Abstract
Nontypeable Haemophilus influenzae (NTHi)-associated disease is a major health problem globally. Whole-genome sequence analysis identified the absence of hpd genes encoding Haemophilus protein D in 3 of 16 phylogenetically distinct NTHi isolates. This novel finding is of potential clinical significance, as protein D and hpd represent important NTHi vaccine antigen and diagnostic targets, respectively.
TEXT
Nontypeable Haemophilus influenzae (NTHi)-associated disease represents a major health burden for young children worldwide and, also, for adults with chronic pulmonary disorders. The most commonly reported manifestations relate to chronic middle ear and lung disease, with particularly high rates among Australian Aboriginal children (1, 2). An effective vaccine against NTHi is required, but its development remains a challenge. Nevertheless, an 11-valent prototype pneumococcal-H. influenzae protein D conjugated vaccine (PHiD-CV11), relying upon the conserved outer membrane lipoprotein, protein D, as its sole Haemophilus-specific antigen, reduced NTHi-associated acute otitis media by 35% (3). The effect of its successor, PHiD-CV10, on NTHi carriage and NTHi-associated middle ear and chronic lung disease (4), however, is still under investigation in several different populations. Interestingly, the data published so far indicate either no or, at best, an inconsistent effect upon nasopharyngeal NTHi colonization in children up to 2 years of age (5, 6).
H. influenzae protein D is important for several reasons. First, as described above, it is a potential vaccine antigen. Second, the gene encoding protein D, hpd, is a diagnostic target for differentiating NTHi from phenotypically indistinguishable nonhemolytic strains of the respiratory commensal H. haemolyticus (which only rarely acts as a pathogen) (7). Differentiating between these bacteria is important, as it will help to reduce unnecessary antibiotic prescribing for misidentified H. influenzae disease. Consequently, since standard microbiological testing cannot distinguish the two species, complementary genotypic testing is necessary. Compared to other methods, a PCR targeting the hpd gene provided superior discrimination between H. influenzae and H. haemolyticus. However, the species cutoff was selected arbitrarily (7), and a more meaningful diagnostic is likely required.
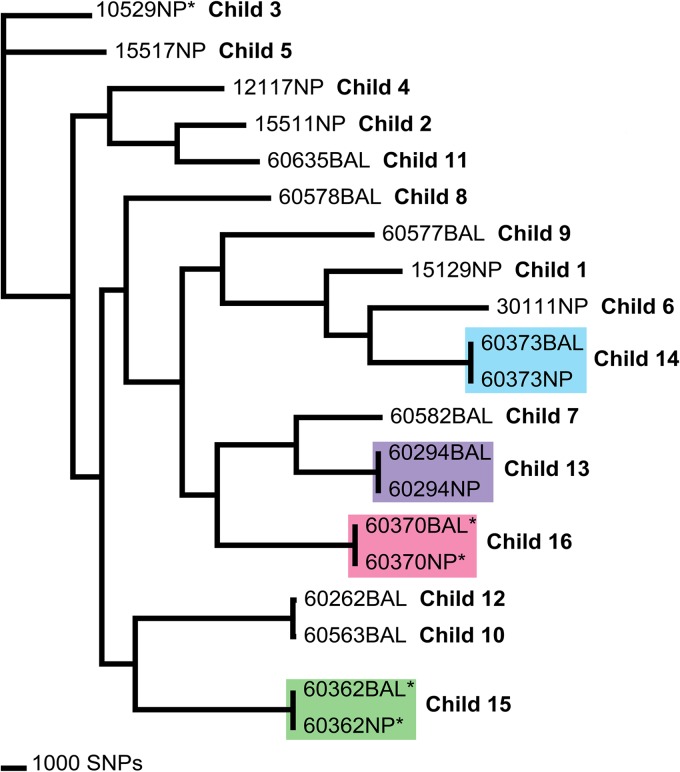
As part of a larger study to investigate the relatedness of common Australian pediatric carriage genotypes and those associated with lower airway infection in Australian Aboriginal children with non-cystic fibrosis bronchiectasis (Human Research Ethics Committee of the Northern Territory Department of Health and Menzies School of Health Research, approvals 07/63 and 07/85), we performed whole-genome sequencing of 20 Haemophilus isolates. These were nasal or nasopharyngeal carriage isolates representing common NTHi genotypes (based on PCR-ribotyping [8] of over 3,000 isolates) from our previous carriage studies of Northern Territory and Western Australian children. Additionally, NTHi and presumptive H. haemolyticus isolates from nasopharyngeal swabs and bronchoalveolar lavage from children with bronchiectasis were selected from children with high airway neutrophilia and clinically significant NTHi infection (>104 CFU/ml) in the lower airways (1). A hybrid de novo assembly using data from the Ion Torrent and Illumina sequencing platforms was undertaken on one nasopharyngeal NTHi isolate, 60294NP, and subsequently used as a reference for 19 additional genomes generated using Ion Torrent. A neighbor-joining maximum likelihood tree was constructed using 50,172 orthologous single-nucleotide polymorphisms (SNPs) across the 20 genomes (Fig. 1). This method provides a robust measure for strain relatedness for highly recombinogenic bacteria, such as NTHi (9).
FIG 1.
Genomic analysis of 20 nontypeable Haemophilus influenzae isolates from 16 Australian children. The neighbor-joining maximum likelihood tree was constructed using 50,172 orthologous SNPs across the 20 genomes (consistency index, 0.46). Isolates marked with an asterisk lack the hpd gene. Isolates in colored boxes were obtained from a single child. BAL, bronchoalveolar lavage; NP, nasal/nasopharyngeal.
The sequenced isolates are listed in Table 1. For the children assigned identifications (ID) 1 to 5, nasal NTHi isolates representing five common genotypes were sequenced. One PCR-ribotype each was performed for isolates from an Aboriginal child (ID 3) and four non-Aboriginal children attending three different child care centers in 2001 in Darwin, Australia (10). The nasopharyngeal NTHi isolate representing another common genotype is from an Aboriginal child (ID 6) who was enrolled in a randomized controlled trial of antibiotics for acute otitis media in 2003 (11).
TABLE 1.
Details on the 20 analyzed NTHi isolates from 16 Australian children
| Child ID | Age (yr) | Site of isolationa | Yr of isolation | Clinical status | No. of doses of PHiD-CV10 | hpd gene |
|---|---|---|---|---|---|---|
| 1 | 1.6 | Nasal cavity | 2001 | Asymptomatic carriage | 0 | + |
| 2 | 4.0 | Nasal cavity | 2001 | Otitis media with effusion | 0 | + |
| 3 | 1.4 | Nasal cavity | 2001 | Asymptomatic carriage | 0 | − |
| 4 | 2.5 | Nasal cavity | 2001 | Asymptomatic carriage | 0 | + |
| 5 | 4.4 | Nasal cavity | 2001 | Asymptomatic carriage | 0 | + |
| 6 | 1.5 | Nasopharynx | 2003 | Acute otitis media | 0 | + |
| 7 | 2.5 | BAL fluid | 2009 | Bronchiectasis | 0 | + |
| 8 | 4.1 | BAL fluid | 2009 | Bronchiectasis | 0 | + |
| 9 | 6.8 | BAL fluid | 2009 | Bronchiectasis | 0 | + |
| 10 | 0.4 | BAL fluid | 2010 | Bronchiectasis | 2 | + |
| 11 | 1.7 | BAL fluid | 2010 | Bronchiectasis | 1 | + |
| 12 | 1.8 | BAL fluid | 2008 | Bronchiectasis | 0 | + |
| 13 | 4.2 | BAL fluid | 2008 | Bronchiectasis | 0 | + |
| 13 | 4.2 | Nasopharynx | 2008 | Bronchiectasis | 0 | + |
| 14 | 2.0 | BAL fluid | 2009 | Bronchiectasis | 0 | + |
| 14 | 2.0 | Nasopharynx | 2009 | Bronchiectasis | 0 | + |
| 15 | 1.1 | BAL fluid | 2009 | Bronchiectasis | 0 | − |
| 15 | 1.1 | Nasopharynx | 2009 | Bronchiectasis | 0 | − |
| 16 | 2.1 | BAL fluid | 2009 | Bronchiectasis | 0 | − |
| 16 | 2.1 | Nasopharynx | 2009 | Bronchiectasis | 0 | − |
BAL, bronchoalveolar lavage.
The children assigned ID 7 to 16 were Aboriginal children who underwent bronchoscopy at Royal Darwin Hospital between 2008 and 2010 (1), and they provided 14 Haemophilus isolates, including NTHi isolates from bronchoalveolar lavage (BAL) specimens (ID 7 to 12), two NTHi nasopharyngeal isolates with contemporaneously paired BAL fluid isolates (ID 13 and 14), and similarly, two pairs of BAL fluid and nasopharyngeal isolates identified as presumptive H. haemolyticus based on an hpd-targeted PCR assay (ID 15 and 16). The Haemophilus isolates were identified by colony morphology and X and V factor dependence, and all failed to react with capsular antisera using the Phadebact Haemophilus coagglutination test. Although concurrent colonization/infection with multiple genotypes is common in Aboriginal (8) and non-Aboriginal Australian children, only one colony from each specimen was sequenced in this project.
The two pairs of BAL fluid and nasopharyngeal isolates identified as presumptive H. haemolyticus clustered with NTHi using an orthologous SNP phylogeny (Fig. 1, ID 15 and 16) and were found to have been misidentified due to the absence of the hpd gene. Screening of the remaining genomes revealed that a further nasal NTHi isolate lacked hpd (Fig. 1, ID 3). These findings confirmed negative PCR results using hpd#3 primers (7). Overall, 5 of 20 NTHi genomes from 3 of 16 unrelated children lacked hpd. Furthermore, the orthologous SNP phylogeny demonstrated that the three independent isolates were phylogenetically unrelated (Fig. 1) and, thus, did not represent expansion of a single hpd-negative clone. The hpd-negative isolates were detected in 2001 and 2009, and none were detected in children who had received PHiD-CV10 (Table 1).
To our knowledge, this is the first report on the absence of hpd in clinical NTHi strains. Despite involving only a small number of isolates, our findings have implications for hpd-based diagnostic tests currently recommended for differentiating between NTHi and H. haemolyticus. Our findings also identify a potential limitation in the efficacy of PHiD-CV11 (3) and PHiD-CV10 (5) against NTHi-related carriage and disease. They highlight the challenges inherent in developing single-target NTHi diagnostics and vaccines, given the high degree of heterogeneity in the NTHi population. Screening NTHi populations for hpd in diverse patient groupings and geographic settings will provide information on whether alternative NTHi diagnostic tests are needed to predict the likely effectiveness of PHiD-CV10 for NTHi-related disease. The significance of the absence of hpd for NTHi pathogenicity and host immune responses also remains to be discovered.
ACKNOWLEDGMENTS
This work was supported by Channel 7 Children's Research Foundation (grant number 13699) and the Australian National Health and Medical Research Council (grants 1023781, 1040830, and 1024175 to H.C.S.-V., 1020561 to A.J.L., 1034703 to R.L.M., and 545216 to A.B.C.).
We thank the families who participated in these studies. We also thank the Menzies Ear Health Research Team, Respiratory Team, and Child Health Laboratory Team, particularly Gabrielle McCallum, Jemima Beissbarth, Kim Hare, and Elizabeth Nosworthy, for clinical swabs, clinical data, and laboratory support.
Footnotes
Published ahead of print 27 November 2013
REFERENCES
- 1.Hare KM, Grimwood K, Leach AJ, Smith-Vaughan H, Torzillo PJ, Morris PS, Chang AB. 2010. Respiratory bacterial pathogens in the nasopharynx and lower airways of Australian indigenous children with bronchiectasis. J. Pediatr. 157:1001–1005. 10.1016/j.jpeds.2010.06.002 [DOI] [PubMed] [Google Scholar]
- 2.Leach A, Wood Y, Gadil E, Stubbs E, Morris P. 2008. Topical ciprofloxin versus topical framycetin-gramicidin-dexamethasone in Australian aboriginal children with recently treated chronic suppurative otitis media: a randomized controlled trial. Pediatr. Infect. Dis. J. 27:692–698. 10.1097/INF.0b013e31816fca9d [DOI] [PubMed] [Google Scholar]
- 3.Prymula R, Peeters P, Chrobok V, Kriz P, Novakova E, Kaliskova E, Kohl I, Lommel P, Poolman J, Prieels JP, Schuerman L. 2006. Pneumococcal capsular polysaccharides conjugated to protein D for prevention of acute otitis media caused by both Streptococcus pneumoniae and non-typable Haemophilus influenzae: a randomised double-blind efficacy study. Lancet 367:740–748. 10.1016/S0140-6736(06)68304-9 [DOI] [PubMed] [Google Scholar]
- 4.O'Grady KA, Grimwood K, Cripps A, Mulholland EK, Morris P, Torzillo PJ, Wood N, Smith-Vaughan H, Revell A, Wilson A, Van Asperen AP, Richmond P, Thornton R, Rablin S, Chang AB. 2013. Does a 10-valent pneumococcal-Haemophilus influenzae protein D conjugate vaccine prevent respiratory exacerbations in children with recurrent protracted bacterial bronchitis, chronic suppurative lung disease and bronchiectasis: protocol for a randomised controlled trial. Trials 14:282. 10.1186/1745-6215-14-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Bergh MR, Spijkerman J, Swinnen KM, Francois NA, Pascal TG, Borys D, Schuerman L, Ijzerman EP, Bruin JP, van der Ende A, Veenhoven RH, Sanders EA. 2013. Effects of the 10-valent pneumococcal nontypeable Haemophilus influenzae protein D-conjugate vaccine on nasopharyngeal bacterial colonization in young children: a randomized controlled trial. Clin. Infect. Dis. 56:e30–e39. 10.1093/cid/cis922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prymula R, Habib A, Francois N, Borys D, Schuerman L. 2013. Immunological memory and nasopharyngeal carriage in 4-year-old children previously primed and boosted with 10-valent pneumococcal non-typeable Haemophilus influenzae protein D conjugate vaccine (PHiD-CV) with or without concomitant prophylactic paracetamol. Vaccine 31:2080–2088. 10.1016/j.vaccine.2013.01.044 [DOI] [PubMed] [Google Scholar]
- 7.Binks MJ, Temple B, Kirkham LA, Wiertsema SP, Dunne EM, Richmond PC, Marsh RL, Leach AJ, Smith-Vaughan HC. 2012. Molecular surveillance of true nontypeable Haemophilus influenzae: an evaluation of PCR screening assays. PLoS One 7:e34083. 10.1371/journal.pone.0034083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith-Vaughan HC, McBroom J, Mathews JD. 2001. Modelling of endemic carriage of Haemophilus influenzae in Aboriginal infants in Northern Australia. FEMS Immunol. Med. Microbiol. 31:137–143. 10.1111/j.1574-695X.2001.tb00510.x [DOI] [PubMed] [Google Scholar]
- 9.Pearson T, Giffard P, Beckstrom-Sternberg S, Auerbach R, Hornstra H, Tuanyok A, Price EP, Glass MB, Leadem B, Beckstrom-Sternberg JS, Allan GJ, Foster JT, Wagner DM, Okinaka RT, Sim SH, Pearson O, Wu Z, Chang J, Kaul R, Hoffmaster AR, Brettin TA, Robison RA, Mayo M, Gee JE, Tan P, Currie BJ, Keim P. 2009. Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol. 7:78. 10.1186/1741-7007-7-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith-Vaughan HC, Byun R, Nadkarni M, Jacques NA, Hunter N, Halpin S, Morris PS, Leach AJ. 2006. Measuring nasal bacterial load and its association with otitis media. BMC Ear Nose Throat Disord. 6:10. 10.1186/1472-6815-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris PS, Gadil G, McCallum GB, Wilson CA, Smith-Vaughan HC, Torzillo P, Leach AJ. 2010. Single-dose azithromycin versus seven days of amoxycillin in the treatment of acute otitis media in Aboriginal children (AATAAC): a double blind, randomised controlled trial. Med. J. Aust. 192:24–29 [DOI] [PubMed] [Google Scholar]