Abstract
Since its first description, Chromera velia has attracted keen interest as the closest free-living relative of parasitic Apicomplexa. The life cycle of this unicellular alga is complex and involves a motile biflagellate form. Flagella are thought to be formed in the cytoplasm, a rare phenomenon shared with Plasmodium in which the canonical mode of flagellar assembly, intraflagellar transport, is dispensed with. Here we demonstrate the expression of intraflagellar transport components in C. velia, answering the question of whether this organism has the potential to assemble flagella via the canonical route. We have developed and characterized a culturing protocol that favors the generation of flagellate forms. From this, we have determined a marked shift in the mode of daughter cell production from two to four daughter cells per division as a function of time after passage. We conduct an ultrastructural examination of the C. velia flagellate form by using serial TEM and show that flagellar biogenesis in C. velia occurs prior to cytokinesis. We demonstrate a close association of the flagellar apparatus with a complex system of apical structures, including a micropore, a conoid, and a complex endomembrane system reminiscent of the apical complex of parasitic apicomplexans. Recent work has begun to elucidate the possible flagellar origins of the apical complex, and we show that in C. velia these structures are contemporaneous within a single cell and share multiple connections. We propose that C. velia therefore represents a vital piece in the puzzle of the origins of the apical complex.
INTRODUCTION
The eukaryotic flagellum is a microtubule-based organelle that protrudes from the cell surface and was a feature of the last ancestor common to all eukaryotes (1–4). The flagellum plays a vital role for many protists in at least one stage of their life cycles, serving specialized roles in processes such as cellular locomotion, dispersal, reproduction, cell division, and signal transduction (5–7).
Canonically, the flagellum is assembled outward from the cell surface within its own membrane-bound compartment. The flagellar building blocks are transported by specialized motor-driven protein complexes from the cytoplasm to the distal flagellar tip, where assembly occurs. This process of cargo delivery is known as intraflagellar transport (IFT) (8). The IFT system is composed of IFT particles (multiprotein complexes A and B), the heterotrimeric anterograde IFT motor kinesin II, and the retrograde IFT motor cytoplasmic dynein 2 (8, 9). IFT also plays a role in maintaining flagellar length by continually recycling the turnover products of the dynamic flagellar tip back to the cell body, coupled with the delivery of building blocks from the cytoplasm back to the assembly site (10–12). While the characterization of IFT was pioneered in the model alga Chlamydomonas reinhardtii, this system is now known to be widely conserved across the majority of flagellate eukaryotes, with IFT protein orthologues detected in organisms ranging from protists to metazoans (1, 13).
Apicomplexan parasites are significant pathogens, causing huge health and economic burdens. Malaria, caused by Plasmodium spp., infects over 250 million people annually (14). Several other apicomplexan parasites are also important in terms of veterinary medicine and agriculture. One of the defining features of the Apicomplexa is the apical complex, which plays a role in host cell invasion (15). The apical complex of all apicomplexans contains an apical polar ring that serves as an organizing center for the subpellicular microtubules. Rhoptries and micronemes—secretory organelles that deliver the enzymes necessary for host cell invasion—extend from the apical polar ring into the cell body (16, 17). In some apicomplexans (for example, Toxoplasma), a conoid—a cone-shaped arrangement of tubules made up of unusual polymers of tubulin (18)—extends from the apical polar ring into the extreme anterior (apical) end of the cell. The conoid is topped by preconoidal rings, and a pair of short intraconoidal microtubules traverses the conoidal lumen.
The mode of flagellar biogenesis among the various lineages of the Apicomplexa is something of a mixed bag (13, 19–22). There are several independent losses of flagella and seemingly at least two independent adoptions of a wholly cytoplasmic approach to flagellar biogenesis. In all lineages where the flagellum is retained, it is restricted to male microgametes (22, 23). Plasmodium, which produces uniflagellate microgametes, has dispensed with IFT completely and genes encoding IFT-related proteins are entirely absent from the genome (13). Instead, axonemes are constructed at an exceptional speed completely within the cytoplasm of the gametocyte and emerge as a complete structure during exflagellation (22). The mechanisms involved in the construction and patterning of axonemes in this way are unknown. The Coccidia, such as Toxoplasma spp. and Eimeria spp., employ compartmentalized (external) flagellum assembly and possess genes encoding IFT-related proteins (24), albeit with some surprising absences, such as the retrograde dynein motor from Toxoplasma (13, 25).
Insights into flagellum biogenesis in apicomplexans may be provided by Chromera velia, a recently discovered sister of the Apicomplexa (26). C. velia is a photosynthetic microalga that was isolated from Australian hard corals and shows a range of genetic and morphological similarities to Apicomplexa (26–28). The life cycle of C. velia alternates between nonflagellate coccoid cells and motile biflagellate cells (26, 28, 29). The two flagella emerge from the anterior end of the cell, with the shorter (anterior) flagellum located to the anterior part of the longer (posterior) flagellum. In culture, the emergence of flagellate forms occurs with a predictable diurnal rhythm across a 24-h light-dark cycle. This emergence is also influenced by light intensity, salinity, and nutrient levels (28, 30). In an initial ultrastructural analysis of C. velia, internal, non-membrane-bound axonemes were noted (26). By analogy to its malaria-causing relative Plasmodium, it was proposed that C. velia assembles its axoneme completely within the cytoplasm, most likely via an IFT-independent mechanism (26, 28).
Here we present molecular and morphological evidence of the existence of an IFT system in C. velia. By mining C. velia expressed sequence tags (ESTs), we have identified orthologues of several IFT- and flagellum-associated components. Further, we show that flagellate forms arise through a cytokinetic process and not by the transformation of individual cells. Finally, we conduct an ultrastructural examination of the flagellar apparatus of C. velia. We provide new evidence of the presence of an apical complex in C. velia and demonstrate its intimate association with components of the flagellar apparatus, providing a novel potential avenue for investigation of the evolution, organization, and biogenesis of this iconic and important structure.
MATERIALS AND METHODS
Culture.
C. velia isolated from the stony coral Leptastrea purpurea from One Tree Island Great Barrier Reef, Queensland, Australia, was used throughout this study (26). The original isolate was subcultured in 2008 and maintained as a unicellular culture, CvLp_vc08/1; it is available from the us upon request. Cells were cultured in f/2 medium under previously described temperature and light conditions (29). To promote the emergence of flagellate forms, cultures were maintained at cell densities between 1 × 106 and 5 × 106 cells ml−1.
Sequence analysis.
Public databases were queried (BLAST) for known flagellar proteins (GenBank, www.ncbi.nlm.nih.gov/GenBank/; Swiss-Prot, http://www.expasy.org/; ToxoDB, www.toxodb.org). Putative homologues of C. velia ESTs were retrieved together with known representatives, including C. reinhardtii and Toxoplasma gondii. BLAST results were confirmed by reciprocal BLAST, multiple-sequence alignments, gene ontology, and protein domain searches in CLC Main Workbench 6.7.1 (CLC bio, Aarhus, Denmark).
RNA isolation and RT-PCR.
Total RNA was isolated from cultures containing flagellate cells with the FastRNA Pro Green kit (MP Biomedicals, Solon, OH). DNase-treated RNA was checked for absence of genomic DNA contamination via PCR as a control without reverse transcription (RT). cDNA was synthesized with the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, Australia) primed with random hexamers. Gene-specific PCR primer pairs targeting IFT and flagellar genes were designed in CLC Main Workbench 6.7.1 (CLC bio) on the basis of EST sequences (see Table S1 in the supplemental material). Template cDNA was PCR amplified with each primer pair with an annealing temperature of 52°C. Each set of PCR mixtures included a no-template negative control, and primers were tested for the ability to amplify a template genomic DNA. PCR amplicons were visualized by agarose gel electrophoresis and directly sequenced at Macrogen Ltd. (Seoul, South Korea).
Live-cell imaging.
C. velia cultures were transferred to six-well plates and continuously monitored with either a Nikon Eclipse TiE (Nikon) or an Olympus CKX41 (Olympus). Images were captured at 10-s intervals and analyzed in ImageJ.
TEM.
For transmission electron microscopy (TEM), samples were fixed by high-pressure freezing in a Leica EMPACT2 HPF (2,050 × 105 to 2,065 × 105 Pa); frozen pellets were transferred under liquid nitrogen to a Leica EM AFS2, where they were substituted in 1% OsO4 in anhydrous acetone at −90°C for 96 h, and then heated at 3° per h to room temperature over a 36-h period. Samples were infiltrated at room temperature in an ascending series of Spurr's resin (soft formulation) over 2 days and polymerized overnight at 60°C. Serial thin (∼70-nm) sections were cut (Leica Ultracut UC7), mounted on Formvar-coated slot grids, and stained (uranyl acetate and lead citrate). Sections were examined on a JEOL 1400 (120 kV), and images were acquired with an UltraScan charge-coupled device camera. Image processing was performed in ImageJ v1.47 (rsbweb.nih.gov/ij/).
Tomography.
Samples for tomography were prepared and sectioned the same way as for TEM. Two-hundred-nm sections were examined on a JEOL 2100 (200 kV). Images were captured for sequential 2° tilts between 60° and −60° with SerialEM software (31). Tomograms were calculated and analyzed in IMOD (32).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in GenBank under accession no. KC590349 to KC590354.
RESULTS
Live-cell time-lapse imaging of C. velia captures rapid formation of two and four daughter progeny with flagella.
In order to examine flagellar biogenesis in C. velia, we first developed a protocol to improve both the magnitude and frequency of the periodic appearance of the motile flagellate form.
By maintaining cultures at densities between 1 × 106 and 5 × 106 cells ml−1 (which in our experiments necessitated dilution every 3 to 4 days) at room temperature under light and medium conditions as previously described (29), we were able to eliminate the previously reported lag time in the appearance of flagellate cells. The production of flagellate forms occurred daily, beginning around 2 h after first exposure to light and persisting throughout the light cycle (since this was an investigation only of the presence of flagellate cells in culture, we did not investigate the fate or longevity of individual cells). Previous studies have involved weekly or biweekly dilution of cultures to a low density (∼1 × 105 cpm) and incubation under a 12-h light–12-h dark regimen (28). This results in the appearance of flagellate forms after 2 to 3 days and the continued production of flagellate forms for up to a week. Flagellate forms appear to be produced in response to light, and various other factors, such as medium nutrient levels and salinity, have also been implicated in this process (30).
We made time-lapse movies of live cells under our modified growth conditions and recognized two cyst-like forms that we named types II and IV; the former releases two daughter cells, and the latter releases four (Fig. 1; see Movie S1 in the supplemental material). These cyst types can be further divided into those producing flagellate progeny and those producing nonflagellate progeny, i.e., types IIF, IVF, IIN, and IVN. We did not observe any cases of mixed (flagellate and nonflagellate) progeny emerging from a single cyst. The production of flagellate forms in all cases occurred through the production of multiple daughter cells from a cyst and not by the transformation of a single cell from nonflagellate to flagellate.
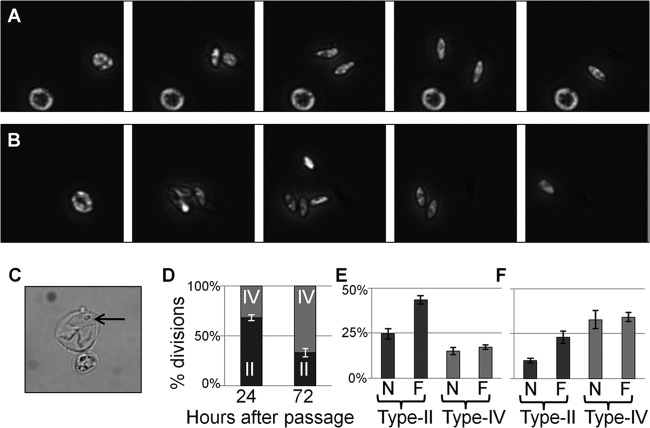
FIG 1.
Dark-field imaging of live, dividing C. velia cells. (A, B) Stills taken from Movie S1 in the supplemental material showing type IIF (A) and IVF (B) division. Daughter cells show the distinctive bullet-shaped morphology of C. velia flagellate cells and rapidly swim out of the field of view. Each series represents a 350-s time period. (C) An empty cyst shell and residuum (arrow) following the emergence of daughter cells. (D) Proportion of division events producing two or four daughter cells 24 and 72 h after passage. (E, F) Proportions of division events broken down by type at 24 h (E) and 72 h (F) after passage. n = 3; error bars represent standard deviations.
Immediately prior to the emergence of flagellate daughter progeny, rapid movement occurs within the cyst for a period of around 60 s (see Movie S1 in the supplemental material). Following the emergence of daughter cells, a cyst wall-like shell remains that often contains a small spherical structure reminiscent of the residuum described previously (Fig. 1C) (28).
We assessed the mode of daughter cell production across a field of view for 100 min beginning 2 h after the cells were first exposed to light (Fig. 1D to F). Three independent observations were made of cultures at both 24 and 72 h after passage. The following observations were of a single representative replicate. At 24 h, in 1,013 cells examined, 61 division events were observed during the monitoring period. Most of the daughter cell production was via the type II pathway, accounting for 68.9% of the division events. Type IIF cysts were almost twice as common as type IIN cysts (45.9% of the divisions, compared to 23.0%). For type IV cysts, the numbers of F and N forms were approximately equal (14.8 and 16.4%, respectively). At 72 h after passage, 2,130 cells were examined (the cell number in culture had doubled in the intervening period) and 59 division events occurred within the monitoring period. In an almost complete reversal of the situation at 24 h, 71.2% of the division events observed were via the type IV pathway. The frequency of F and N forms of both cyst types was similar to that observed at 24 h. Type IIF cysts were twice as frequent as type IIN cysts (20.3% of the division events, compared to 8.5%), while type IVF and IVN cysts occurred with the same frequency (each accounted for 35.6% of the division events).
C. velia expresses IFT-like and flagellum-associated genes.
We aligned C. reinhardtii flagellar proteome sequences (33) limited to those identified by five or more unique peptides (n = 361) with C. velia EST sequences (34, 35) by tBLASTn. We detected 124 candidate flagellum-related transcripts with BLAST E values below our cutoff (10−10) (see Table S2 in the supplemental material). Within these matches were transcripts putatively encoding proteins identified by homology as tubulins, inner and outer dynein arm proteins, radial spoke proteins, and IFT components.
The IFT system comprises anterograde motors (kinesin II), retrograde motors (cytoplasmic dynein 2), two core complexes (A and B), and several accessory proteins. We were able to identify BLAST hits with E values of 10−10 or less for a putative kinesin II motor (JO793544) and a putative cytoplasmic dynein chain (JO804161). IFT complex A was represented by a putative match to IFT139, and complex B was represented by matches to IFT20, IFT22, IFT25, IFT27, IFT70, IFT80, IFT81, IFT88, and IFT172 (Table 1). Putative homologues to the IFT accessory proteins TLP1 and DYF13 were also noted (Table 1).
TABLE 1.
Identification of putative IFT particle- and flagellum-associated protein components in C. velia
| Name |
C. reinhardtii accession no. |
C. velia |
T. gondii |
|||
|---|---|---|---|---|---|---|
| GenBank | Swiss-Prot | Accession no.a | E value | Accession no.b | E value | |
| IFT20 | XP_001701966 | Q8LLV9 | HO865979 | 8.00E-04 | —d | |
| JO809038 | 2.00E-02 | |||||
| IFT22 | XP_001689669 | A8HME3 | Not found | — | ||
| JO792196 | 9.00E-14 | |||||
| IFT25 | ABU90455 | B8LIX8 | Not found | — | ||
| JO793390 | 6.00E-08 | |||||
| IFT27 | XP_001689745 | A8HN58 | Not found | — | ||
| JO811700 | 4.00E-30 | |||||
| IFT70 | XP_001692406 | A8ITN7 | Not found | TGME49_283500 | 2.00E-96 | |
| JO798397 | 6.00E-47 | |||||
| IFT80 | XP_001693341 | A8IXE2 | HO866580 | 2.00E-57 | TGME49_293890 | Annotatede |
| JO804036 | 9.00E-29 | |||||
| IFT81 | XP_001697224 | Q68RJ5 | Not found | TGME49_305570 | Annotated | |
| JO797612 | 2.00E-22 | |||||
| IFT88 | XP_001700100 | A8JCJ2 | HO865102 | 1.00E-61 | TGME49_207410 | 9.00E-81 |
| JO806909 | 1.00E-02 | |||||
| IFT172 | XP_001691740 | IF172 | Not found | TGME49_288050 | Annotated | |
| JO793301 | 1.00E-35 | |||||
| IFT139 | ABU95018 | A9XPA6 | Not found | — | ||
| JO800827 | 1.00E-24 | |||||
| TLP1 | XP_001697058 | A8J5Y5 | Not found | — | ||
| JO794992 | 2.00E-34 | |||||
| DYF13 | XP_001698769 | A8JA42 | HO865228 | 5.00E-86 | TGME49_255500 | 8.00E-68 |
| JO810674 | 2.00E-166 | |||||
| RSP9 | XP_001690441 | Q27YU5 | HO865818 | 2.00E-03 | TGME49_255200 | Annotated |
| JO797974 | 3.00E-01 | |||||
| Tctex1c | XP_001702138 | O64980 | HO866591 | 7.00E-26 | TGME49_247560 | 4.00E-22 |
| JO808746 | 3.00E-26 | |||||
Accession numbers are from ToxoDB 8.2.
Flagellar inner-arm dynein light-chain TcTex1.
—, no homolog annotated in database and no homologous hits supported by our analysis.
Annotated, the identity of the T. gondii protein homolog is annotated in the database.
In cultures producing flagellate forms, expression at the transcriptional level was investigated for a subset of intraflagellar transport proteins (IFT80, IFT88, IFT20, DYF13) and two flagellar proteins, radial spoke RSP9 and inner arm dynein Tctex1. Gene-specific primers were designed from EST data and in each case were able to amplify cDNA generated from total RNA, producing amplicons of the expected sizes (Fig. 2), and identity was confirmed by sequencing (KC590349 to KC590354). The housekeeping gene (for glyceraldehyde 3-phosphate dehydrogenase [GAPDH]) served as a control. Only primer pairs for IFT80 and TcTex1 resulted in the amplification of template genomic DNA; the others failed possibly because one or both of the EST-derived primers spanned exon-exon boundaries. Sequencing of the IFT80 amplicon from template genomic DNA confirmed exon identity and revealed a single intron (KC590353).
FIG 2.
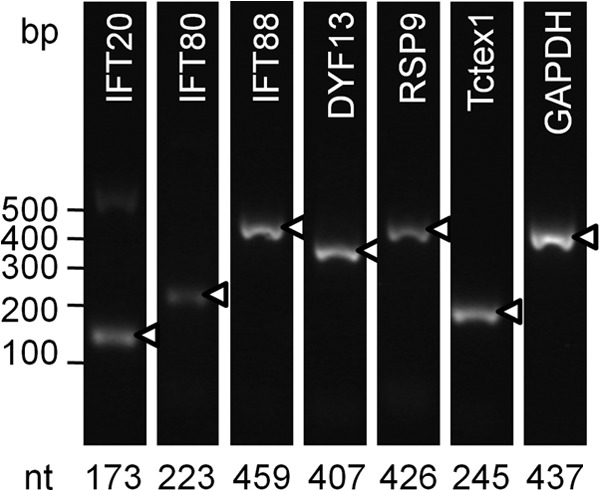
C. velia contains and expresses IFT- and flagellum-related genes. Equal amounts of cDNA were used for PCR amplification with each primer pair. The housekeeping gene for GAPDH acted as a nonflagellar control. The expected size of the product is indicated below each lane, and marked amplicons (triangles) were confirmed by DNA sequencing.
Flagellar biogenesis in C. velia occurs prior to cytokinesis.
There is an apparent incongruity between our observations of an apparently extremely rapid emergence of flagellate forms from the mother cyst, coupled with previous descriptions of intracytoplasmic axonemes (28), and our discovery of IFT-related transcripts within C. velia. In order to determine the events within the cyst that lead to exflagellation, we next conducted an ultrastructural examination of C. velia cells at the very beginning of the phase of flagellate production.
C. velia cells, harvested 48 h after passage and between 90 min and 2 h after the beginning of the light cycle, were fixed by rapid high-pressure freezing and subsequent freeze substitution. Multiple adjacent sections from four independent preparations were examined by thin-section TEM. Although flagella were readily detectable (Fig. 3A and B), we did not observe a single instance of an intracytoplasmic axoneme (Table 2). All of the flagella observed were surrounded by a flagellar membrane and were external to the main cell body (Fig. 3A to C).
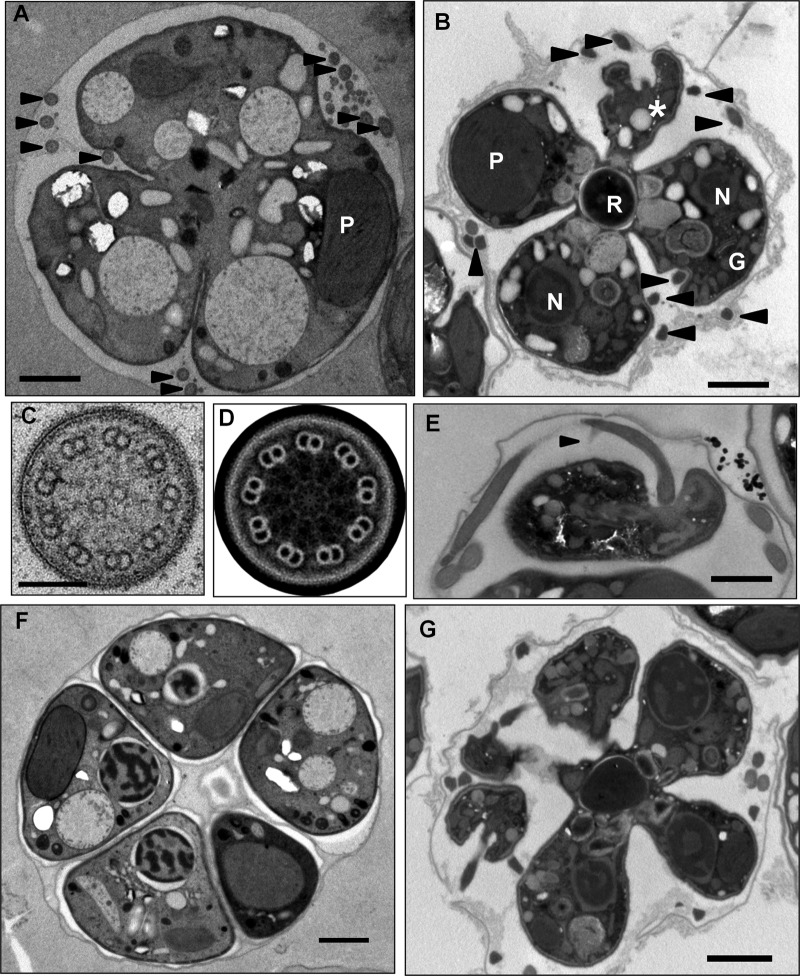
FIG 3.
(A) Probable type IVF cell within a cyst. Cleavage furrow ingression and multiple external flagellar profiles are apparent (arrowheads). P, plastid; bar, 1 μm. (B) Late-stage type IVF cell. The cell adopts a characteristic cruciform morphology with the four daughter cells containing plastids (P), nuclei (N), and a Golgi apparatus (G) arrayed around the residuum (R). Multiple external flagellar profiles are apparent (arrowheads), and a flagellar emergence point can be seen (*). Bar, 1 μm. (C, D) Transverse section of a C. velia flagellum. The canonical nine-plus-two microtubular arrangement is present, and 9-fold rotation around the nine planes of symmetry, followed by image averaging, highlights the radial spokes and dynein arms. Bar in panel C, 100 nm. (E) The anterior flagellum attains its full length within the cyst. The flagellum tapers at the distal tip and already possesses the curved appendage (arrowhead). This image was taken from a series through the cell that encompassed the entire anterior flagellum (not shown). Bar, 1 μm. (F, G) C. velia division can produce more than four daughter cells from a single cyst. Cysts containing more than four nonflagellate (F) or flagellate (G) progeny were observed. The flagellate-type cysts in particular demonstrate that multiple rounds of mitosis occur prior to the completion of cytokinesis. Bars, 1 μm.
TABLE 2.
Frequencies of C. velia cyst types in TEM preparationsa
| No. of cells in cyst | % of cysts with: |
Total %b | ||
|---|---|---|---|---|
| Internal axonemes | External axonemes | No axonemes | ||
| 1 | 0.0 | 2.4 | 49.2 | 51.6 |
| 2 | 0.0 | 6.1 | 13.4 | 19.5 |
| 3 | 0.0 | 4.0 | 16.8 | 20.9 |
| 4 | 0.0 | 1.1 | 5.3 | 6.4 |
| >4 | 0.0 | 0.3 | 1.3 | 1.6 |
| Total | 0.0 | 13.9 | 86.1 | |
Counts were made from three sections cut from randomly selected blocks, and the results were pooled. Assessments of the number of cells in a cyst are likely to be underestimates because the full diameter of cysts (∼5 μm) exceeded the thickness of the section (∼70 nm).
Total n = 374.
Flagellar profiles exhibited the canonical eukaryotic nine-plus-two microtubular arrangement with nine outer doublet microtubules surrounding a central pair of singlet microtubules (Fig. 3C). Rotation of flagellar profiles through the nine planes of symmetry generated by the outer doublet microtubules and averaging of the resulting images highlighted the presence of inner and outer dynein arms and radial spokes (Fig. 3D), consistent with our detection of transcripts associated with these structures in EST databases.
As with our live-cell imaging, we recognized two predominant cyst types associated with flagellar profiles which we interpret as corresponding to types IIF and IVF described above (Table 2). In type IVF cysts (Fig. 3B), cleavage furrow ingression results in a cruciform morphology with the four daughter cells connected via the residuum at the center. In all of the flagellate-type cysts examined, axonemes were compartmentalized within their own membrane and external to the daughter cell body while still being contained within the cyst volume, inside the cyst wall. We found examples of cells at various stages of division in which external flagellar profiles could be observed, suggesting that the emergence of flagella onto the cell surface, by whatever means this occurs, happens relatively early in the cell division process. The short anterior flagellum reaches its full length of around 5.5 μm (29) within the cyst (Fig. 3E). This flagellum distinctly tapers toward the tip, and the intriguing curved appendage described previously in scanning electron microscopic (SEM) images (28, 29) is also already present within the cyst.
We were also able to readily detect both flagellate-type and non-flagellate-type cysts containing more than four nascent daughter cells (Fig. 3F and G). Cysts of these types made up only around 1% (4 out of 374, Table 2) of the total cyst profiles but were reproducibly present in all of the preparations examined. Cysts containing five, six, and seven daughter cells of both the flagellate and nonflagellate types were confirmed.
The C. velia flagellar basal bodies are associated with microtubular rootlets.
As previously determined by SEM techniques (29), the cell body of the flagellate form has a broad, rounded posterior that tapers to a narrow anterior tip (Fig. 4A). The two flagella emerge subapically into depressions or grooves in the plasma membrane on the ventral side of the cell.
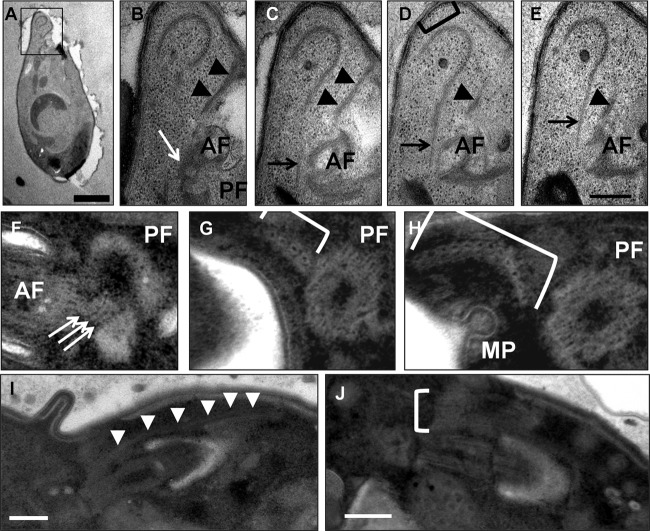
FIG 4.
Flagellar basal bodies. (A) A flagellate cell that has emerged from the cyst. The cell tapers toward the anterior tip (top), which contains a crescent-shaped arrangement of microtubules. Bar, 1 μm. (B to E) Serial sections through the anterior end of the flagellate cell shown in panel A. The apical microtubules are distinct from the population of subpellicular microtubules (bracket in panel D) and are connected via a set of rootlet microtubules (black arrows) to the basal body of the anterior flagellum (AF). Another set of fibers (black arrowheads) that appear to be microtubular and that we interpret as being part of the subpellicular microtubule array lie alongside the anterior flagellar groove. A third set of fibers connects the anterior basal body to the posterior one (white arrow). PF, posterior flagellum; bar, 200 nm. (F to H). Serial transverse sections through the proximal end of the posterior basal body. (F) The three-layered structure (white arrows) connects the proximal extremes of the posterior (PF) and anterior (AF) basal bodies. (G, H) A set of 12 rootlet microtubules arranged in a crescent (bracket) nucleate adjacent to the posterior basal body. A micropore (MP) associated with the anterior flagellar groove can also be observed. (I, J) Longitudinal negative staining of the posterior flagellum from two cells showing rootlet microtubules (white arrowheads in panel I, white bracket in panel J) originating near the posterior basal body and extending around the posterior flagellar groove. Bars, 200 nm.
Microtubular roots are associated with the basal body of each flagellum. The first rootlet associated with the anterior basal body extends from ca. 100 nm proximal to the basal body, around the dorsal face of the anterior flagellar groove into the tapered anterior tip of the cell. Here the rootlet lies alongside a set of apical microtubules reminiscent of the structure previously described as the pseudoconoid (28) (Fig. 4C to E). The microtubular structure of the pseudoconoid is distinct from the population of subpellicular microtubules that underlie the cortical alveoli (Fig. 4D). A second set of microtubules extends anterodorsally from the top of the anterior basal body, along the edge of the cell membrane that forms the flagellar groove; we interpret these as subpellicular microtubules arising next to the alveoli at the anterior tip of the cell (Fig. 4B to E). An additional fiber extends from the anterior basal body toward the posterior basal body and appears as a trilaminar structure connecting the two basal bodies (Fig. 4B and F). The posterior basal body rootlet consists of 12 microtubules arranged in a shallow, horizontal crescent (Fig. 4F to H) arising on the distal left edge of the basal body. This rootlet extends parallel to the basal body along the posterior flagellar groove, beneath the plasma membrane and cortical alveoli, toward the posterior of the cell (Fig. 4I and J).
The C. velia flagella are part of an extensive apical complex.
The fibers of the pseudoconoid of C. velia appear to be microtubular and to form a tight ring at the apical tip of the cell that broadens and opens up as it extends into the cell body, thus forming a cone shape that is open on the side adjacent to the anterior flagellar groove (Fig. 5A to I). The microtubules of the first anterior flagellar rootlet run alongside and slightly outside the pseudoconoid, adjacent to the open side of the cone, between this and the anterior flagellar groove (Fig. 5C to F). Two additional intraconoidal microtubules extend from the apex of the pseudoconoid and through the lumen of the cone (Fig. 5B to H). The apical microtubules are associated with projections oriented away from the lumen of the cone that likely represent the presence of microtubule-associated proteins (Fig. 5J). Two membrane-bound compartments are also associated with the pseudoconoid. The first occupies the lumen of the pseudoconoid, and we refer to it here as the lumenal endomembrane system (Fig. 5C to I). The second lies alongside the pseudoconoid between the apical microtubules and the plasma membrane on the side of the cone opposite the anterior flagellar groove. We refer to this system here as the extralumenal endomembrane system (Fig. 5E to I). In longitudinal section, the lumenal system can be seen to extend beyond the pseudoconoid, where it nests within a distinctive, noose-shaped extension of the extralumenal system (Fig. 6). The extralumenal system has multiple lobes and appears to cross between the microtubules of the pseudoconoid at several points. Observations of this nested, noose-shaped morphology are highly reproducible between individual cells.
FIG 5.
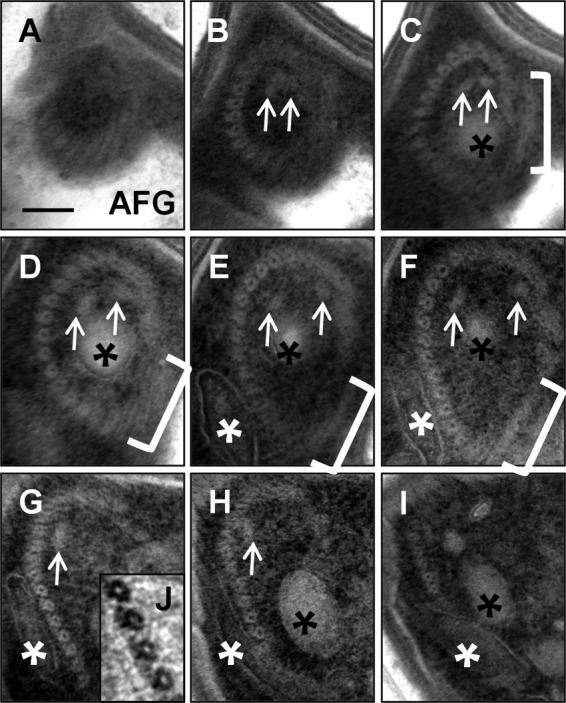
The pseudoconoid from top to bottom. (A to I) Negatively stained serial sections through the apical complex, where panel A is at the apical tip of the cell and the series moves down through the cell body. The apical microtubules form a tight ring at the tip of the cell that broadens and opens up toward its base. The intraconoidal microtubular pair (white arrows) and the lightly staining endomembrane system (black asterisks) occupy the lumen of the cone. The anterior rootlet microtubules (brackets) are positioned between the apical microtubules and the anterior flagellar groove (AFG) and partially cover the open side of the cone at its tip. A lobe of the darkly staining endomembrane system (white asterisks) is present on the outside face of the cone. Bar, 100 nm. (J) Blown-up and inverted view of the apical complex showing projections from the outside face of the microtubules.
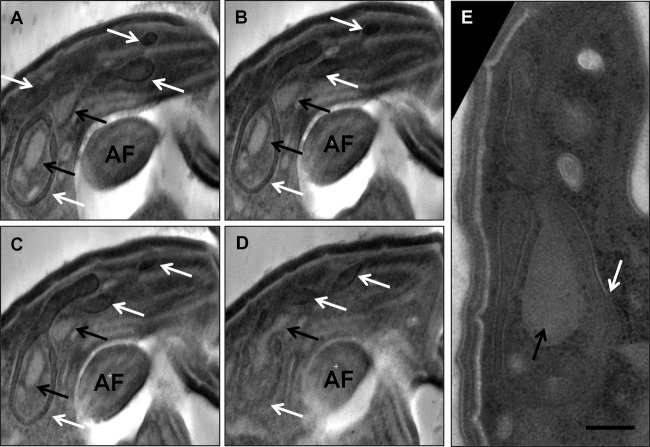
FIG 6.
The apical endomembrane system. (A to D) Pseudosections from a negatively stained tomogram of the apical tip of a flagellate cell. Two nested endomembrane systems are present. The noose-shape extralumenal system (white arrows) has several lobes that extend into and around the pseudoconoid. The lumenal system (black arrows) is nested within the noose of the extralumenal system and extends into the lumen of the apical complex. AF, anterior flagellum. (E) Negatively stained section of a different cell showing the same distinctive morphology. This endomembrane system occupies a position equivalent to that of the rhoptries and micronemes of Apicomplexa in relation to the apical microtubules. Bar, 200 nm.
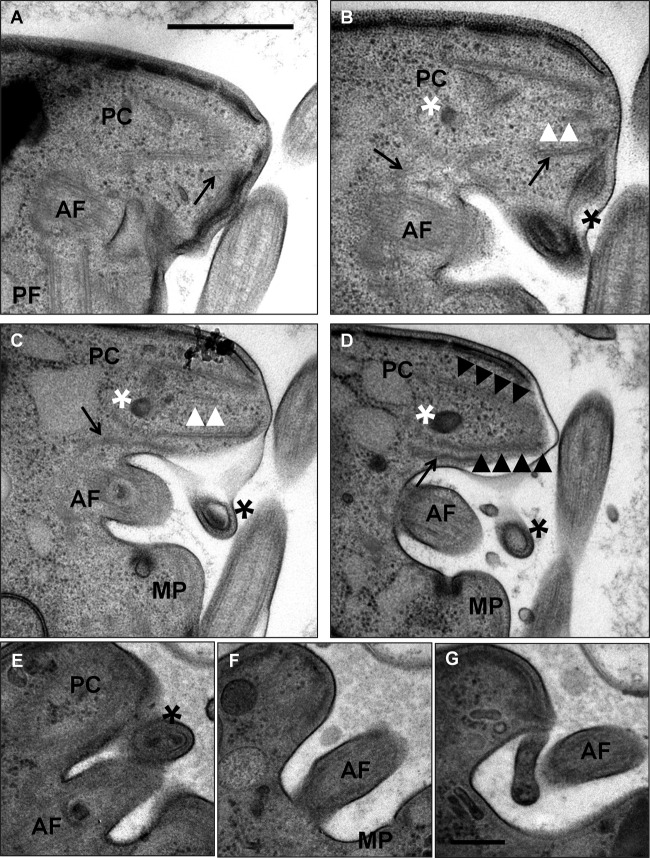
The anterior flagellar groove always contains a micropore, a structure unique to alveolates and characterized by an electron-dense ring of material constricting the lumen, where it opens onto the cell surface (Fig. 7C, D, and F). This flagellar groove micropore is always positioned on the posterior face of the anterior flagellar groove (it can also be seen in Fig. 4H).
FIG 7.
The anterior flagellum and apical complex. (A to D) Adjacent sections showing the relative positional context of the anterior flagellum (AF) and pseudoconoid (PC). The apical microtubules can be seen in longitudinal section displaying the distinctive cone-shaped configuration. The cone contains an endomembrane system (white asterisk) and a set of intraconoidal microtubules (white arrowheads). A projection of the plasma membrane at the emergence point of the anterior flagellum contains a coiled, electron-dense structure (black asterisk) that is connected via a fiber (most clearly visible in panel B) to the pseudoconoid. The anterior rootlet microtubules (black arrows) extend alongside the apical microtubules. The invariantly positioned micropore (MP) is located within the flagellar groove. PF, posterior flagellum; bar, 500 nm. (E to G) Alternate sections through a second cell, where panel E roughly corresponds to panel C and panel F continues from panel D. Panel G shows a finger-like projection covering the proximal portion of the anterior flagellum distinct from the projection containing the coiled structure shown in panel E. Labeling is the same as in panels A to D. Bar, 200 nm.
An electron-dense structure that appears to be a coiled fiber is positioned adjacent to the apical cone at the anteroventral entrance to the anterior flagellar groove (Fig. 7 B to E). An uncharacterized fiber connects this coiled structure to the anterior end of the apical microtubules. A flap of the plasma membrane (previously described in our SEM study of the flagellate form [29]) emerges next to the coiled fiber and partially covers the anterior flagellar groove (Fig. 7G).
DISCUSSION
C. velia occupies an intriguing position as the closest extant sister group of Apicomplexa and has been thought of as representing a more ancestral state because of its functioning chloroplast and presumed autotrophic lifestyle (36). The presence of cytoplasmic axonemes was noted in the original type description, and it was hypothesized that C. velia, like Plasmodium, may also lack IFT (26, 28), although given the phylogenetic relationships, this would be overwhelmingly likely to represent another independent loss of IFT rather than an ancestral state.
C. velia encodes components for both anterograde and retrograde IFT.
In this work, we compared some of the best available descriptions of the protein composition of eukaryotic flagella to EST databases generated from C. velia. We focused particularly on the identification of transcripts associated with a role in IFT. We found putative homologues for components of both the anterograde kinesin motor—kinesin II—and the retrograde dynein motor—cytoplasmic dynein 2, although because of the conserved modular nature of proteins of these types, a final confirmation of these identifications must await the completion of the C. velia genome project. We also detected a number of transcripts with significant homology to components of the two main IFT complexes, A and B (9, 37). Although the IFT cohort that we detected is not comprehensive, it does include components associated with both anterograde and retrograde transport and includes candidates for proteins absent from Toxoplasma (IFT20, IFT22, IFT25, IFT27, and IFT139). Given these data and the fact that we were sampling from a limited database of ESTs, we predict that C. velia does possess a complete or nearly complete cohort of IFT proteins.
Our live-cell and ultrastructural examinations of C. velia showed that biogenesis of the flagellate form occurs through a process of cell division and not by the transformation of a discrete cell. The decision to construct flagella is made in the mother cyst and applies to all of the progeny of that cyst; we did not observe any instance of mixed progeny emerging from a single cyst. As has been previously reported (26, 28), we observed cysts that gave rise to two or four progeny (which we have classified as type II and IV cysts, respectively), but our ultrastructural studies also revealed the existence of cysts containing more than four nascent daughter cells. In several examples of these cysts, particularly those giving rise to flagellate progeny, all of the observable daughter cells remain attached to one another at the residuum. This suggests that the mother cell undergoes multiple rounds of mitosis prior to the completion of cytokinesis.
The case for intracytoplasmic axoneme formation.
We were unable to detect any intracytoplasmic axonemes in our ultrastructural examination of flagellate-form-producing cultures. Given our observations of cells more advanced in their cell cycle, if flagella were constructed internally, we would assume that at least 4 and perhaps as many as 16 intracytoplasmic axonemes in total would be present within flagellate-type cysts. It is possible that axoneme formation and exflagellation occur so rapidly that very few cells will exhibit internal axonemes at any given time. This rapid deployment of flagella is certainly the case in Plasmodium (22). However, there are marked differences in the production of flagellate forms between C. velia and Plasmodium. Exflagellation in Plasmodium occurs essentially in order to generate a motile nucleus with few, if any, additional organelles (22). The assembly process is a somewhat messy affair, and mistakes in axoneme patterning are common (22). Although the role of the C. velia flagellate form in the life cycle is not yet known, these cells contain a broad complement of organelles and structures, including a chloroplast, a Golgi apparatus, an apical complex, and probable endo- or exocytotic machinery (i.e., micropores and possibly the membranous compartments associated with the pseudoconoid), all indicative of a self-contained, metabolically active cell. In contrast to the rather violent emergence of Plasmodium male microgametes from the gametocyte, daughter cells of C. velia are generated via cytokinesis following an orderly cleavage furrow ingression. In these terms, the emphasis appears to be on quality over speed in a way that flagellate production in Plasmodium is not. The presence of a large cohort of IFT components also argues for a compartmentalized mode of flagellum biogenesis in C. velia; however, we acknowledge than inasmuch as we did not detect internal axonemes, we were also unable to provide direct evidence of flagellum extension occurring outside the cell. There are certainly other roles that IFT could be fulfilling in the cell that do not involve flagellum biogenesis. IFT is involved in the maintenance of existing flagella, as well as being increasingly implicated in a range of cellular processes, including signal transduction and exocytosis (38–40). C. velia also has accessory structures associated with its flagella, most notably, the arm-like appendage of the short anterior flagellum. It is possible that IFT would have a role in the assembly of structures like these, irrespective of the means by which the axoneme itself was assembled.
Given the lack of observable internal axonemes in our flagellate-form-producing cultures and the presence of IFT-related transcripts, we favor the model whereby the flagellum is assembled in a compartmentalized fashion in line with the canonical IFT-dependent pathway. This does not preclude a role for internal axonemes in other life cycle stages. The life cycle of C. velia is poorly understood. As we show here, even over the course of 72 h in culture, the favored mode of daughter cell production changes drastically. There are a number of life cycle stages that have previously been reported that we did not observe in our preparations, most notably, the stage with the enigmatic chromerosome (28). It is possible that axonemes have been repurposed to perform internal structural roles in other life cycle stages (26), or perhaps cells with internal axonemes represent a transitional stage between the flagellate and nonflagellate forms.
C. velia possesses elements reminiscent of an apicomplexan conoid.
In a recent examination of the ultrastructure of various putative life cycle stages of C. velia, Oborník and colleagues (28) presented the first evidence of the presence of a pseudoconoid, a structure found in predatory Colpodella and other groups of alveolates with morphological similarity to the apical complex of Apicomplexa. In this work, we have provided three-dimensional ultrastructural and positional detail of this structure by using serial-section TEM to place it within the context of an extensive apical complex that includes elements of the flagellar apparatus. Figure 8 shows a cartoon representation of our findings.
FIG 8.
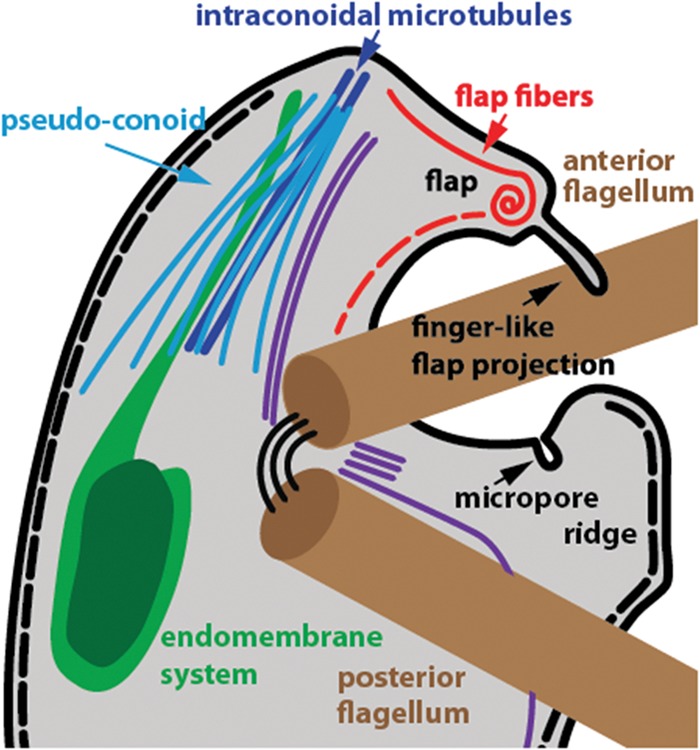
Cartoon representation of the apical complex and flagella of C. velia. Representations of the microtubules of the pseudoconoid (light blue) and flagellar rootlets (purple) are indicative of relative positions only and not the numbers of individual microtubules. The alveoli (inner membrane complex) are represented by a dotted black line beneath the plasma membrane (thick black line). The laminar connector joining the two basal bodies is shown as three thin black lines.
The apicomplexan conoid, most extensively studied in Toxoplasma, is formed from fibers composed of an unusual polymer of tubulin, tightly coiled into a closed-cone-shaped structure. The fibers of the C. velia conoid appear to be microtubules and are arranged as an open-sided cone. These fibers are distinct from the subpellicular microtubules that underlie the cortical alveoli, and we therefore suggest that they represent an ancestral state of the conoid itself and are not homologous to the subpellicular microtubules associated with the apical complex in Toxoplasma. Further morphological homology arises from the presence of a pair of intraconoidal microtubules within the lumen of the conoid of C. velia—a feature of the Toxoplasma conoid not previously described in pseudoconoids present in other nonapicomplexan alveolates—and a complex, nested endomembrane system that occupies a position equivalent to that of the rhoptries of Apicomplexa and is morphologically highly conserved between individual cells.
Coincidence of the flagellum and the apical complex in the closest relative of Apicomplexa.
Recent work on Toxoplasma has begun to elucidate an intriguing evolutionary link between the apical complex and the flagellum (41, 42). Fibers homologous to the basal-body-associated striated fibers of green algae have been shown to play an essential role in matching organelles to daughter buds (42). This is achieved by tethering nuclei to the apical end of the nascent daughter buds via a connection between the apical complex and the centrioles. Centrioles and basal bodies are generally considered to be the same structure, although in Apicomplexa, the centriole (but not the basal body) has an unusual arrangement (15). Further evidence of the involvement of flagellar components in the apical complex was the characterization of a protein that locates to the basal plate of basal bodies in Trypanosoma brucei and to the apical end of the conoid in Toxoplasma (41). Significantly, all Apicomplexa have dispensed with flagella in life cycle stages that possess an apical complex.
The most intriguing feature of the apical complex of C. velia, given the recent findings on Toxoplasma (41, 42), is that it is contemporaneous with the flagella in a single cell; in fact, there are multiple connections between the pseudoconoid and the anterior flagellum. A set of so-far-unidentified fibers connect the anterior flagellar groove to the apex of the pseudoconoid via an enigmatic coiled structure that resides beneath a protrusion of the plasma membrane. It should be noted that this protrusion underlies the anterior flagellum and is not the flap covering the proximal end of the anterior flagellum described in our previous SEM analysis of C. velia (29). The second and most striking of these connections is a set of rootlet microtubules that originate at the anterior basal body and extend along the open side of the pseudoconoid. Striated fibers with components homologous to those involved in the organization of daughter cell buds in Toxoplasma are associated with basal body rootlets in algae (42, 43). We hypothesize that C. velia also encodes an orthologue of striated-fiber assemblin and that this protein will be associated with the anterior basal body rootlet, forming a link between the pseudoconoid and the basal body. We suggest that the association of rootlet microtubules with the pseudoconoid in C. velia represents an ancestral state in the evolution of the striated-fiber-based organizing mechanism employed by Toxoplasma during daughter cell development that occurs via a link between the centriole and the apical complex.
Conclusion.
Flagella coexist with ancestral elements of a conoidal apical complex in C. velia, and this fascinating organism can be maintained in culture and induced to produce flagellate forms with relative ease. We propose that C. velia presents a novel opportunity to investigate the remarkable repurposing of flagellar structures that has apparently occurred during the evolution of the apicomplexan apical complex.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Australian Research Council, Discovery Project DP0986372, and in part by the Faculty of Veterinary Science, University of Sydney, and an ARC/NH&MRC Network for Parasitology travel grant. Support for N.P. is provided by the University of Sydney Postdoctoral Research Fellowship scheme. C.F. is funded by the Jean Walker Trust Fellowship Scholarship, Faculty of Veterinary Science, University of Sydney. G.W. was supported by an ARC/NHMRC Network for Parasitology Travel Award.
The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
We acknowledge the expert assistance of Naveena Gokoolparsadh and the facilities at the Australian Microscopy & Microanalysis Research Facility node (Sydney Microscopy and Microanalysis) at the University of Sydney and Renee Whan and Iveta Šlapetová at the Biomedical Imaging Facility, University of New South Wales. We also thank Arnab Pain (KAUST, Saudi Arabia) for insightful discussion, encouragement, and access to unpublished data.
Footnotes
Published ahead of print 20 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00155-13.
REFERENCES
- 1.Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. 2004. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell 117:527–539. 10.1016/S0092-8674(04)00412-X [DOI] [PubMed] [Google Scholar]
- 2.Cavalier-Smith T. 2002. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 52:297–354. 10.1099/ijs.0.02058-0 [DOI] [PubMed] [Google Scholar]
- 3.Jékely G, Arendt D. 2006. Evolution of intraflagellar transport from coated vesicles and autogenous origin of the eukaryotic cilium. Bioessays 28:191–198. 10.1002/bies.20369 [DOI] [PubMed] [Google Scholar]
- 4.Mitchell DR. 2007. The evolution of eukaryotic cilia and flagella as motile and sensory organelles. Adv. Exp. Med. Biol. 607:130–140. 10.1007/978-0-387-74021-8_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginger ML, Portman N, McKean PG. 2008. Swimming with protists: perception, motility and flagellum assembly. Nat. Rev. Microbiol. 6:838–850. 10.1038/nrmicro2009 [DOI] [PubMed] [Google Scholar]
- 6.Kohl L, Robinson D, Bastin P. 2003. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 22:5336–5346. 10.1093/emboj/cdg518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell DR. 2000. Chlamydomonas flagella. J. Phycol. 36:261–273. 10.1046/j.1529-8817.2000.99218.x [DOI] [Google Scholar]
- 8.Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. 1993. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. U. S. A. 90:5519–5523. 10.1073/pnas.90.12.5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole DG. 2003. The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic 4:435–442. 10.1034/j.1600-0854.2003.t01-1-00103.x [DOI] [PubMed] [Google Scholar]
- 10.Marshall WF, Rosenbaum JL. 2001. Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J. Cell Biol. 155:405–414. 10.1083/jcb.200106141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan J, Snell WJ. 2005. Chlamydomonas shortens its flagella by activating axonemal disassembly, stimulating IFT particle trafficking, and blocking anterograde cargo loading. Dev. Cell 9:431–438. 10.1016/j.devcel.2005.07.010 [DOI] [PubMed] [Google Scholar]
- 12.Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. 2004. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164:255–266. 10.1083/jcb.200308132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briggs LJ, Davidge JA, Wickstead B, Ginger ML, Gull K. 2004. More than one way to build a flagellum: comparative genomics of parasitic protozoa. Curr. Biol. 14:R611–R612. 10.1016/j.cub.2004.07.041 [DOI] [PubMed] [Google Scholar]
- 14.WHO 2012. World malaria report 2012 World Health Organization, Geneva, Switzerland [Google Scholar]
- 15.Morrissette NS, Sibley LD. 2002. Cytoskeleton of apicomplexan parasites. Microbiol. Mol. Biol. Rev. 66:21–38. 10.1128/MMBR.66.1.21-38.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradley PJ, Sibley LD. 2007. Rhoptries: an arsenal of secreted virulence factors. Curr. Opin. Microbiol. 10:582–587. 10.1016/j.mib.2007.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Proellocks NI, Coppel RL, Waller KL. 2010. Dissecting the apicomplexan rhoptry neck proteins. Trends Parasitol. 26:297–304. 10.1016/j.pt.2010.02.012 [DOI] [PubMed] [Google Scholar]
- 18.Hu K, Roos DS, Murray JM. 2002. A novel polymer of tubulin forms the conoid of Toxoplasma gondii. J. Cell Biol. 156:1039–1050. 10.1083/jcb.200112086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuriyama R, Besse C, Geze M, Omoto CK, Schrével J. 2005. Dynamic organization of microtubules and microtubule-organizing centers during the sexual phase of a parasitic protozoan, Lecudina tuzetae (Gregarine, Apicomplexa). Cell Motil. Cytoskeleton 62:195–209. 10.1002/cm.20092 [DOI] [PubMed] [Google Scholar]
- 20.Prensier G, Dubremetz J-F, Schrével J. 2008. The unique adaptation of the life cycle of the coelomic gregarine Diplauxis hatti to its host Perinereis cultrifera (Annelida, Polychaeta): an experimental and ultrastructural study. J. Eukaryot. Microbiol. 55:541–553. 10.1111/j.1550-7408.2008.00361.x [DOI] [PubMed] [Google Scholar]
- 21.Prensier G, Vivier E, Goldstein S, Schrével J. 1980. Motile flagellum with a “3 + 0” ultrastructure. Science 207:1493–1494. 10.1126/science.7189065 [DOI] [PubMed] [Google Scholar]
- 22.Sinden RE, Talman A, Marques SR, Wass MN, Sternberg MJ. 2010. The flagellum in malarial parasites. Curr. Opin. Microbiol. 13:491–500. 10.1016/j.mib.2010.05.016 [DOI] [PubMed] [Google Scholar]
- 23.Ferguson DJP. 2002. Toxoplasma gondii and sex: essential or optional extra? Trends Parasitol. 18:351–355. 10.1016/S1471-4922(02)02330-9 [DOI] [PubMed] [Google Scholar]
- 24.Ferguson DJ, Hutchison WM, Dunachie JF, Siim JC. 1974. Ultrastructural study of early stages of asexual multiplication and microgametogony of Toxoplasma gondii in the small intestine of the cat. Acta Pathol. Microbiol. Scand. B Microbiol. Immunol. 82:167–181 [DOI] [PubMed] [Google Scholar]
- 25.Wickstead B, Gull K. 2007. Dyneins across eukaryotes: a comparative genomic analysis. Traffic 8:1708–1721. 10.1111/j.1600-0854.2007.00646.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore RB, Oborník M, Janouškovec J, Chrudimský T, Vancová M, Green DH, Wright SW, Davies NW, Bolch CJS, Heimann K, Šlapeta J, Hoegh-Guldberg O, Logsdon JM, Carter DA. 2008. A photosynthetic alveolate closely related to apicomplexan parasites. Nature 451:959–963. 10.1038/nature06635 [DOI] [PubMed] [Google Scholar]
- 27.Janouškovec J, Horák A, Oborník M, Lukeš J, Keeling PJ. 2010. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc. Natl. Acad. Sci. U. S. A. 107:10949–10954. 10.1073/pnas.1003335107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oborník M, Vancová M, Lai DH, Janouškovec J, Keeling PJ, Lukeš J. 2011. Morphology and ultrastructure of multiple life cycle stages of the photosynthetic relative of apicomplexa, Chromera velia. Protist 162:115–130. 10.1016/j.protis.2010.02.004 [DOI] [PubMed] [Google Scholar]
- 29.Weatherby K, Murray S, Carter D, Šlapeta J. 2011. Surface and flagella morphology of the motile form of Chromera velia revealed by field-emission scanning electron microscopy. Protist 162:142–153. 10.1016/j.protis.2010.02.003 [DOI] [PubMed] [Google Scholar]
- 30.Guo JT, Weatherby K, Carter D, Šlapeta J. 2010. Effect of nutrient concentration and salinity on immotile-motile transformation of Chromera velia. J. Eukaryot. Microbiol. 57:444–446. 10.1111/j.1550-7408.2010.00495.x [DOI] [PubMed] [Google Scholar]
- 31.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- 32.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- 33.Pazour GJ, Agrin N, Leszyk J, Witman GB. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170:103–113. 10.1083/jcb.200504008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woehle C, Dagan T, Martin WF, Gould SB. 2011. Red and problematic green phylogenetic signals among thousands of nuclear genes from the photosynthetic and apicomplexa-related Chromera velia. Genome Biol. Evol. 3:1220–1230. 10.1093/gbe/evr100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan H, Šlapeta J, Carter D, Chen M. 2012. Phylogenetic analysis of the light-harvesting system in Chromera velia. Photosynth. Res. 111:19–28. 10.1007/s11120-011-9710-9 [DOI] [PubMed] [Google Scholar]
- 36.Okamoto N, McFadden GI. 2008. The mother of all parasites. Future Microbiol. 3:391–395. 10.2217/17460913.3.4.391 [DOI] [Google Scholar]
- 37.Pedersen LB, Geimer S, Rosenbaum JL. 2006. Dissecting the molecular mechanisms of intraflagellar transport in Chlamydomonas. Curr. Biol. 16:450–459. 10.1016/j.cub.2006.02.020 [DOI] [PubMed] [Google Scholar]
- 38.Baldari CT, Rosenbaum J. 2010. Intraflagellar transport: it's not just for cilia anymore. Curr. Opin. Cell Biol. 22:75–80. 10.1016/j.ceb.2009.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoder BK. 2006. More than just the postal service: novel roles for IFT proteins in signal transduction. Dev. Cell 10:541–542. 10.1016/j.devcel.2006.04.010 [DOI] [PubMed] [Google Scholar]
- 40.Goetz SC, Ocbina PJ, Anderson KV. 2009. The primary cilium as a Hedgehog signal transduction machine. Methods Cell Biol. 94:199–222. 10.1016/S0091-679X(08)94010-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Leon JC, Scheumann N, Beatty W, Beck JR, Tran JQ, Yau C, Bradley PJ, Gull K, Wickstead B, Morrissette NS. 2013. A SAS-6-like protein suggests that the Toxoplasma conoid complex evolved from flagellar components. Eukaryot. Cell 12:1009–1019. 10.1128/EC.00096-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Francia ME, Jordan CN, Patel JD, Sheiner L, Demerly JL, Fellows JD, Cruz de Leon JC, Morrissette NS, Dubremetz JF, Striepen B. 2012. Cell division in apicomplexan parasites is organized by a homolog of the striated rootlet fiber of algal flagella. PLoS Biol. 10:e1001444. 10.1371/journal.pbio.1001444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lechtreck KF, Melkonian M. 1998. SF-assemblin, striated fibers, and segmented coiled coil proteins. Cell Motil. Cytoskeleton 41:289–296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.