nd Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC) and 23rd European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) conferences (12, 14, 15).
MATERIALS AND METHODS
Bacterial strains and growth and antibiotics.
Reference strains were obtained from the American Type Culture Collection (ATCC), and the hypervirulent and fluoroquinolone-resistant ribotype 027 strain (NCTC 13366) was obtained from the National Collection of Type Cultures. Other clinical isolates of C. difficile used in this study, including linezolid-resistant strains, were kindly provided by M. Wilcox (Leeds, United Kingdom) and D. Gerding (Hines, IL). Experiments were performed in an anaerobic glove box (Coy Laboratory) in an atmosphere of 85% N2–10% CO2–5% H2 unless specified differently in the text.
Cadazolid (ACT-179811; purity, 98.8%) and moxifloxacin base were synthesized at Actelion Pharmaceuticals Ltd. Fidaxomicin (lipiarmycin A3) (16) was obtained from Biofocus DPI by fermentation of Actinoplanes deccanens is DSM 43806 and subsequent isolation of the target natural product. Other antibiotics were obtained from commercial sources, as follows: vancomycin, Sigma V2002; rifaximin, Sigma R9904; ciprofloxacin, Fluka 17850; and linezolid, AK scientific, catalog no. 70412.
Determination of the MIC.
The MICs of C. difficile were determined using the Clinical and Laboratory Standards Institute (CLSI)-recommended agar dilution method for anaerobes (17). MICs were determined at least in duplicates, and ranges are given when values were different. Due to limited water solubility, cadazolid was dissolved and serially diluted in dimethyl sulfoxide (DMSO) before incorporation into supplemented Brucella agar plates (ref. 211086; Beckton Dickinson and Company). The final DMSO concentration was 1% (vol/vol). DMSO concentrations of up to 2.5% (vol/vol) in the test medium were found to have no effect on growth or susceptibility of C. difficile. To prepare the inoculum, bacterial colonies were suspended in supplemented Brucella broth (0.5 mg/liter vitamin K1, 5 mg/liter hemin, and 5% [vol/vol] laked sheep blood) and adjusted to the equivalent of a 0.5 McFarland standard, resulting in approximately 104 to 105 CFU per spot after application with a Steers replicator. The plates were incubated under anaerobic conditions (GENbox anaer, ref. 96124; bioMérieux) for 48 h at 37°C. The MIC was defined as the lowest concentration that completely inhibited visible growth compared to the drug-free control. Strain ATCC 700057 was used as the quality control strain, and MICs of reference antibiotics were compared to quality control limits suggested by the CLSI.
Macromolecular labeling studies.
Clostridium strains were grown anaerobically overnight on supplemented Brucella agar. Turbid suspensions of the strains were prepared in 5 ml preanaerobized supplemented Brucella broth and then incubated 2 h in the anaerobic glove box at 37°C. Cells were then washed with 1 ml of 1:5 diluted Brucella (37°C) medium and resuspended in one-half of the original volume (2.5 ml) of 1:5 diluted Brucella broth (37°C). Stock solutions of test antibiotics were prepared to a 20× working solution and then dispensed into assay plates, leading to final concentrations of 0.03, 0.125, 0.5, 2, 8, 32, and 128 μg/ml.
For the labeling of C. difficile proteins and peptidoglycans, l-[3,4,5-3H(N)]-leucine (150 Ci/mmol; PerkinElmer) and N-acetyl-d-[6-3H]glucosamine (30 Ci/mmol; Anawa, Wangen, Switzerland) were used at 20 μCi/ml, respectively (18). A 120-μl volume of cells was added to the label and incubated under anaerobic conditions for 2 h at 37°C. To analyze DNA synthesis in C. difficile, [2,8-3H]-adenine (35 to 50 Ci/mmol; PerkinElmer) was used at a final concentration of 10 μCi/ml, as thymidine was not incorporated into macromolecules of a variety of C. difficile strains available in the laboratory. Incubation was done under anaerobic conditions for 1 h at 37°C. To extract base labile nucleic acids (i.e., RNA), an additional step was performed (15). Labeling reaction samples were treated with 1.3 M NaOH at 65°C for 1 h and, after a short spin, further incubated on ice for 30 min. Finally, macromolecules were precipitated on ice with 10% tricarboxylic acid (TCA) and read in a TopCount scintillation counter (Packard). In control experiments, 4,900 to 6,300 cpm was measured in proteins, 13,500 to 58,300 cpm was found in peptidoglycans, and 2,000 to 7,000 cpm was found in nucleic acids after extraction.
CFTA.
For C. difficile cell-free coupled transcription/translation assay (CFTA), bacterial S12 extracts were prepared according to a simplified Escherichia coli procedure (19) using a FastPrep-24 cell disruptor instrument from Lucerna Chem AG for lysing the cells. Bacteria were grown overnight at 37°C in 50 ml supplemented Brucella broth under anaerobic conditions. After centrifugation, bacteria were resuspended in 1 ml buffer B (10 mM Tris-acetate [pH 8.2], 14 mM Mg-acetate, 60 mM K-glutamate, 1.0 mM dithiothreitol [DTT]) and transferred into 2 ml-tubes containing lysing matrix B (silica spheres measuring <100 μm). Cells were lysed in the presence of a protease inhibitor cocktail (Thermo Scientific). Suspensions were centrifuged for 10 min at 12,000 × g, and aliquots of the supernatants were incubated at 37°C for 30 min and then stored at −80°C. In order to improve expression from extracts derived from C. difficile, the firefly luciferase reporter gene was resynthesized with codons optimized for C. difficile at Geneart AG (Regensburg, Germany) and cloned into plasmid pSP-luc+NF Fusion (Promega), replacing the existing luciferase gene. In addition, a 180-bp-long BglII-HindIII promoter fragment of gene abrB310 derived from Clostridium acetobutylicum was synthesized and cloned in front of the codon-optimized luciferase gene for efficient translation (20). The cell-free protein synthesis reactions were carried out based on the procedure published for E. coli (19) using optimized buffer conditions (57 mM HEPES [pH 7.5], 1.2 mM ATP, 0.85 mM CTP/GTP/UTP, 0.64 mM cyclic AMP [cAMP], 2 mM DTT, 0.175 mg/ml E. coli tRNA mix, 200 mM K-glutamate, 27.5 mM NH4-acetate, 10.7 mM Mg-acetate, 68 μM folinic acid, 4% polyethylene glycol [PEG] 8000, 80 mM creatine phosphate, 0.25 mg/ml creatine kinase, and 0.5 mM each amino acid). Serial dilutions of compounds (in 1% DMSO, final concentration) were mixed in half-area-white 96-well microtiter plates (Costar) with extracts, buffer, and 5 units of SP6 RNA polymerase (Roche Applied Science) in a total volume of 15 μl. Reaction mixtures were incubated at 37°C for 10 min. One microliter (500 ng) reporter plasmid DNA was added, and incubations were continued at 37°C for 30 min (or at room temperature for 120 min), followed by addition of 15 μl Bright Glo (Promega) luciferase substrate solution. The luminescence was immediately measured using a Tecan SpectralFluor Plus Reader (GeniosPro) instrument. Fifty percent inhibitory concentrations (IC50s) were calculated from the data using the proprietary software IC50 Witch.
In vitro RNA production and in vitro translation assays.
XhoI-digested and purified plasmid DNA pSP-luc+NF fusion vector (30 μl [6 μg]) was used in the RiboMAX large-scale RNA production system-SP6 (Promega). Twenty microliters of 5× SP6 transcription buffer, 20 μl rNTP mix (25 mM ATP, CTP, GTP, UTP), 20 μl H2O, and 10 μl SP6 enzyme mix were added. After a 4-h incubation at 37°C, 6 μl RQ1 (RNase-free DNase) was added, and the mixture was incubated for an additional 15 min at 37°C. The in vitro-produced luciferase mRNA was purified using the RNeasy Plus Minikit (Qiagen), and the concentration of the purified mRNA was determined as optical density (OD) at 260 nm.
IC50s were determined in an E. coli S30 in vitro translation system containing a modified buffer solution without CTP, GTP, or UTP (preventing transcription) and with purified luciferase mRNA. Rifaximin, a bacterial RNA polymerase inhibitor, and mitramycin A, a DNA-interacting compound, were used as controls. Additionally, IC50s were determined in an E. coli S30 in vitro transcription/translation system using plasmid pT7-luc (luciferase gene under the control of a phage T7 promoter; 400 ng/reaction volume; Promega) and phage T7 RNA polymerase (4 units/reaction volume; Roche Applied Science). Rifampin (50 ng) was added to the reaction mixtures to inhibit the E. coli RNA polymerase.
DNA topoisomerase assays.
gyrA and gyrB genes were cloned from Clostridium difficile strain ATCC 43602, expressed in E. coli BL21(DE3), and purified using standard protocols. An equimolar mixture of the two subunits GyrA and GyrB was preincubated for 30 min on ice to reconstitute gyrase holoenzyme. C. difficile DNA gyrase (4 nM) was sufficient to convert 0.1 μg of substrate DNA in 1 h in a DNA supercoiling assay in the absence of drug, whereas 16 nM enzyme was needed in a decatenation assay (data not shown). DNA supercoiling activity was assayed with relaxed pBR322 (Inspiralis, United Kingdom) as a DNA substrate. The reaction mixture (20 μl) contained 50 mM Tris-HCl (pH 7.5), 20 mM KCl, 5 mM MgCl2, 5 mM DTT, 3 mM ATP, 700 mM K-glutamate, 50 μg/ml bovine serum albumin (BSA), 0.1 μg relaxed plasmid, and 4 nM reconstituted C. difficile DNA gyrase. Decatenation activity was assayed using 0.1 μg kinetoplast DNA (Topogen Inc., USA) in 50 mM Tris-HCl (pH 7.5), 20 mM KCl, 5 mM MgCl2, 1.5 mM ATP, 5 mM spermidine, 5 μg/ml BSA, 100 mM K-glutamate, and 16 nM reconstituted C. difficile DNA gyrase. E. coli DNA gyrase activity was measured using a standard supercoiling assay (21). E. coli topoisomerase IV assays were performed with supercoiled pBR322 DNA as a substrate as described previously (22) and according to the manufacturer's instructions (Inspiralis, United Kingdom). All reactions were carried out at 37°C for 1 h and stopped by adding a mixture of EDTA, bromophenol blue, and glycerol. DNA molecules were separated by gel electrophoreses (1% agarose in TAE buffer [40 mM Tris-acetate, 1 mM EDTA, pH 8.0]) and stained in water with Gel Red (Biotium), and DNA bands were visualized (Fluochem System 5500 using AlphaEase Stand Alone Software, Alpha Innotech). For supercoiling inhibition assays, 50% inhibitory concentration (IC50) was set at the compound concentration at which the formation of the supercoiled gel band was reduced by 50%. For decatenation inhibition assays, IC50 was defined as the concentration of inhibitor necessary to inhibit the formation of 50% DNA minicircles from kinetoplast DNA.
In vitro resistance development studies.
To determine the spontaneous resistance frequencies, C. difficile cultures grown in brain heart infusion (BHI) broth were concentrated by centrifugation and then plated (108 to 109 CFU/plate) on supplemented Brucella agar containing a range of different concentrations of cadazolid and other antibiotics representing 2× to 16× the MICs. Colonies growing on drug-containing agar were counted after 5 days of anaerobic incubation at 37°C, and the resistance frequency was calculated by dividing the total number of colonies growing on drug-containing agar by the total number of CFU plated as determined by CFU count on drug-free agar. For selection of resistance in multiple steps, colonies from the plate with the highest antibiotic concentration were isolated with a swab and used to prepare the inoculum for the next resistance selection step, as detailed above. This procedure was repeated for up to three independent steps. At every step, single colony isolates growing on agar plates at the highest drug concentration were purified by three passages on drug-free agar and assessed by the standard CLSI MIC agar dilution method.
To test the effect of continuous, gradually increasing exposure at sub-MICs, bacterial strains were serially passaged in supplemented Brucella broth. Tubes containing 2 ml of 2-fold serial dilutions of the test compound were inoculated with approximately 5 × 106 to 2 × 107 CFU. After incubation for 48 h at 37°C, the tube with the highest antibiotic concentration permitting growth (equal to or higher than a 1.0 McFarland standard), was used to inoculate a new series of tubes (1%, vol/vol). This procedure was repeated for 13 passages, and the lowest antibiotic concentration inhibiting growth in the tubes (MICt) was recorded at the end of every passage. Finally, single colony isolates were purified by two passages on drug-free agar and subjected to standard MIC analysis.
RESULTS
Macromolecular labeling studies.
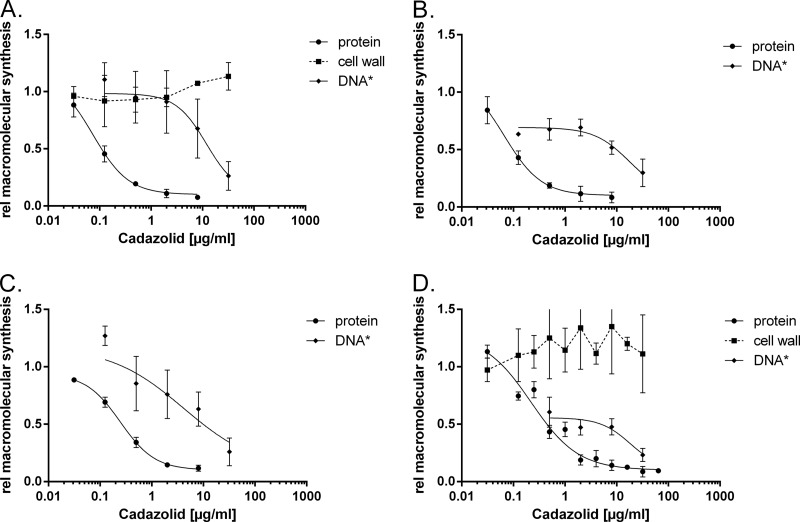
The effects of cadazolid and reference antibiotics on incorporation of l-leucine (protein synthesis), adenine (nucleic acid synthesis), and N-acetyl-d-glucosamine (cell wall synthesis) were investigated in four strains of C. difficile (Table 1). Cadazolid showed potent inhibition of protein synthesis in wild-type and quinolone-resistant as well as in linezolid-resistant strains (Fig. 2). Half-maximal inhibition (IC50) was achieved with 0.08 to 0.31 μg/ml, whereas the IC50s for linezolid ranged from 1.7 to 68 μg/ml. Cadazolid retained its potency even in linezolid-resistant clinical isolates, while the IC50s for linezolid shifted 5- to 35-fold, which is consistent with the observed shift in MIC. DNA synthesis was inhibited with cadazolid with an IC50 of 12.0 to 18.6 μg/ml (Table 1 and Fig. 2); in contrast, the IC50s ranged from 2.3 to 43 μg/ml for moxifloxacin. Cadazolid retained its potency in quinolone-resistant clinical isolates, while the IC50s for moxifloxacin shifted at least 5- to 20-fold, which is again consistent with the shift in MIC. Linezolid did not inhibit DNA synthesis in any of the strains tested up to 128 μg/ml. These results show that cadazolid is a weak inhibitor of DNA synthesis in C. difficile, with concentrations needed to observe half-maximal inhibition at least 60-fold higher than the concentrations needed for protein synthesis inhibition.
TABLE 1.
Results of macromolecular labeling studies for effects of cadazolid and reference antibiotics on incorporation of [3H]-l-leucine (protein synthesis), [3H]-l-adenine (nucleic acid synthesis), and N-acetyl-d-[6-3H]glucosamine (cell wall synthesis) in different C. difficile strainsa
| Test | Strain (phenotype) | Cadazolid | Linezolid | Moxifloxacin | Vancomycin |
|---|---|---|---|---|---|
| Inhibition of protein synthesis IC50 | ATCC 43602 (LZDs, FQs) | 0.09 (0.08–0.10) | 1.7 (1.4–2.0) | >64 | ND |
| NCTC 13366 (LZDs, FQr) | 0.08 (0.07–0.09) | 1.8 (1.5–2.1) | >64 | ND | |
| A-1291 (LZDr, FQs) | 0.19 (0.17–0.21) | 11.8 (9.2–15) | >64 | ND | |
| A-1410 (LZDr, FQr) | 0.31 (0.27–0.36) | 68.0 (51–89) | >32 | ND | |
| Inhibition of nucleic acid synthesis IC50 | ATCC 43602 (LZDs, FQs) | 12.0 (7.0–20.4) | >128 | 2.4 (1.9–2.9) | ND |
| NCTC 13366 (LZDs, FQr) | 17.6 (12.7–24.3) | >128 | 46 (31.6–68) | ND | |
| A-1291 (LZDr, FQs) | 14.3 (8.4–24.5) | >128 | 6.0 (4.5–7.9) | ND | |
| A-1410 (LZDr, FQr) | 18.6 (9.4–36.9) | >128 | 43.0 (8.0–231) | ND | |
| Inhibition of cell wall synthesis IC50 | ATCC 43602 (LZDs, FQs) | >32 | ND | ND | 1.4 (0.89–2.3) |
| A-1410 (LZDr, FQr) | >32 | ND | ND | 4.7 (3.2–6.8) | |
| MIC | ATCC 43602 (LZDs, FQs) | 0.125–0.25 | 2 | 2 | 1 |
| NCTC 13366 (LZDs, FQr) | 0.125–0.25 | 1 | 32 | 1 | |
| A-1291 (LZDr, FQs) | 0.25–0.5 | 16–32 | 1 | 2 | |
| A-1410 (LZDr, FQr) | 0.5 | 32–64 | 32 | 1 |
Values are in μg/ml; for IC50s, 95% confidence intervals are given in parentheses. LZDs, linezolid susceptible; LZDr, linezolid resistant; FQs, fluoroquinolone susceptible; FQr, fluoroquinolone resistant; ND, not determined.
FIG 2.
Concentration-dependent inhibition of macromolecular labeling by cadazolid in C. difficile strains. The inhibition of protein (•), DNA (◆), and cell wall (■) synthesis was measured as the decrease of acid-precipitable 3H labels with different concentrations of inhibitors. The C. difficile strains used were ATCC 43602 (wild type) (A), NCTC 13366 (quinolone resistant) (B), A-1291 (linezolid resistant) (C), and A-1410 (quinolone and linezolid resistant) (D). Curves were generated using a log (inhibitor) versus response model with variable slope and a least-squares fit. If no convergence was achieved, then a connection dashed line was drawn.
No inhibition of peptidoglycan synthesis was detected, in agreement with the proposed specific mechanism of action (Fig. 2). No experiments to assay RNA synthesis inhibition could be performed, as neither uracil nor thymidine was incorporated efficiently in C. difficile macromolecules (data not shown).
Activities in C. difficile cell-free coupled transcription/translation assay.
We developed a CFTA using extracts from different C. difficile strains to measure the interaction of cadazolid with protein biosynthesis. Cadazolid potently inhibited in vitro translation in C. difficile cell extracts derived from linezolid-susceptible and -resistant strains (IC50, 0.12 to 0.5 μg/ml; Table 2). In contrast, linezolid was significantly less potent in extracts from linezolid-resistant strains (IC50, 2 to 12 μg/ml). IC50s correlated well with MICs, which suggests that inhibition of protein synthesis is the primary mode of action in these strains, in agreement with macromolecular labeling studies. As expected, ciprofloxacin was not active in the translation inhibition assay.
TABLE 2.
Activities in C. difficile cell-free translation assay (CFTA) and corresponding MICsa
| C. difficile strain (resistance profile)b | Cadazolid |
Linezolid |
Ciprofloxacin |
|||
|---|---|---|---|---|---|---|
| MIC | CFTA (IC50 range) | MIC | CFTA (IC50 range) | MIC | CFTA (IC50 range) | |
| ATCC 43602 (LZDs, FQs) | 0.125–0.25 | 0.12–0.4 | 2 | 0.4–0.5 | 16 | 9.4 |
| ATCC 700057 (LZDs, FQs) | 0.125–0.25 | 0.23–0.35 | 2–4 | 0.17–0.5 | 16 | >20 |
| ATCC 9689 (LZDs, FQs) | 0.125–0.25 | 0.18 | 2 | 0.17 | 16 | NTc |
| A-1290 (LZDr, FQs) | 0.25–0.5 | 0.35–0.52 | 16–32 | 2.2–3.2 | 16 | 20 |
| A-1291 (LZDr, FQs) | 0.25–0.5 | 0.35–0.47 | 16–32 | 2.5–3.5 | 16 | >20 |
| A-1410 (LZDr, FQr) | 0.5 | 0.3–0.47 | 32–64 | 11.7–12.2 | >32 | >20 |
| A-1412 (LZDr, FQr) | 0.5 | 0.41–0.47 | 32–64 | 4.4–10.0 | >32 | >20 |
CFTA IC50s were determined at least in triplicates. MICs were determined at least in duplicates; when replicates were different, ranges are shown. All values are in μg/ml.
LZDs, linezolid susceptible; LZDr, linezolid resistant; FQs, fluoroquinolone susceptible; FQr, fluoroquinolone resistant.
NT, not tested.
CFTA is a coupled transcription/translation system with RNA polymerase transcribing the luciferase gene and the extract translating the mRNA into the luciferase enzyme. Therefore, substances interfering with this process can interfere not only with the translation step but also with the transcription reaction, or they can be DNA-interacting compounds. To discriminate between these possibilities, in vitro translation experiments were performed with added in vitro-produced luciferase mRNA. The DNA-interacting compound mitramycin A inhibited transcription of the bacterial and phage T7 RNA polymerases but did not inhibit the pure translation reaction with luciferase mRNA (Table 3). The transcription inhibitor rifaximin inhibited only transcription of the bacterial RNA polymerase, but not transcription by T7 RNA polymerase or the pure translation reaction. Cadazolid and linezolid, on the other hand, inhibited the pure translation reaction, indicating that these compounds are interfering mainly with the translation machinery (Table 3).
TABLE 3.
E. coli in vitro translation system: addition of mRNA
| Process or reporter gene | Assay conditions and result |
||
|---|---|---|---|
| Transcription | E. coli RNA polymerase | T7 RNA polymerase | No transcription |
| Translation | E. coli ribosomesa | E. coli ribosomesb | E. coli ribosomesc |
| Luciferase gene | pBestluc | pT7-luc | Purified luciferase mRNA |
| IC50 (μM) of antibiotic | |||
| Cadazolid | 0.37 | 1.0 | 2.9 |
| Linezolid | 2–6 | 7.3 | 10.3 |
| Rifaximin (transcription inhibitor) | 0.10 | >40 | >40 |
| Mitramycin A (DNA-interacting compound) | <1.0 | <1.0 | >40 |
E. coli S30 Extract System for Circular DNA (catalog no. L1020; Promega); E. coli RNA polymerase transcribing the luciferase gene from plasmid pBestluc.
E. coli S30 Extract System for Circular DNA with T7 RNA polymerase added; T7 RNA polymerase transcribing the luciferase gene from plasmid pT7-luc.
E. coli S30 Extract System for Circular DNA with purified luciferase mRNA added; all four nucleotides were omitted to prevent transcription.
Activity in DNA topoisomerase assays.
In order to investigate whether cadazolid, with its fluoroquinolone moiety, is acting as a DNA topoisomerase inhibitor, DNA supercoiling and decatenation assays were performed with bacterial DNA gyrases. Analysis of C. difficile genomes has indicated that this bacterium lacks genes for topoisomerase IV as was already described for other species such as Mycobacterium tuberculosis and Helicobacter pylori (23). It was proposed that C. difficile DNA gyrase would assume both the supercoiling and the relaxation/decatenation activities normally performed by DNA gyrase and topoisomerase IV in bacteria containing the two enzymes. Indeed, the C. difficile DNA gyrase exhibited supercoiling as well as decatenation activities in vitro that could compensate for the absence of a topoisomerase IV enzyme (results not shown). Cadazolid demonstrated no inhibition in C. difficile DNA gyrase assays up to its maximum solubility in the assay buffers (50 μM) (Table 4). Ciprofloxacin also did not exhibit any inhibition at the higher tested concentrations, but moxifloxacin, a generally more potent quinolone antibiotic, did show inhibition of both Clostridium difficile supercoiling and decatenation DNA gyrase activities. Still, with E. coli DNA gyrase and topoisomerase IV, cadazolid showed measurable inhibition.
TABLE 4.
Activity of cadazolid and comparators in DNA topoisomerase assays
| Enzyme | IC50 (μM) |
|||
|---|---|---|---|---|
| Cadazolidc | Linezolid | Ciprofloxacin | Moxifloxacin | |
| DNA gyrase (E. coli)a | 8–32 | >256 | 0.125–2 | 0.5 |
| DNA topoIV (E. coli)b | 32 to >50 | >256 | 2–8 | 8 |
| DNA gyrase (C. difficile)a | >50 | >256 | >256 | 32–128 |
| DNA gyrase (C. difficile)b | >50 | >256 | >256 | 32–128 |
Supercoiling.
Decatenation.
Maximal solubility of cadazolid in assay buffer (2 to 5% DMSO), 50 μM.
Propensity for in vitro resistance development.
Spontaneous resistance frequencies were measured in four strains of C. difficile, including strains with preexisting resistance to oxazolidinones (linezolid) and/or fluoroquinolones (moxifloxacin). Results obtained with cadazolid were compared to those obtained with linezolid, fluoroquinolones, vancomycin, and fidaxomicin (lipiarmycin A3), as appropriate (Table 5). No colonies were detected after plating high bacterial numbers (generally >109 bacteria/plate) on plates containing cadazolid at 2 to 4× the MIC (frequencies, <10−10). Results obtained with strains with preexisting resistance to fluoroquinolones and or linezolid were not different from those of susceptible strains. For vancomycin and linezolid, resistance frequencies were comparably low, whereas for fidaxomicin and moxifloxacin frequencies were moderate to high (10−7 to 10−8 [Table 5]).
TABLE 5.
Spontaneous resistance frequencies of cadazolid and comparator antibiotics in strains of C. difficile
| Bacterial straina | Antibiotic (fold MIC) | Spontaneous resistance frequency |
|---|---|---|
| C. difficile ATCC 9689 | Cadazolid (2) | <7.8 × 10−10 |
| Cadazolid (4) | <7.8 × 10−10 | |
| Linezolid (4) | <3.3 × 10−10 | |
| Moxifloxacin (4) | 2.7 × 10−8 | |
| Vancomycin (4) | 1.1 × 10−9 | |
| Fidaxomicin (2) | 1.3 × 10−8 | |
| Fidaxomicin (8) | 3.9 × 10−9 | |
| C. difficile NCTC 13366 (Qr, 027 ribotype) | Cadazolid (2) | <3.5 × 10−10 |
| Cadazolid (4) | <3.5 × 10−10 | |
| Linezolid (4) | 1.3 × 10−9 | |
| Vancomycin (4) | <6.1 × 10−9 | |
| Fidaxomicin (4) | 7.2 × 10−7 | |
| Fidaxomicin (8) | 3.3 × 10−8 | |
| C. difficile A-1291 (LZDr) | Cadazolid (2) | <7.1 × 10−10 |
| Cadazolid (4) | <7.1 × 10−10 | |
| Moxifloxacin (4) | 2.2 × 10−8 | |
| Vancomycin (8) | <5.2 × 10−10 | |
| C. difficile A-1410 (Qr and LZDr) | Cadazolid (2) | <4.8 × 10−9 |
| Cadazolid (4) | <4.8 × 10−9 | |
| Vancomycin (8) | <4.8 × 10−9 | |
| Fidaxomicin (8) | 1.1 × 10−6 |
Qr, quinolone resistant; LZDr, linezolid resistant.
Colonies or lawns from residual growth obtained after one-step resistance development experiments were subjected to further selection steps. MICs of single-colony isolates grown at the highest drug concentration were determined for individual steps and are provided in Table 6. Colonies isolated from plates with cadazolid after 1 to 3 selection steps had a maximally 2-fold-increased MIC for cadazolid and linezolid, and no increased MICs for any of the other antibiotics tested (moxifloxacin, fidaxomicin). Strains selected with linezolid in 1 to 3 steps had moderately increased MICs for linezolid (from 2 to 8 μg/ml) but only 2-fold-increased MICs for cadazolid. In contrast, 2- to 3-step selection with fidaxomicin or moxifloxacin resulted in strains with high MICs for the selecting antibiotic (>32 μg/ml) (Table 6). Cadazolid's activity against fidaxomicin- and moxifloxacin-nonsusceptible strains was unchanged, indicating absence of cross-resistance.
TABLE 6.
MICs of selected clones of C. difficile obtained in single-step and multistep resistance development experiments compared to parent strains
| C. difficile strain and selecting antibiotic | Selection step, medium | MIC (μg/ml) |
|||
|---|---|---|---|---|---|
| Cadazolid | Linezolid | Moxifloxacin | Fidaxomicin | ||
| NCTC 13366 (parent) | |||||
| None | 0.25 | 2 | 32 | 0.25 | |
| Cadazolid | Step 1, agar | 0.25 | 2 | 32 | 0.25 |
| Cadazolid | Step 2, agar | 0.5 | 2 | 32 | 0.25 |
| Cadazolid | Step 3, agar | 0.25 | 2 | 32 | 0.25 |
| Linezolid | Step1, agar | 0.5 | 8 | 32 | 0.25 |
| Linezolid | Step 2, agar | 0.5 | 8 | 32 | 0.25 |
| Linezolid | Step 3, agar | 0.5 | 8 | 32 | 0.25 |
| Fidaxomicin | Step 1, agar | 0.125 | 2 | 32 | 4 |
| Fidaxomicin | Step 2, agar | 0.125 | 2 | 32 | 128 |
| ATCC 9689 (parent) | |||||
| None | 0.25 | 2 | 2 | ≤0.031 | |
| Cadazolid | Step 1, agar | 0.25 | 2 | 2 | ≤0.03 |
| Cadazolid | Step 2, agar | 0.5 | 4 | 2 | ≤0.03 |
| Cadazolid | Step 3, agar | 0.5 | 4 | 2 | ≤0.03 |
| Cadazolid | Passage 13, liquid medium | 0.25 | 2 | 2 | ≤0.03 |
| Moxifloxacin | Step 1, agar | 0.06 | 2 | 4 | 0.008 |
| Moxifloxacin | Step 2, agar | 0.125 | 2 | 64 | 0.008 |
| Linezolid | Step 1, agar | 0.5 | 8 | 2 | ≤0.03 |
| Linezolid | Step 2, agar | 0.5 | 8 | 2 | ≤0.03 |
| Fidaxomicin | Step 1, agar | 0.25 | 4 | 2 | 2 |
| Fidaxomicin | Step 2, agar | 0.25 | 4 | 2 | 32 |
| Fidaxomicin | Step 3, agar | 0.125 | 4 | 2 | >128 |
| ATCC 700057 (quality control) | 0.25 | 2 | 2 | 0.06 | |
In another set of experiments, attempts to select resistance were done by serial passages in liquid medium in tubes (Fig. 3). Cadazolid MICs recorded in the tubes (MICt) increased only very slowly upon 13 passages in the three strains tested, i.e., maximally 1 or 2 MIC dilution steps, and MICs for linezolid, moxifloxacin, and fidaxomicin were not significantly changed in the final isolate (Table 6). In comparison, MICts of moxifloxacin increased significantly (up to 16-fold) in the passage experiments, while linezolid MICts hardly increased, similar to those of cadazolid.
FIG 3.
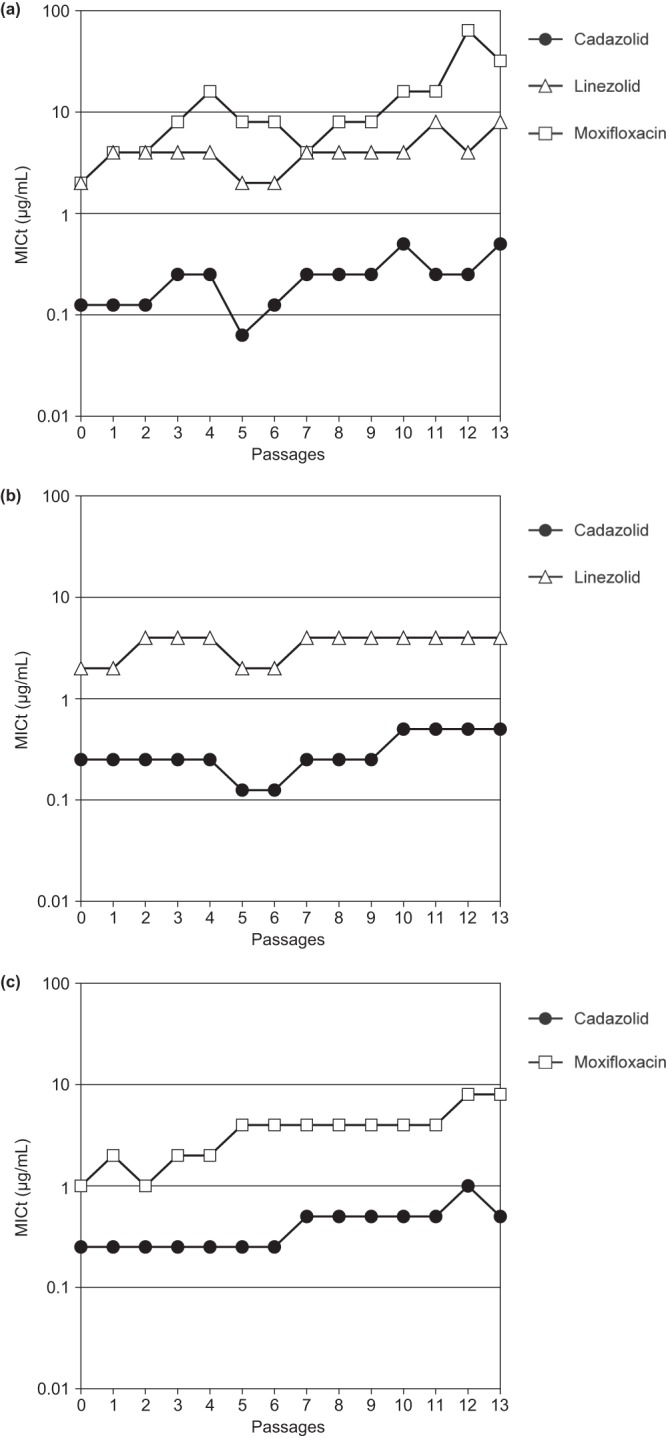
Test of development of resistance to cadazolid and comparator antibiotics by serial passages in liquid medium. C. difficile strains ATCC 9689 (a), NCTC 13366 (fluoroquinolone resistant) (b), and A-1290 (linezolid resistant) (c). Tubes containing 2 ml of 2-fold serial dilutions of the test antibiotics were inoculated with approximately 5 × 106 to 2 × 107 CFU. After incubation for 48 h at 37°C, the tube with the highest antibiotic concentration permitting growth was used to inoculate the next series of tubes (1%, vol/vol). This procedure was repeated for 13 passages, and the lowest antibiotic concentration inhibiting growth in the tubes (MICt) was recorded at the end of every passage.
DISCUSSION
Several experimental approaches were used to determine the mechanism of action of cadazolid in C. difficile. Macromolecular labeling, in vitro transcription/translation, and topoisomerase assays indicate that cadazolid acts primarily on protein synthesis as a translation inhibitor with weak inhibition of DNA synthesis as a second effect.
Even though incorporation of thymidine into DNA was not sufficient to evaluate DNA biosynthesis, it could still be assayed in C. difficile indirectly after labeling of all newly synthesized nucleic acids and removal of RNA under basic conditions. Macromolecular labeling data indicate that cadazolid also acts as a potent inhibitor of protein synthesis in strains resistant or nonsusceptible to linezolid and suggest that it also has the capacity to inhibit DNA synthesis in C. difficile, albeit at higher concentrations than needed to inhibit protein synthesis. Surprisingly, quinolone resistance did not result in any significant shift of the IC50 with cadazolid, whereas the IC50 with moxifloxacin increased more than 20-fold. Therefore, the inhibition of DNA synthesis contributes only marginally to the antibacterial activity in C. difficile strains. Finally, the absence of inhibition of cell wall biosynthesis with cadazolid further supports the hypothesis that the potent inhibition observed is the result of specific inhibition of protein synthesis rather than unspecific effects. This is supported by our CFTA data indicating that cadazolid strongly inhibits in vitro translation in extracts derived from C. difficile strains. Experiments with the goal to discriminate between effects on transcription and translation have shown that cadazolid is interfering mainly with the translation machinery. Notably, CFTA IC50s of cadazolid were similar in extracts derived from linezolid-susceptible and -resistant strains, in agreement with MIC values, whereas linezolid lost its potency substantially in extracts from linezolid-resistant strains.
We also attempted to measure the effect of cadazolid in bacterial topoisomerase assays. While a significant inhibition was obtained in the E. coli gyrase assay, no inhibitory activity could be observed in the C. difficile gyrase assay. This is in contrast to the macromolecular labeling data in which inhibition of DNA synthesis in C. difficile was detected, albeit at relatively high concentrations. However, this discrepancy may be explained by the low sensitivity of the topoisomerase assays and the limited water solubility of cadazolid. Taken together, our data suggest that cadazolid can overcome linezolid resistance because of more-potent translational inhibition activity rather than by interaction with DNA synthesis. Consistently with the proposed mechanism of action, i.e., inhibition of protein synthesis, cadazolid was also shown to be a potent inhibitor of C. difficile toxin and spore formation, even in the absence of bacterial killing (15). Further studies involving ribosomal binding and structure-activity relationship experiments are needed to investigate the detailed mode of action of cadazolid.
The frequency of spontaneous resistance development of cadazolid in strains of C. difficile was very low (<10−10), and the selection of clones with significantly increased MICs for cadazolid was not possible even in strains with preexisting resistance to fluoroquinolones (such as the hypervirulent NAP1/027/BI clone) and to linezolid. This also included multiple-passage experiments. No mutations in the quinolone resistance-determining region of DNA gyrase A and B subunits as well as in the 23S ribosomal genes were detected in C. difficile strains selected in vitro with cadazolid in multiple-passage experiments (P. Caspers, unpublished data). The mechanism of LZD resistance in C. difficile has not been characterized so far, but LZD resistance in a Clostridium perfringens strain has recently been linked to a mutation in ribosomal protein L4 (24).
Overall, our results suggest that cadazolid has a low in vitro propensity of spontaneous resistance development, similar to or better than that of linezolid and lower than those of moxifloxacin and fidaxomicin. Furthermore, cadazolid retained activity against quinolone-resistant as well as linezolid-resistant strains and did not select for strains with significantly increased MICs for fluoroquinolones or linezolid, indicating an absence of cross-resistance. In an in vitro human gut model of C. difficile infection, no evidence for selecting quinolone- or linezolid-resistant gut bacteria was obtained after treatment with cadazolid, and the compound had a very limited impact on the indigenous gut microflora (25). Importantly, cadazolid showed also no cross-resistance with antibiotics currently used to treat CDAD and retained potent activity against strains nonsusceptible to metronidazole (25) and fidaxomicin (this study). However, clinical data are needed to confirm the overall low propensity of resistance development of cadazolid. Results of in vitro and in vivo evaluations of cadazolid are reported in a companion article by Locher et al. (26).
ACKNOWLEDGMENTS
We thank M. Wilcox, Leeds (United Kingdom), and D. Gerding, Chicago (IL), for providing C. difficile strains and for helpful advice and discussions.
Potential conflicts of interest: H. H. Locher, P. Caspers, T. Bruyère, S. Schroeder, P. Pfaff, A. Knezevic, W. Keck, and D. Ritz are or have been employees and stockholders of Actelion Pharmaceuticals Ltd.
Footnotes
Published ahead of print 25 November 2013
REFERENCES
- 1.Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536. 10.1038/nrmicro2164 [DOI] [PubMed] [Google Scholar]
- 2.Dubberke E. 2012. Clostridium difficile infection: the scope of the problem. J. Hosp. Med. 7(Suppl 3):S1–S4. 10.1002/jhm.1916 [DOI] [PubMed] [Google Scholar]
- 3.Razavi B, Apisarnthanarak A, Mundy LM. 2007. Clostridium difficile: emergence of hypervirulence and fluoroquinolone resistance. Infection 35:300–307. 10.1007/s15010-007-6113-0 [DOI] [PubMed] [Google Scholar]
- 4.Gerding DN, Johnson S. 2011. Clostridium difficile infection in 2010: advances in pathogenesis, diagnosis and management of CDI. Nat. Rev. Gastroenterol. Hepatol. 8:67–68. 10.1038/nrgastro.2010.215 [DOI] [PubMed] [Google Scholar]
- 5.Ananthakrishnan AN. 2011. Clostridium difficile infection: epidemiology, risk factors and management. Nat. Rev. Gastroenterol. Hepatol. 8:17–26. 10.1038/nrgastro.2010.190 [DOI] [PubMed] [Google Scholar]
- 6.Koo HL, Garey KW, Dupont HL. 2010. Future novel therapeutic agents for Clostridium difficile infection. Expert Opin. Invest. Drugs 19:825–836. 10.1517/13543784.2010.495386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardesty JS, Juang P. 2011. Fidaxomicin: a macrocyclic antibiotic for the treatment of Clostridium difficile infection. Pharmacotherapy 31:877–886. 10.1592/phco.31.9.877 [DOI] [PubMed] [Google Scholar]
- 8.Louie TJ, Miller MA, Mullane KM, Weiss K, Lentnek A, Golan Y, Gorbach S, Sears P, Shue YK, OPT-80-003 Clinical Study Group 2011. Fidaxomicin versus vancomycin for Clostridium difficile infection. N. Engl. J. Med. 364:422–431. 10.1056/NEJMoa0910812 [DOI] [PubMed] [Google Scholar]
- 9.Cornely OA, Crook DW, Esposito R, Poirier A, Somero MS, Weiss K, Sears P, Gorbach S, OPT-80-004 Clinical Study Group 2012. Fidaxomicin versus vancomycin for infection with Clostridium difficile in Europe, Canada, and the USA: a double-blind, non-inferiority, randomised controlled trial. Lancet Infect. Dis. 12:281–289. 10.1016/S1473-3099(11)70374-7 [DOI] [PubMed] [Google Scholar]
- 10.Rashid MU, Martinez Lozano H, Weintraub A, Nord CE. 2013. In vitro activity of cadazolid against Clostridium difficile strains isolated from primary and recurrent infections in Stockholm, Sweden. Anaerobe 20:32–35. 10.1016/j.anaerobe.2013.02.003 [DOI] [PubMed] [Google Scholar]
- 11.Hecht DW, Osmolski JR, Sambol S, Cheknis A, Gerding DN. 2012. In vitro activity of cadazolid against 209 toxigenic isolates of Clostridium difficile, abstr E-808 Abstr. 52nd Intersci. Conf. Antimicrob. Agents Chemother., San Francisco, CA. American Society for Microbiology, Washington, DC [Google Scholar]
- 12.Locher HH, Pfaff P, Schroeder S, Specklin JL, Hubschwerlen C, Keck W. 2012. Cadazolid, a novel quinolonyl-oxazolidinone antibiotic with potent activity against Clostridium difficile: in vitro antibacterial activity and propensity for resistance development, abstr C1-1346 Abstr. 52nd Intersci. Conf. Antimicrob. Agents Chemother., San Francisco, CA. American Society for Microbiology, Washington, DC [Google Scholar]
- 13.Drlica K, Malik M. 2003. Fluoroquinolones: action and resistance. Curr. Top. Med. Chem. 3:249–282. 10.2174/1568026033452537 [DOI] [PubMed] [Google Scholar]
- 14.Caspers P, Ritz D, Locher HH, Bruyère T, Schroeder S, Pfaff P, Knezevic A, Dos Santos M, Hubschwerlen C, Keck W. 2013. Cadazolid, a new antibiotic with potent activity against Clostridium difficile inhibits protein synthesis also in linezolid-resistant strains, abstract/poster P-1657 Abstr. 53rd Eur. Congr. Clin. Microbiol. Infect. Dis., Berlin, Germany [Google Scholar]
- 15.Locher HH, Ritz D, Pfaff P, Schroeder S, Knezevic A, Hubschwerlen C, Keck W. 2012. Cadazolid, a novel quinolonyl-oxazolidinone antibiotic: mode of action and effect on Clostridium difficile toxin and spore formation, abstr C1-1347 Abstr. 52nd Intersci. Conf. Antimicrob. Agents Chemother., San Francisco, CA. American Society for Microbiology, Washington, DC [Google Scholar]
- 16.Srivastava A, Talaue M, Liu S, Degen D, Ebright RY, Sineva E, Chakraborty A, Chatterje S, Zozula A, Shen J, Xin C, Kaneko T, Jansen R, Donadio S, Connell N, Ebright RH. 2011. New target for inhibition of bacterial RNA polymerase: ‘switch region.’ Curr. Opin. Microbiol. 14:532–543. 10.1016/j.mib.2011.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CLSI 2007. Methods for antimicrobial susceptibility testing of anaerobic bacteria: approved standard—7th ed, M11-A7. Clinical and Laboratory Standards Institute, Wayne, PA: [PubMed] [Google Scholar]
- 18.Mathur T, Kumar M, Barman TK, Kumar GR, Kalia V, Raj VS, Das B, Bhatnagar PK. 2011. Activity of RBx 11760, a novel biaryl oxazolidinone, against Clostridium difficile. J. Antimicrob. Chemother. 66:1087–1095. 10.1093/jac/dkr033 [DOI] [PubMed] [Google Scholar]
- 19.Kim TW, Keum JW, Oh IS, Choi CJ, Park CG, Kim DM. 2006. Simple procedures for the construction of a robust and cost-effective cell-free protein synthesis system. J. Biotechnol. 126:554–561. 10.1016/j.jbiotec.2006.05.014 [DOI] [PubMed] [Google Scholar]
- 20.Scotcher MC, Rudolph FB, Bennett GN. 2005. Expression of abrB310 and SinR, and effects of decreased abrB310 expression on the transition from acidogenesis to solventogenesis in Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 71:1987–1995. 10.1128/AEM.71.4.1987-1995.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett JF, Bernstein JI, Krause HM, Hilliard JJ, Ohemeng KK. 1993. Testing potential gyrase inhibitors of bacterial DNA gyrase: a comparison of the supercoiling inhibition assay and “cleavable complex” assay. Anal. Biochem. 214:313–317. 10.1006/abio.1993.1493 [DOI] [PubMed] [Google Scholar]
- 22.Fisher LM, Pan XS. 2008. Methods to assay inhibitors of DNA gyrase and topoisomerase IV activities. Methods Mol. Med. 142:11–23. 10.1007/978-1-59745-246-5_2 [DOI] [PubMed] [Google Scholar]
- 23.Dridi L, Tankovic J, Burghoffer B, Barbut F, Petit JC. 2002. GyrA and gyrB mutations are implicated in cross-resistance to ciprofloxacin and moxifloxacin in Clostridium difficile. Antimicrob. Agents Chemother. 46:3418–3421. 10.1128/AAC.46.11.3418-3421.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holzel CS, Harms KS, Schwaiger K, Bauer J. 2010. Resistance to linezolid in a porcine Clostridium perfringens strain carrying a mutation in the rplD gene encoding the ribosomal protein L4. Antimicrob. Agents Chemother. 54:1351–1353. 10.1128/AAC.01208-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baines S, Crowther G, Todhunter S, Freeman J, Wilcox M. 2012. In vitro activity of cadazolid (ACT-179811) against Clostridium difficile and in an in vitro gut model of C. difficile infection, abstr B-662 Abstr. 52nd Intersci. Conf. Antimicrob. Agents Chemother., San Francisco, CA. American Society for Microbiology, Washington, DC [Google Scholar]
- 26.Locher HH, Seiler P, Chen X, Schroeder S, Pfaff P, Enderlin M, Klenk A, Fournier E, Hubschwerlen C, Ritz D, Kelly CP, Keck W. 2014. In Vitro and In Vivo Antibacterial Evaluation of Cadazolid, a New Antibiotic for Treatment of Clostridium difficile Infections. Antimicrob. Agents Chemother. 58:892–900 [DOI] [PMC free article] [PubMed] [Google Scholar]