Abstract
Escherichia coli (328 isolates), Klebsiella pneumoniae (296), Klebsiella oxytoca (44), and Proteus mirabilis (33) isolates collected during 2012 from the nine U.S. census regions and displaying extended-spectrum-β-lactamase (ESBL) phenotypes were evaluated for the presence of β-lactamase genes, and antimicrobial susceptibility profiles were analyzed. The highest ESBL rates were noted for K. pneumoniae (16.0%, versus 4.8 to 11.9% for the other species) and in the Mid-Atlantic and West South Central census regions. CTX-M group 1 (including CTX-M-15) was detected in 303 strains and was widespread throughout the United States but was more prevalent in the West South Central, Mid-Atlantic, and East North Central regions. KPC producers (118 strains [112 K. pneumoniae strains]) were detected in all regions and were most frequent in the Mid-Atlantic region (58 strains). Thirteen KPC producers also carried blaCTX-M. SHV genes encoding ESBL activity were detected among 176 isolates. Other β-lactamase genes observed were CTX-M group 9 (72 isolates), FOX (10), TEM ESBL (9), DHA (7), CTX-M group 2 (3), NDM-1 (2 [Colorado]), and CTX-M groups 8 and 25 (1). Additionally, 62.9% of isolates carried ≥2 β-lactamase genes. KPC producers were highly resistant to multiple agents, but ceftazidime-avibactam (MIC50/90, 0.5/2 μg/ml) and tigecycline (MIC50/90, 0.5/1 μg/ml) were the most active agents tested. Overall, meropenem (MIC50, ≤0.06 μg/ml), ceftazidime-avibactam (MIC50, 0.12 to 0.5 μg/ml), and tigecycline (MIC50, 0.12 to 2 μg/ml) were the most active antimicrobials when tested against this collection. NDM-1 producers were resistant to all β-lactams tested. The diversity and increasing prevalence of β-lactamase-producing Enterobacteriaceae have been documented, and ceftazidime-avibactam was very active against the vast majority of β-lactamase-producing strains isolated from U.S. hospitals.
INTRODUCTION
The β-lactamase production scenario among Enterobacteriaceae isolates collected in U.S. hospitals differs from that of most other countries due to the late appearance of CTX-M-producing isolates (1–3), high prevalence of KPC producers (4, 5), and low incidence of isolates carrying metallo-β-lactamase genes (4). Within the United States, there are also differences in the prevalence of KPC-producing isolates, and although KPC-mediated resistances have been reported throughout the United States, the New York City area is where these strains seem to be endemic (6), and these strains have recently been isolated in Texas with increasing frequency (7). Furthermore, extended-spectrum-β-lactamase (ESBL) rates have been documented to vary among U.S. regions (8), and the extent of genetic diversity of β-lactamase combinations among ESBL-positive phenotypes has only recently been documented (9). This unique profile raises awareness not only for the need for surveillance of these resistance mechanisms but also for the necessity of therapeutic options to treat multidrug-resistant isolates, including KPC-producing organisms that may be resistant to all classes of clinically available antimicrobial agents, including colistin and/or tigecycline (10, 11).
Avibactam is a novel non-β-lactam β-lactamase inhibitor of enzymes belonging to Ambler structural classes A and C and some class D enzymes (12). In a previous study (13), avibactam combined with ceftaroline was able to extend the activity of the cephalosporin against β-lactamase-producing isolates, including those carrying blaKPC. The vast majority of tested isolates were resistant to ceftaroline alone but were susceptible in the presence of avibactam (13).
We evaluated 701 ESBL-phenotype-positive isolates of Escherichia coli, Klebsiella spp., and Proteus mirabilis, identified by using reference criteria (14, 15), for the prevalence of common β-lactamase-encoding genes and analyzed the distribution of these genes in each U.S. census region. Detection of β-lactamase genes/groups was performed by using a validated commercial methodology for U.S. isolates (9). Additionally, we analyzed the activities of ceftazidime-avibactam and comparator agents tested against isolates with the most prevalent, characterized β-lactamases.
MATERIALS AND METHODS
Bacterial isolates.
A total of 5,739 isolates of E. coli (n = 2,767), Klebsiella spp. (n = 2,289 [1,847 K. pneumoniae and 442 K. oxytoca isolates]), and P. mirabilis (n = 683) were consecutively collected from 72 U.S. hospitals during 2012 for analysis. Only one isolate per patient was included in the study, from the following sites of infection: bloodstream (n = 991), respiratory tract from hospitalized patients (n = 1,047), intra-abdominal sources (n = 285), skin and skin structure sources (n = 1,565), urinary tract (n = 1,558), and other or unknown sources (n = 293). Species identification was confirmed when necessary by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) using the Bruker Daltonics (Billerica, MA, USA) MALDI Biotyper according to the manufacturer's instructions.
Antimicrobial susceptibility testing.
All isolates were tested for susceptibility by using dry-form Sensititre panels from ThermoFisher Scientific (formerly Trek Diagnostic Systems, Inc., Cleveland, OH) and the reference broth microdilution method as described by the Clinical and Laboratory Standards Institute (CLSI) (14). Categorical interpretations for all antimicrobials were those found in method M100-S23 (15) and those reported by the EUCAST (16). Quality control (QC) was performed with E. coli ATCC 25922, K. pneumoniae ATCC 700603, and Pseudomonas aeruginosa ATCC 27853, and all QC results were within specified ranges reported in CLSI documents (15).
Screening for β-lactamases.
Isolates displaying the CLSI criteria for an ESBL phenotype (MIC of >1 μg/ml for aztreonam, ceftazidime, and/or ceftriaxone [15]) were tested for β-lactamase-encoding genes by using the microarray-based assay Check-MDR CT101 kit (Check-points; Wageningen, Netherlands). The assay was performed according to the manufacturer's instructions. This kit has the capability to detect CTX-M groups 1, 2, 8 plus 25, and 9; TEM WT and ESBL; SHV WT and ESBL; ACC; ACT/MIR; CMYII; DHA; FOX; KPC; and NDM-1.
Multilocus sequence typing.
NDM-1-producing K. pneumoniae isolates were evaluated by multilocus sequence typing (MLST) according to methods of the Institut Pasteur (Paris, France) (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html).
RESULTS AND DISCUSSION
ESBL phenotype rates and U.S. census regions.
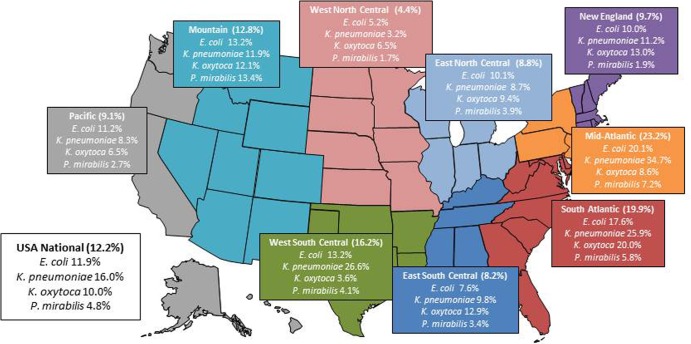
Among 5,739 E. coli, Klebsiella species, and P. mirabilis isolates collected in U.S. hospitals, 701 (12.2%) met the CLSI epidemiological screening criteria for an ESBL phenotype (15) and underwent further analysis by using microarray-based technology for detection of β-lactamase-encoding genes. Isolates with an ESBL phenotype included 328 E. coli (11.9% of the overall samples for this species), 296 K. pneumoniae (16.0%), 44 K. oxytoca (10.0%), and 33 P. mirabilis (4.8%) isolates. ESBL-phenotype-positive isolates from the four targeted bacterial species were noted in all U.S. census regions, with rates varying according to region and pathogen (Fig. 1). Overall, the West North Central region had the lowest rates (1.7% [P. mirabilis] to 6.5% [K. oxytoca]), and the South Atlantic (5.8% [P. mirabilis] to 25.9% [K. pneumoniae]) and Mid-Atlantic (7.2% [P. mirabilis] to 34.7% [K. pneumoniae]) regions had the highest ESBL rates. K. pneumoniae had the highest ESBL rates among the species analyzed in five of nine census regions.
FIG 1.
ESBL rates among Enterobacteriaceae isolates collected in 72 U.S. hospitals located in the nine U.S. census regions. (Courtesy of Ian Critchley, reproduced with permission.)
Prevalence of β-lactamase genes detected.
CTX-M group 1 was the most prevalent positive result in the microarray-based assay and was detected in 303 (43.2%) isolates. CTX-M group 1, here named CTX-M-15-like, has been documented to be the most prevalent CTX-M type in U.S. hospitals (2, 3, 9, 17). This group included CTX-M-15, CTX-M-3, and CTX-M-1, which have been observed in the United States (3, 9). This gene/family was detected among all bacterial species evaluated and across all U.S. census regions (Table 1), being more prevalent in the West South Central (67 strains; 61.5% of the isolates from region), East North Central (50 strains; 50.5%), and Mountain (27; 48.2%) regions (Table 1). Other regions had 14.2 to 24.3% CTX-M-15-like-producing isolates among the ESBL-producing strains identified.
TABLE 1.
Occurrences of ESBL, acquired cephalosporinases (AmpC), and carbapenemase enzymes detected among 701 ESBL-phenotype-positive strains collected in U.S. hospitals by U.S. census regions
| Region (no. of isolates tested) | No. of isolates (% of isolates tested by region) positive fora: |
|||||||
|---|---|---|---|---|---|---|---|---|
| CTX-M-15-like | SHV ESBL | KPC | CTX-M-14-like | CMY-2-like | FOX | TEM ESBL | DHA | |
| Overall (701) | 303 (43.2) | 176 (25.1) | 118 (16.8) | 72 (10.3) | 64 (9.1) | 10 (1.4) | 9 (1.3) | 7 (1.0) |
| New England (47) | 19 (40.4) | 9 (19.1) | 1 (2.1) | 5 (10.6) | 6 (12.8) | 2 (4.3) | ||
| Mid-Atlantic (143) | 50 (35.0) | 42 (29.4) | 58 (40.6) | 11 (7.7) | 15 (10.5) | 2 (1.4) | 2 (1.4) | 1 (0.7) |
| East North Central (99) | 50 (50.5) | 18 (18.2) | 13 (13.1) | 7 (7.1) | 6 (6.1) | |||
| West North Central (24) | 9 (37.5) | 1 (4.2) | 1 (4.2) | 4 (16.7) | 5 (20.8) | 2 (8.3) | ||
| South Atlantic (110) | 45 (40.9) | 37 (33.6) | 18 (16.4) | 10 (9.1) | 4 (3.6) | 3 (2.7) | 2 (1.8) | |
| East South Central (39) | 10 (25.6) | 11 (28.2) | 1 (2.6) | 3 (7.7) | 2 (5.1) | 4 (10.3) | 1 (2.6) | |
| West South Central (109) | 67 (61.5) | 30 (27.5) | 14 (12.8) | 9 (8.3) | 12 (11.0) | 1 (0.9) | 2 (1.8) | |
| Mountain (56) | 27 (48.2) | 10 (17.9) | 8 (14.3) | 9 (16.1) | 6 (10.7) | |||
| Pacific (74) | 26 (35.1) | 18 (24.3) | 4 (5.4) | 14 (18.9) | 8 (10.8) | 2 (2.7) | 2(2.7) | |
Various isolates were positive for one or more β-lactamase-encoding genes tested.
A CTX-M-15-like phenotype was observed in combination with up to 4 other β-lactamase-encoding genes or gene families included in the applied microarray in 181 isolates. The most common combination was CTX-M-15-like plus TEM WT or narrow-spectrum enzymes (83 isolates), followed by CTX-M-15-like plus SHV WT plus TEM WT (42 isolates). However, 53 strains harboring CTX-M-15-like genes also carried determinants encoding other β-lactamases having broad-spectrum activity, including carbapenemases (NDM-1 and KPC), ESBLs (CTX-M group 9, SHV, and TEM), and plasmid-mediated AmpCs.
The second most prevalent group/family was the SHV enzymes with an extended spectrum of activity (SHV ESBL), which were detected among 176 (25.1%) isolates from all bacterial species and census regions (Table 1). From our previous studies (9), SHV-12, SHV-5, SHV-7, SHV-2, and SHV-30 (in that order) were the most common SHV ESBL types found in U.S. hospitals (3, 9). SHV ESBLs were the only enzymes detected in 68 isolates and were noted in various combinations with other β-lactamases among other isolates.
KPC serine carbapenemases were observed in 118 (16.8%) isolates, most of them K. pneumoniae (112 isolates; 94.9%). Three isolates each of E. coli and K. oxytoca also harbored this gene (two of each species were from the Mid-Atlantic region). KPC-producing isolates were detected in all census regions and were most prevalent in the Mid-Atlantic region (58 isolates; 16.4% of the ESBL-phenotype-positive strains), which includes the New York City area, where KPC-carrying isolates were first reported to be endemic (18, 19). The West South Central, South Atlantic, East North Central, and Mountain regions had occurrences of KPC producers of 7.2 to 18.6%. In the remaining census regions, the prevalence of KPC producers was <3.5% (1 to 4 isolates). KPC-producing isolates often harbored TEM WT and SHV WT (ubiquitous in K. pneumoniae), but 13 isolates with CTX-M-15-like-encoding genes were also observed. In two independent surveillance initiatives from 2007, KPC-producing isolates did not harbor genes encoding CTX-M-15-like enzymes, whereas in more recent evaluations, numbers of isolates carrying genes encoding both types of enzymes seem to be increasing. Isolates having blaKPC and blaCTX-M-15 detected in this study and in a 2010 data set (9) were in all instances K. pneumoniae isolates, suggesting that this bacterial species is able to accumulate multiple genetic elements harboring resistance genes.
The most common CTX-M group 9 enzymes found among isolates from the United States were CTX-M-14 and CTX-M-27 (3, 9) (here named CTX-M-14-like). Most of these 72 isolates (10.3%) were E. coli (57 isolates). However, this enzyme group was also detected in K. pneumoniae and P. mirabilis (6 and 9 isolates, respectively). CTX-M-14-like-producing isolates were obtained from all census regions, and the prevalence varied from 7.1 to 18.9% of the isolates tested in each region, with the highest incidence in the Pacific region and the lowest in the East North Central region (Table 1). A total of 43 CTX-M group 9-positive isolates were also positive for the probes targeting DHA, CMYII, TEM, and SHV, in addition to two isolates harboring genes encoding CTX-M-15-like enzymes.
Sixty-four isolates were CMYII probe positive on the Check-MDR CT101 kit, which has the ability to detect the majority of the nonintrinsic genes encoding CMY-2-like variants (Check-points, personal communication). CMY-2 has been demonstrated to be the most commonly detected acquired cephalosporinase (AmpC) gene in U.S. hospitals (3, 9). CMY-2-like-positive isolates included 50 E. coli isolates, 13 P. mirabilis isolates, and 1 K. pneumoniae isolate, which were observed in all U.S. census regions and were noted to be most prevalent in the West North Central region (20.8%; other regions, 3.6 to 11.0% of the isolates tested of each region).
Two NDM-1-producing K. pneumoniae isolates were detected from one medical center in the Mountain region. These isolates were clonally related (MLST type 147), also carried CTX-M group 1 genes, and were recently reported by the U.S. Centers for Disease Control and Prevention (CDC) (20).
Other ESBLs screened by the Check-points methodology were also detected in smaller numbers, as follows: FOX (10 isolates, 4 regions), TEM ESBL (9 isolates, 5 regions), DHA (7 isolates, 4 regions), CTX-M group 2 (3 K. pneumoniae isolates, Mid-Atlantic region only [1 hospital]), and CTX-M groups 8 and 25 (1 P. mirabilis isolate, Mid-Atlantic region). Additionally, TEM enzymes with limited-spectrum activity (TEM WT) were quite prevalent and were detected in 341 isolates from all four screened species. SHV WT enzymes are considered ubiquitous among K. pneumoniae isolates, but they were also noted in four isolates from other bacterial species (E. coli and K. oxytoca).
Seventy isolates yielded a negative result for all β-lactamase-encoding genes/families tested. The majority of these β-lactamase-negative isolates displayed borderline MIC values (≤2 μg/ml) (Table 2) for the β-lactams used for ESBL screening (15). K. oxytoca strains that yielded negative results had the characteristic profile of OXY hyperproducers, as recently demonstrated (9), displaying elevated aztreonam and ceftriaxone and low ceftazidime MICs.
TABLE 2.
Activities of ceftazidime-avibactam and comparator antimicrobial agents tested against 701 ESBL-phenotype-positive Enterobacteriaceae isolates collected from 72 U.S. hospitals located in the nine U.S. census regions
| Isolate group (no. of isolates tested) and antimicrobial agent | MIC50 (μg/ml) | MIC90 (μg/ml) | MIC range (μg/ml) | % S/% I/% R according toa: |
|
|---|---|---|---|---|---|
| CLSI | EUCAST | ||||
| KPC producers (118) | |||||
| Ceftazidime-avibactam | 0.5 | 2 | 0.06–4 | –/–/– | –/–/– |
| Ceftazidime | >32 | >32 | 16–>32 | 0.0/0.0/100.0 | 0.0/0.0/100.0 |
| Ceftriaxone | >8 | >8 | 8–>8 | 0.0/0.0/100.0 | 0.0/0.0/100.0 |
| Meropenem | >8 | >8 | 2–>8 | 0.0/4.2/95.8 | 4.2/24.6/71.2 |
| Piperacillin-tazobactam | >64 | >64 | >64 | 0.0/0.0/100.0 | 0.0/0.0/100.0 |
| Levofloxacin | >4 | >4 | ≤0.12–>4 | 10.2/1.7/88.1 | 5.9/4.3/89.8 |
| Gentamicin | 8 | >8 | ≤1–>8 | 47.5/17.8/34.7 | 32.2/15.3/52.5 |
| Tetracycline | 4 | >32 | 1–>32 | 63.6/15.2/21.2 | –/–/– |
| Tigecyclineb | 0.5 | 1 | 0.06–8 | 98.3/0.8/0.9 | 93.2/5.1/1.7 |
| CTX-M-15-like producers (288)c | |||||
| Ceftazidime-avibactam | 0.12 | 0.5 | ≤0.015–2 | –/–/– | –/–/– |
| Ceftazidime | 16 | >32 | 0.5–>32 | 14.6/14.2/71.2 | 2.8/11.8/85.4 |
| Ceftriaxone | >8 | >8 | 8–>8 | 0.0/0.0/100.0 | 0.0/0.0/100.0 |
| Meropenem | ≤0.06 | ≤0.06 | ≤0.06–2 | 99.7/0.3/0.0 | 100.0/0.0/0.0 |
| Piperacillin-tazobactam | 16 | >64 | ≤0.5–>64 | 67.2/17.3/15.5 | 48.3/18.9/32.8 |
| Levofloxacin | >4 | >4 | ≤0.12–>4 | 12.5/4.2/83.3 | 11.5/1.0/87.5 |
| Gentamicin | 2 | >8 | ≤1–>8 | 53.3/2.1/44.6 | 51.6/1.7/46.7 |
| Tetracycline | >32 | >32 | 0.5–>32 | 30.2/0.7/69.1 | –/–/– |
| Tigecyclineb | 0.12 | 1 | 0.06–4 | 98.3/1.7/0.0 | 94.8/3.5/1.7 |
| CTX-M-14-like producers (70)d | |||||
| Ceftazidime-avibactam | 0.12 | 0.25 | 0.03–1 | –/–/– | –/–/– |
| Ceftazidime | 2 | 16 | 0.12–>32 | 74.3/11.4/14.3 | 35.7/38.5/25.7 |
| Ceftriaxone | >8 | >8 | >8 | 0.0/0.0/100.0 | 0.0/0.0/100.0 |
| Meropenem | ≤0.06 | ≤0.06 | ≤0.06–0.25 | 100.0/0.0/0.0 | 100.0/0.0/0.0 |
| Piperacillin-tazobactam | 2 | 8 | ≤0.5–>64 | 92.9/2.8/4.3 | 91.4/1.5/7.1 |
| Levofloxacin | >4 | >4 | ≤0.12–>4 | 17.1/2.9/80.0 | 17.1/0.0/82.9 |
| Gentamicin | 2 | >8 | ≤1–>8 | 55.7/0.0/44.3 | 54.3/1.4/44.3 |
| Tetracycline | >32 | >32 | 0.5–>32 | 22.9/0.0/77.1 | –/–/– |
| Tigecyclineb | 0.12 | 1 | 0.06–4 | 94.3/5.7/0.0 | 94.3/0.0/5.7 |
| SHV ESBL producers (83)e | |||||
| Ceftazidime-avibactam | 0.12 | 0.25 | ≤0.015–0.5 | –/–/– | –/–/– |
| Ceftazidime | >32 | >32 | 1–>32 | 12.0/7.3/80.7 | 2.4/9.6/88.0 |
| Ceftriaxone | >8 | >8 | 0.12–>8 | 9.6/7.3/83.1 | 9.6/7.3/83.1 |
| Meropenem | ≤0.06 | 0.12 | ≤0.06–2 | 98.8/1.2/0.0 | 100.0/0.0/0.0 |
| Piperacillin-tazobactam | >64 | >64 | 1–>64 | 45.8/3.6/50.6 | 36.1/9.7/54.2 |
| Levofloxacin | >4 | >4 | ≤0.12–>4 | 39.8/7.2/53.0 | 37.3/2.5/60.2 |
| Gentamicin | 2 | >8 | ≤1–>8 | 61.4/10.9/27.7 | 51.8/9.6/38.6 |
| Tetracycline | 4 | >32 | 0.5–>32 | 55.4/9.7/34.9 | –/–/– |
| Tigecyclineb | 0.5 | 1 | 0.06–2 | 100.0/0.0/0.0 | 94.0/6.0/0.0 |
| CMY-2-like producers (54)f | |||||
| Ceftazidime-avibactam | 0.12 | 0.5 | 0.03–1 | –/–/– | –/–/– |
| Ceftazidime | 16 | >32 | 2–>32 | 13.0/24.0/63.0 | 0.0/13.0/87.0 |
| Ceftriaxone | >8 | >8 | 0.5–>8 | 1.9/14.8/83.3 | 1.9/14.8/83.3 |
| Meropenem | ≤0.06 | 0.12 | ≤0.06–0.25 | 100.0/0.0/0.0 | 100.0/0.0/0.0 |
| Piperacillin-tazobactam | 4 | >64 | ≤0.5–>64 | 81.5/7.4/11.1 | 72.2/9.3/18.5 |
| Levofloxacin | ≤0.12 | >4 | ≤0.12–>4 | 70.4/1.8/27.8 | 64.8/5.6/29.6 |
| Gentamicin | ≤1 | >8 | ≤1–>8 | 79.6/1.9/18.5 | 77.8/1.8/20.4 |
| Tetracycline | >32 | >32 | 0.5–>32 | 33.3/1.9/64.8 | –/–/– |
| Tigecyclineb | 0.12 | 2 | 0.06–4 | 96.3/3.7/0.0 | 85.2/11.1/3.7 |
| ESBL microarray-negative isolates (70) | |||||
| Ceftazidime-avibactam | 0.12 | 0.5 | ≤0.015–1 | –/–/– | –/–/– |
| Ceftazidime | 1 | 4 | 0.06–8 | 94.3/5.7/0.0 | 50.0/44.3/5.7 |
| Ceftriaxone | 1 | >8 | 0.12–>8 | 55.7/4.3/40.0 | 55.7/4.3/40.0 |
| Meropenem | ≤0.06 | ≤0.06 | ≤0.06–0.5 | 100.0/0.0/0.0 | 100.0/0.0/0.0 |
| Piperacillin-tazobactam | >64 | >64 | 4–>64 | 30.0/7.1/62.9 | 22.9/7.1/70.0 |
| Levofloxacin | ≤0.12 | >4 | ≤0.12–>4 | 78.6/0.0/21.4 | 75.7/2.9/21.4 |
| Gentamicin | ≤1 | 2 | ≤1–>8 | 92.9/0.0/7.1 | 92.9/0.0/7.1 |
| Tetracycline | 2 | >32 | 0.5–>32 | 57.1/10.0/32.9 | –/–/– |
| Tigecyclineb | 0.12 | 1 | 0.03–2 | 100.0/0.0/0.0 | 95.7/4.3/0.0 |
Criteria according to the CLSI (15) and EUCAST (16). S, sensitive; I, intermediate; R, resistant. Dashes were used for categories if breakpoint criteria were not available.
U.S. FDA breakpoints were applied (21).
CTX-M-15-like-producing isolates do not include KPC- or NDM-1-producing isolates.
CTX-M-14-like-producing isolates do not include CTX-M-15-like-producing isolates.
SHV ESBL-producing isolates do not include CTX-M-15-like-, CTX-M-14-like-, KPC-, or NDM-1-producing isolates.
CMY-2-like-producing isolates do not include CTX-M-15-like-, CTX-M-14-like-, KPC-, or NDM-1-producing isolates.
Susceptibility profiles of ESBL-phenotype-positive isolates.
KPC producers were very resistant to all β-lactams tested, with the exception of ceftazidime-avibactam (MIC50/90, 0.5/2 μg/ml) (Table 2). Among other antimicrobial classes, only tigecycline (a glycylcycline) exhibited good activity against these strains (MIC50/90, 0.5/1 μg/ml) (Table 2).
Isolates harboring blaCTX-M-15-like genes that did not carry carbapenemase-encoding genes displayed elevated MIC values for cephalosporins: 14.6 and 0.0% were susceptible to ceftazidime and ceftriaxone, respectively (CLSI criteria) (Table 2). Antimicrobial agents showing the greatest activity against these isolates were piperacillin-tazobactam (67.2% susceptible), tigecycline (98.3%), and meropenem (99.0%). All CTX-M-15-like-producing isolates had ceftazidime-avibactam (MIC50/90, 0.12/0.5 μg/ml) MIC values at ≤2 μg/ml.
Meropenem (MIC50/90, ≤0.06/0.12 μg/ml), ceftazidime-avibactam (MIC50/90, 0.25/1 μg/ml), and tigecycline (MIC50/90, 0.5/1 μg/ml) (Table 2) were the most active agents tested against isolates carrying SHV ESBL without the presence of carbapenemases. Applying CLSI breakpoint criteria, ceftazidime and piperacillin-tazobactam displayed acceptable activity against CTX-M-14-like-carrying isolates, with 74.3 and 92.9% of isolates categorized as being susceptible to these agents, respectively. Meropenem (MIC50/90, ≤0.06/≤0.06 μg/ml), ceftazidime-avibactam (MIC50/90, 0.12/0.25 μg/ml), and tigecycline (MIC50/90, 0.12/1 μg/ml) (Table 2) were the most active agents tested against these strains.
Isolates that were positive for CMY-2-like-encoding genes but not carrying carbapenemases showed slightly lower MIC values against most β-lactams than other groups; however, resistance rates were still elevated for cephalosporins (63.0 to 100.0%) (Table 2). Ceftazidime-avibactam (MIC50/90, 0.06/0.12 μg/ml) was very active against these isolates.
NDM-1 producers were highly resistant to all agents tested, including ceftazidime-avibactam (MIC, >32 μg/ml), but were susceptible to tigecycline (MIC values of 0.5 μg/ml for both strains [data not shown]).
As recently demonstrated by our group and others (3, 9), CTX-M-15-like-producing isolates followed by organisms carrying genes encoding KPC and SHV ESBL were very prevalent among ESBL-phenotype-positive E. coli, Klebsiella species, and P. mirabilis isolates collected during 2012 in U.S. hospitals. Additionally, a very large proportion (62.9%) of the isolates carried multiple β-lactamases in combinations involving all genes/families detected by the microarray assay utilized. These isolates represented all bacterial species assessed and were detected in all U.S. census regions, but despite the β-lactamase enzymes or combinations of enzymes produced, ceftazidime-avibactam displayed potent activity against β-lactamase-producing isolates (except those harboring NDM-1).
Variations were observed in the prevalence of ESBL-phenotype-positive strains and β-lactamase production among isolates collected from monitored U.S. census regions. To our knowledge, this is the first comprehensive study to characterize β-lactamases for a large collection of U.S. Enterobacteriaceae, allowing the analysis of isolates stratified by geographic area. An understanding of the regional epidemiology of β-lactamases seems prudent to guide hospital and public health policies to minimize the further dissemination of these resistance genes and also to establish antimicrobial stewardship guidelines.
ACKNOWLEDGMENTS
We express our appreciation to the JMI staff members for scientific and technical assistance in performing this study.
This study was supported by Cerexa, Inc., a wholly owned subsidiary of Forest Laboratories, Inc. Scientific Therapeutics Information, Inc., provided editorial coordination, which was funded by Forest Research Institute, Inc.
Forest Laboratories, Inc., was involved in the study design, interpretation of data, and decision to present these results. Forest Laboratories, Inc., had no involvement in the collection and analysis of data. JMI Laboratories, Inc., has received research and educational grants in 2011 to 2013 from Aires; the American Proficiency Institute (API); Anacor; Astellas; AstraZeneca; Bayer; bioMérieux; Cempra; Cerexa; Contrafect; Cubist; Dipexium; Furiex; GlaxoSmithKline; Johnson & Johnson; LegoChem Biosciences, Inc.; Meiji Seika Kaisha; Merck; Nabriva; Novartis; Pfizer; PPD Therapeutics; Premier Research Group; Rempex; Rib-X Pharmaceuticals; Seachaid; Shionogi; The Medicines Co.; Theravance; and ThermoFisher Scientific. Some JMI employees are advisors/consultants for Astellas, Cubist, Pfizer, Cempra, Cerexa-Forest, Johnson & Johnson, and Theravance. In regard to speakers' bureaus and stock options, we have none to declare.
Footnotes
Published ahead of print 18 November 2013
REFERENCES
- 1.Bush K. 2008. Extended-spectrum beta-lactamases in North America, 1987-2006. Clin. Microbiol. Infect. 14(Suppl 1):134–143. 10.1111/j.1469-0691.2007.01848.x [DOI] [PubMed] [Google Scholar]
- 2.Lewis JS, II, Herrera M, Wickes B, Patterson JE, Jorgensen JH. 2007. First report of the emergence of CTX-M-type extended-spectrum beta-lactamases (ESBLs) as the predominant ESBL isolated in a U.S. health care system. Antimicrob. Agents Chemother. 51:4015–4021. 10.1128/AAC.00576-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castanheira M, Mendes RE, Rhomberg PR, Jones RN. 2008. Rapid emergence of blaCTX-M among Enterobacteriaceae in U.S. medical centers: molecular evaluation from the MYSTIC Program (2007). Microb. Drug Resist. 14:211–216. 10.1089/mdr.2008.0827 [DOI] [PubMed] [Google Scholar]
- 4.Castanheira M, Deshpande LM, Mendes RE, Rodriguez-Noriega E, Jones RN, Morfin-Otero R. 2011. Comment on: role of changes in the L3 loop of the active site in the evolution of enzymatic activity of VIM-type metallo-beta-lactamases. J. Antimicrob. Chemother. 66:684–685. 10.1093/jac/dkq393 [DOI] [PubMed] [Google Scholar]
- 5.Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin. Infect. Dis. 53:60–67. 10.1093/cid/cir202 [DOI] [PubMed] [Google Scholar]
- 6.Landman D, Babu E, Shah N, Kelly P, Olawole O, Backer M, Bratu S, Quale J. 2012. Transmission of carbapenem-resistant pathogens in New York City hospitals: progress and frustration. J. Antimicrob. Chemother. 67:1427–1431. 10.1093/jac/dks063 [DOI] [PubMed] [Google Scholar]
- 7.Castanheira M, Farrell SE, Wanger A, Rolston KV, Jones RN, Mendes RE. 2013. Rapid expansion of KPC-2-producing Klebsiella pneumoniae isolates in two Texas hospitals due to clonal spread of ST258 and ST307 lineages. Microb. Drug Resist. 19:295–297. 10.1089/mdr.2012.0238 [DOI] [PubMed] [Google Scholar]
- 8.Flamm RK, Sader HS, Farrell DJ, Jones RN. 2012. Ceftaroline potency among 9 US census regions: report from the 2010 AWARE program. Clin. Infect. Dis. 55(Suppl 3):S194–S205. 10.1093/cid/cis562 [DOI] [PubMed] [Google Scholar]
- 9.Castanheira M, Farrell SE, Deshpande LM, Mendes RE, Jones RN. 2013. Prevalence of β-lactamase-encoding genes among Enterobacteriaceae bacteremia isolates collected in 26 U.S. hospitals: report from the SENTRY Antimicrobial Surveillance Program (2010). Antimicrob. Agents Chemother. 57:3012–3020. 10.1128/AAC.02252-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogdanovich T, Adams-Haduch JM, Tian GB, Nguyen MH, Kwak EJ, Muto CA, Doi Y. 2011. Colistin-resistant, Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae belonging to the international epidemic clone ST258. Clin. Infect. Dis. 53:373-376. 10.1093/cid/cir401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spanu T, De Angelis G, Cipriani M, Pedruzzi B, D'Inzeo T, Cataldo MA, Sganga G, Tacconelli E. 2012. In vivo emergence of tigecycline resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli. Antimicrob. Agents Chemother. 56:4516–4518. 10.1128/AAC.00234-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stachyra T, Levasseur P, Pechereau MC, Girard AM, Claudon M, Miossec C, Black MT. 2009. In vitro activity of the β-lactamase inhibitor NXL104 against KPC-2 carbapenemase and Enterobacteriaceae expressing KPC carbapenemases. J. Antimicrob. Chemother. 64:326–329. 10.1093/jac/dkp197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castanheira M, Sader HS, Farrell DJ, Mendes RE, Jones RN. 2012. Activity of ceftaroline-avibactam tested against Gram-negative organism populations, including strains expressing one or more beta-lactamases and methicillin-resistant Staphylococcus aureus carrying various staphylococcal cassette chromosome mec types. Antimicrob. Agents Chemother. 56:4779–4785. 10.1128/AAC.00817-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute 2012. M07-A9. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 15.Clinical and Laboratory Standards Institute 2013. M100-S23. Performance standards for antimicrobial susceptibility testing: 23rd informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 16.EUCAST 2013. Breakpoint tables for interpretation of MICs and zone diameters, version 3.0, January 2013. http://www.eucast.org/clinical_breakpoints/ Accessed 2 January 2013 [Google Scholar]
- 17.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 51:286–294. 10.1086/653932 [DOI] [PubMed] [Google Scholar]
- 18.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151–1161. 10.1128/AAC.45.4.1151-1161.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castanheira M, Mendes RE, Woosley LN, Jones RN. 2011. Trends in carbapenemase-producing Escherichia coli and Klebsiella spp. from Europe and the Americas: report from the SENTRY Antimicrobial Surveillance Programme (2007-09). J. Antimicrob. Chemother. 66:1409–1411. 10.1093/jac/dkr081 [DOI] [PubMed] [Google Scholar]
- 20.Pisney L, Barron M, Janelle S, Bamberg W, MacCannell D, Kallen A, Gould C, Limbago B, Epson E, Wendt J. 2013. Notes from the field: hospital outbreak of carbapenem-resistant Klebsiella pneumoniae producing New Delhi metallo-beta-lactamase—Denver, Colorado, 2012. MMWR Morb. Mortal. Wkly. Rep. 62:108 http://www.cdc.gov/mmwr/preview/mmwrhtml/mm6206a5.htm [PMC free article] [PubMed] [Google Scholar]
- 21.Pfizer, Inc 2012. Tygacil package insert. Pfizer, Inc, New York, NY: http://www.tygacil.com Accessed 2 January 2013 [Google Scholar]