Abstract
The cytosolic iron/sulfur cluster assembly (CIA) machinery is responsible for the assembly of cytosolic and nuclear iron/sulfur clusters, cofactors that are vital for all living cells. This machinery is uniquely found in eukaryotes and consists of at least eight proteins in opisthokont lineages, such as animals and fungi. We sought to identify and characterize homologues of the CIA system proteins in the anaerobic stramenopile parasite Blastocystis sp. strain NandII. We identified transcripts encoding six of the components—Cia1, Cia2, MMS19, Nbp35, Nar1, and a putative Tah18—and showed using immunofluorescence microscopy, immunoelectron microscopy, and subcellular fractionation that the last three of them localized to the cytoplasm of the cell. We then used comparative genomic and phylogenetic approaches to investigate the evolutionary history of these proteins. While most Blastocystis homologues branch with their eukaryotic counterparts, the putative Blastocystis Tah18 seems to have a separate evolutionary origin and therefore possibly a different function. Furthermore, our phylogenomic analyses revealed that all eight CIA components described in opisthokonts originated before the diversification of extant eukaryotic lineages and were likely already present in the last eukaryotic common ancestor (LECA). The Nbp35, Nar1 Cia1, and Cia2 proteins have been conserved during the subsequent evolutionary diversification of eukaryotes and are present in virtually all extant lineages, whereas the other CIA proteins have patchy phylogenetic distributions. Cia2 appears to be homologous to SufT, a component of the prokaryotic sulfur utilization factors (SUF) system, making this the first reported evolutionary link between the CIA and any other Fe/S biogenesis pathway. All of our results suggest that the CIA machinery is an ubiquitous biosynthetic pathway in eukaryotes, but its apparent plasticity in composition raises questions regarding how it functions in nonmodel organisms and how it interfaces with various iron/sulfur cluster systems (i.e., the iron/sulfur cluster, nitrogen fixation, and/or SUF system) found in eukaryotic cells.
INTRODUCTION
The assembly of iron/sulfur (Fe/S) clusters is considered one of the basic biosynthetic functions in all living cells. Both prokaryotic and eukaryotic organisms have at least one pathway dedicated to Fe/S cluster biosynthesis. In eukaryotes, these clusters are assembled by distinct biosynthetic pathways, which are localized in different compartments of the cell. The sulfur utilization factors (SUF) system is typically found in plastid-bearing organisms and ensures maturation of apoproteins within plastids, whereas this function is performed by the iron/sulfur cluster (ISC) machinery in mitochondria and mitochondrion-related organelles (MROs). The ISC system is indirectly functionally linked with the cytosolic iron/sulfur cluster assembly (CIA) machinery, which is involved in the maturation of cytosolic and nuclear apoproteins (1). Many of these Fe/S cluster-bearing proteins are involved in key enzymatic activities, such as DNA replication and repair, rRNA processing, and telomere stability (2, 3). Consequently, depletions of components of the CIA system are lethal in Saccharomyces cerevisiae, corroborating the significant role of this biosynthetic pathway in the cell (4–8).
Fe/S cluster biosynthetic machineries typically consist of five main parts: a desulfurase, an iron donor, an electron transfer mechanism, a scaffold, and Fe/S cluster transfer proteins. It is noteworthy that while the SUF, nitrogen fixation (NIF), and ISC systems share several homologous components, no evolutionary link has yet been reported between CIA system proteins and their counterparts in other Fe/S cluster machineries. Currently, it is known that the CIA pathway involves at least eight proteins in yeast and humans—Dre2, Tah18, Nbp35, Cfd1, Cia1, Cia2, Nar1, and MMS19 (see Fig. S1 in the supplemental material)—and that their function is dependent on the mitochondrial ISC machinery (9). The pathway starts when one of the two Fe/S clusters in Dre2 (human Ciapin1) is reduced by the diflavin reductase Tah18 (human Ndor1) (10). Nbp35 and Cfd1 then form a scaffold complex for assembling transiently bound Fe/S clusters (11). In a later stage, mature Fe/S clusters are transferred to apoproteins via the Cia1 (human CIA01) and Nar1 (human IOP1) proteins, a process facilitated through the formation of a CIA-targeting complex with the recently discovered chaperone protein MMS19 (also known as MET18 in yeast) (9, 12, 13) and Cia2 (9, 12).
In most microbial eukaryotes, the role of the CIA machinery in the maturation of the cytosolic and nuclear apoproteins is unclear, as are its functional interactions with other Fe/S cluster maturation pathways, in particular, in organisms bearing more than one cytosolic Fe/S system. For example, the microaerophiles Entamoeba and Mastigamoeba both possess functionally reduced MROs that do not contain the typical ISC pathway. Instead, their genomes encode NIF system components that were acquired by lateral gene transfer (LGT) from epsilonproteobacteria and that localize both in the cytosol and in their organelles (14–16). Another example is found in Blastocystis, an obligate anaerobic parasite that encodes a functional fused version of the SufC and SufB proteins, SufCB, that was also acquired by LGT, in this case, from a methanoarchaeal lineage, and that functions in the cytosol of the organism (17). The presence of both this SUF protein and components of the CIA Fe/S cluster biosynthetic pathway in Blastocystis (18) raises questions regarding their respective cytosolic roles.
In this study, we focused on the CIA system in Blastocystis sp. We show that this organism expresses homologues of six of the CIA system proteins—Nbp35, Nar1, Cia1, Cia2, Tah18, and MMS19—and illustrate by immunomicroscopy and subcellular fractionation that three of them are localized in the cytosol. Using comparative genomic and phylogenetic approaches, we then investigated the distribution and evolutionary histories of the CIA components among eukaryotes.
MATERIALS AND METHODS
Blastocystis culture and maintenance.
Blastocystis sp. strain NandII cultures were obtained from the American Type Culture Collection (ATCC) and maintained in Locke's medium egg slants at 35.6°C in an anaerobic chamber.
Protein extraction and Western blotting.
Blastocystis cells were pelleted at 800 × g for 10 min and then washed twice in 1× Locke's solution before being suspended in 0.15 M NaCl. Cells were disrupted using ultrasonication (for 30 s at 30-s intervals, 6 cycles) (17, 19) on ice. Following disruption, protease inhibitor cocktail (10 μl; Sigma) was added to the sample, and the mixture was then centrifuged at 10,000 × g for 10 min at 4°C. Afterwards, the supernatant was collected and 10 μl of ice-cold nuclease buffer (20 mM Tris-HCl, pH 8.8, 2 mM CaCl2) along with 10 μl of protease inhibitor cocktail was added. Thirty microliters of a DNase-RNase mix (50 mM MgCl2, 0.5 M Tris-HCl, pH 7.0) was added and the mixture was incubated on ice for 3 min. Subsequently, 10 μl of 3% SDS–10% mercaptoethanol was added and the mix was passed through a fine syringe. Samples were then stored at −20°C in NuPAGE lithium dodecyl sulfate (LDS) sample buffer along with 10× sample reducing agent (Invitrogen). Depending on the amount of protein, 5 to 20 μl of the supernatant (∼10%) was analyzed using a polyacrylamide minigel.
Different cell fractions were isolated following procedures previously described (19, 20). Blastocystis cells (well-grown in 20 egg-slant tubes for 5 days) were harvested by centrifugation at 1,200 × g for 10 min at 4°C. Cells were resuspended in Locke's solution (pH 7.4) and pelleted again at the same speed for the same duration. Cells were then broken with 40 strokes in a 10-ml Potter-Elvehjem tissue homogenizer at 4°C in isotonic buffer (200 mM sucrose, pH 7.2, 30 mM phosphate, 15 mM β-mercaptoethanol, 30 mM NaCl, 0.6 mM CaCl2, 0.6 mM KCl). Broken cells were then diluted with isotonic buffer and then centrifuged at 700 × g for 10 min using a Sorvall RC-2B centrifuge to remove unbroken cells. The supernatant was collected and centrifuged at 5,000 × g for 20 min to pellet the large granular fraction (LGF), where MROs are found (see references 21 and 22). The LGF was resuspended (washed) in isotonic buffer and pelleted as described above. Finally, all fractions were stored at −20°C in NuPAGE LDS sample buffer along with 10× sample reducing agent (Invitrogen). Depending on the amount of protein, 5 to 20 μl of the supernatant was analyzed using a polyacrylamide minigel.
For Western blot analysis, proteins were transferred to immunoblot polyvinylidene difluoride (PVDF) membranes (Bio-Rad), visualized by Ponceau staining, and then blocked for an hour with 5% skimmed milk in TBS (10 mM Tris, 0.2 M NaCl, 0.2% bovine serum albumin [BSA], pH 8.1)–0.1% Tween. Membranes were washed with 0.5% skimmed milk in TBS–0.1% Tween three times for 10 min each time. The primary antibodies (see below) were diluted in 1% skimmed milk in TBS–0.1% Tween (anti-Blastocystis SufCB, 1:500; anti-Saccharomyces Nbp35 [6], 1:500; anti-Saccharomyces Nar1 [8], 1:300; anti-Saccharomyces Tah18 [10], 1:200) and applied to the membrane overnight at 4°C. Membranes were washed as described above and incubated with the secondary antibody diluted in 1% skimmed milk in TBS– 0.1% Tween. Membranes were washed in TBS three times for 10 min each time and incubated with enhanced chemiluminescence (ECL) reagent (GE Healthcare), and fluorescence was monitored by autoradiography.
Immunolocalization of CIA components in Blastocystis.
Blastocystis cells were fixed with 4% paraformaldehyde at 37°C for 15 min, followed by three 10-min washes with 1× TBS. Fixed cells were permeabilized with ice-cold acetone and washed three times for 10 min each time with 1× TBS–0.1% Triton. Fixed cells were incubated for 30 min with a blocking solution of 5% skimmed milk powder and 1× TBS–0.1% Triton solution (wt/vol). They were then rinsed with 0.5% milk–1× TBS–0.1% Triton solution for 30 min. The cells were then incubated with a dilution of the antiserum (anti-SufCB, 1:200; anti-Nbp35, 1:100; anti-Nar1, 1:100; anti-Tah18, 1:50) in 1% milk–TBS–0.1% Triton solution overnight at 4°C. Three different dilutions of each antiserum were tested to determine optimal conditions. After three rinses in 1% milk–TBS–0.1% Triton, the slides were incubated with fluorescent dye (Alexa 488 green and Alexa 594 red)-labeled secondary antibodies at a dilution of 1:200. Finally, the slides were incubated with DRAQ5 (Cell Signaling Technology) stain for 5 min (dilution, 1:1,000) and washed three times for 1 min each time with 1× TBS. Coverslips were mounted with antifade mounting medium (Vectashield) and observed under a laser scanning confocal microscope (Zeiss LSM 510 Meta) using a ×100 oil immersion lens.
For the immunolocalization experiments using transmission electron microscopy, Blastocystis NandII cells were fixed and manipulated using the protocols described previously (17). The prepared grids were incubated in sodium borohydride (1 mg/ml) for 10 min, followed by a 10-min incubation in glycine buffer (30 mM glycine in 0.1 M borate buffer, pH 9.6). The grids were then incubated in blocking solution (TBS buffer with 1% skim milk, 1% BSA) for 45 min, followed by a quick rinse with TBS buffer. The grids were then incubated overnight with a dilution of the antiserum (anti-Nbp35, 1:20; anti-Nar1, 1:15; anti-Tah18, 1:5) in TBS buffer at 4°C. Three different dilutions of each antiserum were tested to determine the optimal conditions. The grids were then rinsed three times (15 min each) in washing buffer (10 mM Tris, 0.3 M NaCl, 0.1% BSA, pH 8.1), followed by an hour incubation with secondary antirabbit antibody conjugated with 10-nm gold particles (Sigma) diluted in TBS buffer. The grids were then rinsed three times (15 min each) in washing buffer, incubated for 15 min in 2.5% glutaraldehyde, rinsed three times (3 min each) in distilled water, and stained with 2% uranyl acetate and lead citrate. Samples were viewed with a JEOL JEM 1230 transmission electron microscope to determine quality before proceeding with immunolabeling.
Transcriptomic data obtained by 454 pyrosequencing of cDNA from Blastocystis sp. NandII.
Total RNA from Blastocystis cells was isolated using the TRIzol reagent according to the manufacturer's specifications with the following modification: following separation of the organic phase, the supernatant was collected and underwent a second round of TRIzol extraction. cDNA was constructed by Vertis Biotechnologies AG (Germany), and 454 pyrosequencing was performed by Genome Quebec in a 4SLX titanium platform. The Mira assembly program (version 3.0) (18) was used to assemble the reads into contigs. A BLAST database was created using the resulting Mira contigs. The CIA proteins of the Blastocystis hominis S7 strain (23) and Saccharomyces cerevisiae were used as seeds to perform a local tblastn search against the Blastocystis NandII contig database and extract the corresponding homologues.
Database searches and data set assembly.
Prokaryotic and eukaryotic homologues of the CIA system protein sequences were retrieved from GenBank using blastp and tblastn searches with S. cerevisiae and Homo sapiens as the initial seed query sequences (24). Identical and highly similar sequences were removed. Additional databases that were searched for eukaryotic homologues included the Joint Genome Institute (JGI) database (http://www.jgi.doe.gov), the Broad Institute database (http://www.broadinstitute.org), the Cyanidioschyzon merolae Genome Project database (http://merolae.biol.s.u-tokyo.ac.jp/), GiardiaDB (http://giardiadb.org), AmoebaDB (http://amoebadb.org), NemaGENETAG (http://elegans.imbb.forth.gr/nemagenetag), and the Wellcome Trust Sanger Institute database (http://www.sanger.ac.uk). For a given protein, the absence of a homologue in a taxon was further verified by employing tblastn on the corresponding genomes using multiple queries as seeds. For organisms where the tblastn search did not result in any hits to a particular protein family, a Hidden Markov Model (HMM) profile of the alignment was built and used to search the predicted proteomes using the hmmbuild and hmmsearch programs of the HMMER package, version 3.0 (http://hmmer.janelia.org).
Protein domain identification.
Protein domains were identified using the SMART program (http://smart.embl-heidelberg.de/) (25) and by performing HMMER searches against the PFAM (version 26.0) database (http://pfam.sanger.ac.uk) (21).
Multiple-sequence alignment and phylogenetic analysis.
Protein sequences were aligned by use of the MAFFT program (version 6.903b) (22). Furthermore, the alignments were inspected by eye to detect cases of obviously misaligned regions. Subsequently, the alignments were masked to remove regions of ambiguous alignment using the Block Mapping and Gathering with Entropy (BMGE) program (version 1.1) with default parameters (26).
After trimming, the final alignments contained 59 taxa and 225 sites for the Cia1 protein, 78 taxa and 96 sites for the Dre2 protein, 87 taxa and 55 sites for the MMS19 protein, 81 taxa and 233 sites for the Nar1 protein, 95 taxa and 191 sites for the Nbp35/Cfd1 proteins, 110 taxa and 157 sites for the Tah18 protein, and 60 taxa and 98 sites for the Cia2 protein. Phylogenetic trees were constructed for each individual protein of the CIA machinery. Maximum likelihood (ML) trees were computed using the RAxML program (version 7.2.8) (27) and the Le and Gascuel (LG) amino acid substitution model (28). To account for rate heterogeneity across amino acid sites, the gamma option was also implemented. For each protein data set, bootstrap support was assessed from 100 bootstrap replicates that were subsequently mapped onto the best-scoring ML tree. Phylogenies for the CIA components that displayed interesting patterns (e.g., Nbp35/Cfd1 and Tah18) are shown in Fig. 2 and 3, whereas those with less notable evolutionary histories are included as figures in the supplemental material.
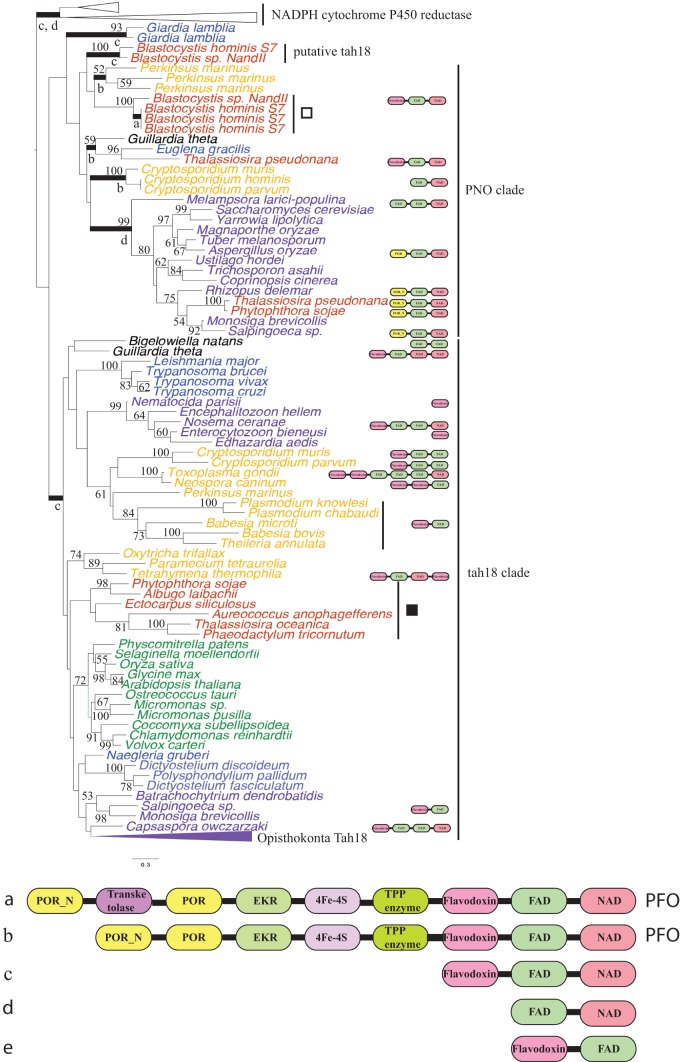
FIG 2.
Tah18 phylogeny. A maximum likelihood phylogeny of the cytosolic Fe/S cluster assembly protein Tah18 and its two closest homologues, NADPH cytochrome P450 reductase and pyruvate ferredoxin oxidoreductase, is shown. Numerical values on the branches represent statistical support in the form of bootstrap values, and the scale bar indicates the branch length in terms of the number of substitutions per site. Only bootstrap support values greater than 50 are shown. Thick branches highlight clades whose members have common protein domain structure. Black and white squares indicate the two alternative phylogenetic branching positions of the putative Blastocystis Tah18 orthologues considered in AU tests (see Materials and Methods). Letters underneath the branches indicate the number and order of the domains identified by letter at the bottom. Protein domains are individually depicted for taxa that possess numbers of domains different from those of most members of their clade. TPP, thiamine pyrophosphate; PFO, pyruvate:ferredoxin oxidoreductase.
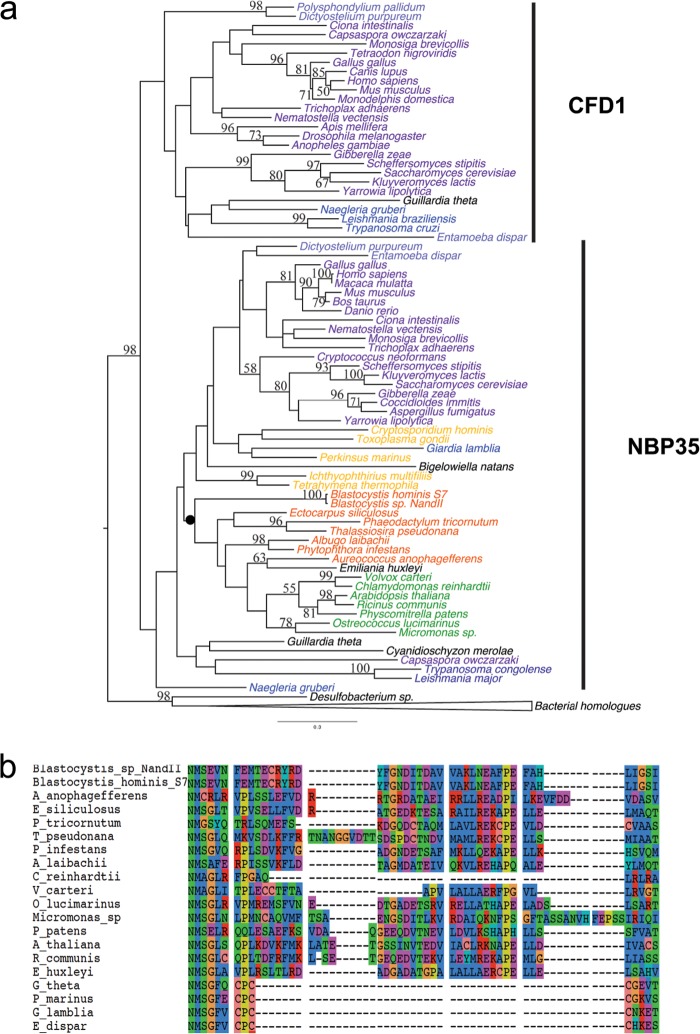
FIG 3.
Nbp35/Cfd1 phylogeny and alignment features. (a) Maximum likelihood phylogeny of the cytosolic Fe/S cluster assembly proteins Nbp35 and Cfd1 and bacterial homologues. The statistical support values shown are as described for Fig. 2. The Nbp35 and Cfd1 groups are indicated and correspond to the presence versus absence of the ferredoxin domain at their N termini, respectively. Members of the clade marked with the black dot share a variable-length insertion in the carboxyl terminus of the Nbp35 protein. (b) A portion of the amino acid sequence alignment depicting the taxa sharing the insertion in the Nbp35 protein.
Topology testing.
In the case of Tah18, we tested whether various phylogenetic hypotheses could be significantly rejected by the data. We tested two alternative phylogenetic placements of putative Blastocystis sp. Tah18 sequences: (i) grouping with the four Blastocystis sequences of the pyruvate-NADPH oxidoreductase (PNO) clade (sequences indicated by a white square in Fig. 2) (case 1) or (ii) grouping as a monophyletic group with other stramenopile Tah18 orthologues (sequences indicated by a black square in Fig. 2) (case 2). We used the approximately unbiased (AU) test implemented in the CONSEL program (29). Since this test requires a large sample of good trees, in addition to the test topologies, to accurately estimate P values (30), 500 bootstrap trees were included in the analyses. The maximum likelihood tree given a specific constraint (i.e., corresponding to a phylogenetic hypothesis) was obtained using the −g RAxML option with all other parameters set, as previously described. Case 1 could not be rejected (P = 0.228, likelihood = −51,718.8), whereas case 2 was rejected (P = 0.036, likelihood = −51,781.3).
Nucleotide sequence accession numbers.
The sequences of the Blastocystis sp. NandII transcripts are deposited in GenBank with the accession numbers KF438229 to KF438233 and KF841440, and the accession numbers of homologous sequences are provided in the supplemental data set (Table S1) in the supplemental material.
RESULTS AND DISCUSSION
Characterization/localization of components of Blastocystis CIA machinery.
In a previous study, we showed that three different Fe/S cluster biosynthetic machineries were likely present in the anaerobic parasite Blastocystis (17). We localized and functionally characterized the mitochondrial Fe/S cluster (ISC) machinery and the cytosolic SUF machinery, but only limited data regarding the presence of the CIA machinery were available. Here, we report homologues of six CIA proteins (Nbp35, Nar1, Cia1, Cia2, Tah18, and MMS19) in our transcriptomic data. The homology of three of these proteins was further confirmed by the identification of conserved features/motifs shared with better-characterized eukaryotic homologues (e.g., human, yeast) (see Fig. S2 to S4 in the supplemental material).
We employed various methods to assess the localization of CIA components in Blastocystis. Using heterologous antibodies raised against the yeast proteins Nbp35 (6), Nar1 (8), and Tah18 (10), we demonstrated their specificity against the Blastocystis homologues in Western blots of total protein extracts (see Fig. S5 in the supplemental material). To further confirm the localization of these proteins, we performed subcellular fractionations employing a two-step centrifugation method followed by a Western blot assay. We used Nbp35, Nar1, and Tah18 antibodies against the corresponding proteins obtained from the fractions. In all cases, the antibodies were localized exclusively in the cytosolic fraction of the cell extracts (see Fig. S5 in the supplemental material).
Immunofluorescence microscopy demonstrated that the Blastocystis Nbp35, Nar1, and Tah18 proteins localized in the cytosol of the cell; these proteins colocalized with the cytosolic protein SufCB as well (17) (Fig. 1), and they did not colocalize with MitoTracker, a dye that labels the MROs of the parasite (see Fig. S6 in the supplemental material). Moreover, immunogold transmission electron microscopy using the same antibodies against the CIA proteins revealed an abundance of gold particles in the cytosol and their virtual absence from the MROs, vacuole, and nucleus (Fig. 1d and e; see Fig. S7 to S9 in the supplemental material). A similar immunogold labeling pattern was demonstrated for the fused cytosolic SufCB protein of the Blastocystis sp. (Fig. 1e) (17).
FIG 1.
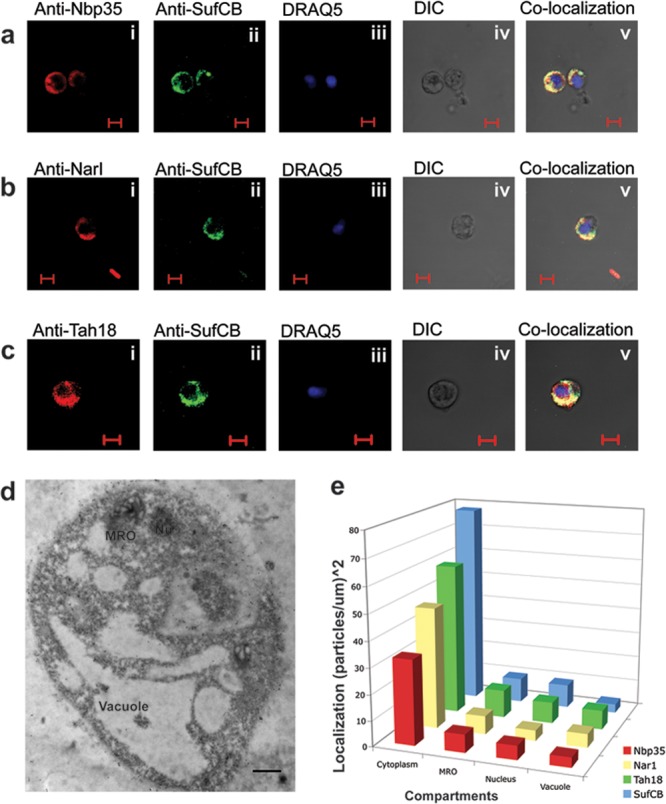
Immunolocalization of Nbp35, Nar1, and Tah18 in the Blastocystis sp. (a) Cellular localization of Nbp35 and SufCB in Blastocystis cells. (i) Rabbit anti-yeast Nbp35 antibody (1:200) detects the Blastocystis Nbp35 protein; (ii) localization of the Blastocystis SufCB antibody; (iii) DRAQ5 staining of the nuclei of Blastocystis; (iv) differential interference contrast (DIC) image of the cells used for immunofluorescence; (v) overlapping of the previous images showing the localization pattern of these proteins. (b) Cellular localization of components of Nar1 and SufCB in Blastocystis cells. (i) Rabbit anti-yeast Nar1 antibody (1:100) detects the Blastocystis Nbp35 protein; (ii) localization of the Blastocystis SufCB antibody; (iii) DRAQ5 staining of the nuclei of Blastocystis; (iv) differential interference contrast image of the cells used for immunofluorescence; (v) overlapping of the previous images showing the localization pattern of these proteins. (c) Cellular localization of components of Tah18 and SufCB in Blastocystis cells. (i) Rabbit anti-yeast Tah18 antibody (1:100) detects the Blastocystis Nbp35 protein; (ii) localization of the Blastocystis SufCB antibody; (iii) DRAQ5 staining of the nuclei of Blastocystis; (iv) differential interference contrast image of the cells used for immunofluorescence; (v) overlapping of the previous images showing the localization pattern of these proteins. (d) Immunogold microscopy image demonstrating the localization pattern of the anti-Nbp35 antibody. A higher-magnification image is shown in Fig. S7 in the supplemental material. (e) The densities of immunogold particle labeling in different compartments of Blastocystis cells suggest that Nbp35, Nar1, and Tah18, along with the previously published SufCB (18) proteins, are mainly localized in the cytosol of the parasite.
This work, along with a previous study (17), demonstrated that key components of the ISC and CIA systems are encoded by genes in the Blastocystis sp. NandII genome and that they localize in this organism's MROs and cytosol, respectively. However, the Blastocystis sp. also has a cytosolic SUF system (17), a machinery never before described in the eukaryotic cytoplasmic compartment (1). With these two cytosolic Fe/S cluster biogenesis systems, the Blastocystis sp. resembles Entamoeba and Mastigamoeba, both of which possess novel cytosolic NIF systems (16, 31), in addition to CIA components (presumed to be cytosolic in these organisms) (Table 1). For these organisms, it is unclear how Fe/S cluster biogenesis is partitioned between the two cytosolic systems and whether or not they function independently or in a coordinated fashion. Currently, it is not possible to genetically manipulate some of these organisms, making it difficult to study these systems in more detail.
TABLE 1.
Distribution of homologues of the CIA machinery among publicly available genomes or expressed sequence tag data from eukaryote taxaa
| Supergroup | Phylum | Species | Distribution |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dre2 | Tah18 | Nbp35 | Cfd1 | Nar1 | Cia1 | Cia2b | MMS19 | |||
| SAR | Apicomplexa | Babesia bovis | + | *+ | + | − | + | + | + | − |
| SAR | Apicomplexa | Cryptosporidium hominis | + | + | + | − | + | + | + | − |
| SAR | Apicomplexa | Cryptosporidium muris RN66 | + | + | + | − | + | + | + | − |
| SAR | Apicomplexa | Cryptosporidium parvum Iowa II | + | + | + | − | + | + | + | − |
| SAR | Apicomplexa | Plasmodium berghei | + | *+ | + | − | + | + | + | # |
| SAR | Apicomplexa | Plasmodium chabaudi | + | *+ | + | − | + | + | + | # |
| SAR | Apicomplexa | Plasmodium falciparum | + | *+ | + | − | + | + | + | # |
| SAR | Apicomplexa | Plasmodium knowlesi strain H | + | *+ | + | − | + | + | + | # |
| SAR | Apicomplexa | Plasmodium vivax | + | *+ | + | − | # | + | + | # |
| SAR | Apicomplexa | Theileria annulata strain Ankara | + | *+ | + | − | + | + | + | − |
| SAR | Apicomplexa | Theileria parva | + | *+ | + | − | + | + | + | − |
| SAR | Apicomplexa | Toxoplasma gondii | + | + | + | − | + | + | + | # |
| SAR | Ciliophora | Paramecium tetraurelia | + | + | + | − | + | + | + | # |
| SAR | Ciliophora | Tetrahymena thermophila SB210 | + | + | + | − | + | + | + | # |
| SAR | Perkinsida | Perkinsus marinus ATCC 50983 | + | *+ | + | − | + | + | + | # |
| SAR | Stramenopila | Aureococcus anophagefferens | + | + | + | − | + | + | + | − |
| SAR | Stramenopila | Hyaloperonospora | + | + | + | − | + | + | + | + |
| SAR | Stramenopila | Phaeodactylum tricornutum CCAP 1055/1 | + | + | + | − | + | + | + | − |
| SAR | Stramenopila | Phytophthora infestans T30-4 | + | + | + | − | + | + | + | + |
| SAR | Stramenopila | Phytophthora ramorum | + | + | + | − | + | + | + | + |
| SAR | Stramenopila | Phytophthora sojae | + | + | + | − | + | + | + | + |
| SAR | Stramenopila | Saprolegnia parasitica CBS 223.65 | + | + | + | − | + | + | + | + |
| SAR | Stramenopila | Nannochloropsis gaditana CCMP526 | + | + | + | − | + | + | + | + |
| SAR | Stramenopila | Thalassiosira pseudonana CCMP1335 | + | + | + | − | + | + | + | − |
| SAR | Stramenopila | Ectocarpus siliculosus | + | + | + | − | + | + | + | + |
| SAR | Stramenopila | Blastocystis sp. strain NandII | − | % | + | − | + | + | + | # |
| SAR | Stramenopila | Blastocystis sp. strain Sub 7 | − | % | + | − | + | + | + | # |
| SAR | Rhizaria | Bigelowiella natans | + | + | + | − | + | + | + | + |
| Hyptophyceae | Emiliania huxleyi | + | − | + | − | # | + | + | + | |
| Cryptophyta | Guillardia theta | + | + | + | + | + | + | + | + | |
| Amoebozoa | Dictyostelium purpureum | + | + | + | + | + | + | + | + | |
| Amoebozoa | Entamoeba dispar SAW760 | − | − | + | + | + | + | + | + | |
| Amoebozoa | Entamoeba histolytica HM-1:IMSS | − | − | + | + | + | + | + | + | |
| Amoebozoa | Mastigamoeba balamuthi | ? | ? | + | + | + | + | ? | ? | |
| Amoebozoa | Polysphondylium pallidum PN500 | + | + | + | + | + | + | + | + | |
| Archaeplastida | Viridiplantae | Chlamydomonas reinhardtii | + | + | + | − | + | + | + | − |
| Archaeplastida | Viridiplantae | Chlorella variabilis | + | + | + | − | + | + | + | + |
| Archaeplastida | Viridiplantae | Coccomyxa subellipsoidea C-169 | + | + | + | − | + | + | + | − |
| Archaeplastida | Viridiplantae | Micromonas pusilla | + | + | + | − | + | + | + | + |
| Archaeplastida | Viridiplantae | Micromonas sp. | + | + | + | − | + | + | + | + |
| Archaeplastida | Viridiplantae | Ostreococcus lucimarinus | + | + | + | − | + | + | + | + |
| Archaeplastida | Viridiplantae | Ostreococcus tauri | + | + | + | − | + | + | + | + |
| Archaeplastida | Viridiplantae | Volvox carteri | + | + | + | − | + | + | + | + |
| Archaeplastida | Rhodophyta | Cyanidioschyzon merolae | + | − | + | − | + | + | + | − |
| Archaeplastida | Viridiplantae | Arabidopsis thaliana | + | + | + | − | + | + | + | + |
| Archaeplastida | Viridiplantae | Brachypodium distachyon | + | + | + | − | + | + | + | + |
| Archaeplastida | Viridiplantae | Glycine max | + | + | + | − | + | + | + | + |
| Excavata | Euglenozoa | Leishmania braziliensis MHOM/BR/75/M2904 | + | + | + | + | + | + | + | # |
| Excavata | Euglenozoa | Leishmania infantum JPCM5 | + | + | + | + | + | + | + | # |
| Excavata | Euglenozoa | Leishmania major strain Friedlin | + | + | + | + | + | + | + | # |
| Excavata | Euglenozoa | Trypanosoma brucei | + | + | + | + | + | + | + | # |
| Excavata | Euglenozoa | Trypanosoma cruzi | + | + | + | + | + | + | + | # |
| Excavata | Fornicata | Giardia intestinalis | − | % | + | − | + | + | + | − |
| Excavata | Parabasalia | Trichomonas vaginalis G3 | − | − | + | + | + | + | + | − |
| Excavata | Heterolobosea | Naegleria gruberi | + | + | + | + | + | + | + | # |
| Opisthokonta | Ascomycota | Saccharomyces cerevisiae | + | + | + | + | + | + | + | + |
| Opisthokonta | Ascomycota | Schizosaccharomyces pombe | + | + | + | + | + | + | + | + |
| Opisthokonta | Ascomycota | Gibberella zeae PH-1 | + | + | + | + | + | + | + | + |
| Opisthokonta | Basidiomycota | Coprinopsis cinerea Okayama7#130 | + | + | + | + | + | + | + | + |
| Opisthokonta | Basidiomycota | Ustilago maydis 521 | + | + | + | + | + | + | + | + |
| Opisthokonta | Chytridiomycota | Batrachochytrium dendrobatidis | + | + | + | + | + | + | + | + |
| Opisthokonta | Microsporidia | Encephalitozoon cuniculi GB-M1 | + | + | + | + | + | + | + | − |
| Opisthokonta | Microsporidia | Enterocytozoon bieneusi H348 | + | *+ | + | + | # | + | + | − |
| Opisthokonta | Microsporidia | Nosema ceranae BRL01 | − | + | + | + | + | + | + | − |
| Opisthokonta | Choanoflagellida | Monosiga brevicollis | + | + | + | + | + | + | + | + |
| Opisthokonta | Metazoa | Apis mellifera | + | + | + | + | + | + | + | + |
| Opisthokonta | Metazoa | Bos taurus | + | + | + | + | + | + | + | + |
| Opisthokonta | Metazoa | Gallus gallus | + | + | + | + | + | + | + | + |
| Opisthokonta | Metazoa | Homo sapiens | + | + | + | + | + | + | + | + |
| Opisthokonta | Metazoa | Trichoplax adhaerens | + | + | + | + | + | + | + | + |
| Opisthokonta | Metazoa | Caenorhabditis elegans | + | + | + | − | + | + | + | + |
The presence (+) and absence (−) of homologues in complete genomes are indicated. ?, absence from an incomplete genome or transcriptomic data; *+, presence of a homologous sequence in which at least one domain is missing; %, Tah18 homologues for which the phylogenetic placement in the tree is unclear (see the text); #, presence of a potential but highly divergent homologue. SAR, Stramenopiles-Alveolates-Rhizaria.
Distribution and evolutionary history of the CIA machinery homologues in other microbial eukaryotes.
Although most of the eight CIA system components present in yeast are essential for its viability (9), our survey of Blastocystis sp. data (23, 32) allowed the identification of only five (potentially six; see the discussion of Tah18 below) of these proteins. Consequently, it is unclear how this pathway functions in Blastocystis and whether other eukaryotes also lack some of the components. To address this question, we carried out a phylogenomic analysis of each individual component.
(i) Dre2.
Dre2 is essential for the assembly of virtually all cytosolic and nuclear target Fe/S proteins in yeast and localizes mostly in the cytosol and partially in the mitochondrial intermembrane space (IMS) (33). The latter finding seems to suggest that Dre2 acts before the rest of the CIA components, an observation further supported by additional experimental evidence (10). Interestingly, Dre2 itself is a Fe/S cluster-containing protein whose maturation seems to occur independently of the CIA system (10). This suggests a putative functional dependence of Dre2 (and, thus, the CIA system) on another Fe/S cluster assembly pathway.
Dre2 had a highly conserved C terminus corresponding to a eukaryote-specific Ciapin1 domain that contained two Fe/S cluster-binding motifs, CX2CXC and CX2CX7CX2C, while the N terminus is more divergent (see Fig. S10 in the supplemental material). In some chlorophytes/embryophytes and metazoans, the N terminus contains a methyltransferase domain (see Fig. S10 in the supplemental material), supporting the recently proposed hypothesis that this part of Dre2 is homologous to the S-adenosylmethionine (SAM) methyltransferase protein family (34). Although the interaction of Dre2 and Tah18 has been shown to be indispensable for yeast survival, a few parasitic taxa (including Blastocystis, Entamoeba, Giardia, and Trichomonas) appear to be lacking a Dre2 homologue (Table 1). Apart from these absences (presumably because of secondary loss), the phylogenetic analysis was consistent with the vertical inheritance of Dre2 from the last eukaryotic common ancestor (LECA) within eukaryotes (see Fig. S11 in the supplemental material). The taxonomic distribution of the SAM domain seems to suggest that Dre2 in LECA had a SAM-Ciapin1 domain composition. It is possible that eukaryotic sequences in which a SAM domain was not detected have evolved beyond recognition, since most of them have an N-terminal sequence that aligns well with (and thus seems to be homologous to) the SAM domain of green plant/metazoan sequences.
(ii) Tah18.
Tah18 is a reductase with binding motifs for flavin mononucleotide (FMN), flavin adenine dinucleotide (FAD), and NAD cofactors (Fig. 2). Tah18 forms a complex with Dre2 that functions as an electron transfer chain by transferring two reducing equivalents from NADPH to Dre2 (10, 34, 35). Experimental evidence suggests that the Tah18/Dre2 complex disassociates under oxidative stress conditions, and as a result, Tah18 retargets to mitochondria in yeast (35), leading to the speculation that Tah18 promotes apoptosis under oxygen stress (34). Phylogenetic analyses show that Tah18 belongs to a large multiprotein family, most members of which share the canonical FMN-FAD-NAD domain organization and are involved in redox reactions. The pyruvate-NADPH oxidoreductase (PNO) and NADPH cytochrome P450 reductase (CPR) subfamilies are the closest homologues of Tah18 (Fig. 2). Our analyses show that the putative Blastocystis Tah18 sequences (Fig. 2) do not group with the rest of the stramenopile Tah18 homologues (Fig. 2, black square) but instead branch with the PNO sequences. While most eukaryotes possess an orthologue of Tah18, the protein seems to be absent in Entamoeba (Table 1) and potentially in Giardia, whose homologues are unclearly related to Tah18 or CPR (Fig. 2). There seems to be a similar phylogenetic profile of the presence/absence of canonical Tah18 and Dre2 (Table 1) that is consistent with them forming a functional module in the CIA system.
(iii) Nbp35/Cfd1.
Nbp35 and Cfd1 are homologous Mrp-like proteins and members of the P-loop NTPase family, which also includes the more distantly related mitochondrial Fe/S apoprotein IND1 (36). Both Nbp35 and Cfd1 proteins possess a Walker A box for ATP hydrolysis and two conserved C-terminal cysteines that serve as the binding sites of Fe/S clusters (11) (see Fig. S4 in the supplemental material). Nbp35 has an N-terminal ferredoxin-like domain that Cfd1 lacks (7) (see Fig. S4 in the supplemental material). While Nbp35 is universally distributed among eukaryotes, Cfd1 has a by far more patchy distribution: it is found only in opisthokonts, amoebozoa, discicristate excavates, and the cryptophyte Guillardia theta (Table 1). These results are in contrast to those presented in a recent publication (37) claiming the universal presence of the Cfd1 protein among eukaryotes (with the exception of the Plantae clade). As the authors of the aforementioned paper used a reciprocal blast approach and not a phylogenetic analysis, we suspect that their approach might have led to a misidentification of the two paralogous families.
In yeast, Cfd1 and Nbp35 form a heterotetramer that serves as the scaffold of cytosolic Fe/S cluster biogenesis (11, 38), and the absence of either of these two proteins in yeast results in cell death (11). However, in the Viridiplantae, which lack Cfd1, it has been shown that Nbp35 forms a homodimer that delivers Fe/S clusters to apoproteins (39). This is possibly how Nbp35 functions in the Blastocystis sp. and in other organisms that lack the Cfd1 orthologue.
Our phylogenetic analysis shows that the Nbp35 and Cfd1 proteins are paralogues and that the duplication event that gave rise to them most likely preceded eukaryotic diversification; thus, these two paralogues existed in the LECA (Fig. 3a). In the Nbp35 subtree, stramenopiles (including Blastocystis) cluster with Viridiplantae and Emiliania huxleyi, albeit with weak support. However, most members of this group share an insertion in the C-terminal region of the protein that ranges in length from 2 to 45 amino acids and that is fairly conserved in sequence (Fig. 3b). A few taxa within this group appear to have lost part or nearly all of the insertion secondarily (e.g., Volvox and Chlamydomonas). The observed branching pattern of Nbp35 is at odds with current conceptions of eukaryotic phylogeny (40) that place stramenopiles with alveolates and Rhizaria. A potential explanation is that the gene has been laterally transferred between stramenopiles, Viridiplantae, and Emiliania, although, if this is correct, the numbers and directions of transfer events could not be determined from the poorly resolved phylogeny. However, the lack of the first of the two conserved central cysteine residues of the C-terminal motif (CPXC) in Blastocystis and Viridiplantae Nbp35 sequences supports the possibility of a recent transfer event involving these two lineages (see Fig. S4 in the supplemental material).
Finally, phylogenetic reconstructions also revealed that bacterial ApbC proteins are the closest prokaryotic homologues of Nbp35/Cfd1. Interestingly, experimental evidence suggests that the ApbC of Salmonella enterica functions in Fe/S cluster biogenesis pathways (41) and homologues from several archaea functionally complement ApbC mutants of S. enterica (42). In addition, biochemical analyses of bacterial ApbC homologues showed that they bind and transfer Fe/S clusters to a yeast apoprotein in vitro (40). Collectively, these results suggest that ApbC/Nbp35/Cfd1 homologues were involved in Fe/S cluster biosynthesis prior to the diversification of the three domains of life.
(iv) Nar1.
Nar1 belongs to the hydrogenase-related protein family; however, it is not involved in hydrogen production (8). Even though Nar1 is predominantly localized in the cytosol, it also appears to be partly membrane associated in yeast (5). The protein acts late in the pathway, where it facilitates the transfer of Fe/S clusters to target proteins. All Nar1 homologues have a highly conserved N terminus ferredoxin-like domain (CXNCX2CX2C) and a CXNCXNCX3C C-terminal motif, the so-called H cluster, which has a likely role in Fe/S cluster coordination (8) (see Fig. S3 in the supplemental material). This protein has a wide distribution among eukaryotes (Table 1), and phylogenetic analysis shows that all Nar1 homologues cluster together with strong bootstrap support (see Fig. S12 in the supplemental material) and represent paralogues of the hydrogenase-like proteins found in anaerobic eukaryotes. In the Nar1 subtree, Blastocystis branches within the alveolates and not with other stramenopiles; however, this relationship is not strongly supported (see Fig. S12 in the supplemental material).
(v) Cia1.
Cia1 is a W40-repeat protein responsible for the last step of the Fe/S cluster assembly process. WD40 proteins are usually known to coordinate the assembly of multiprotein complexes by functioning as a docking site for other proteins. As such, Cia1 interacts with Nar1 and facilitates the transfer of Fe/S clusters to target proteins (4, 43). In yeast, depletion of Cia1 results in the loss of activities of cytosolic and nuclear Fe/S cluster-bearing enzymes (4). The essential role of this protein in the CIA system is further supported by its universal presence in all eukaryotes (Table 1) and by the strict conservation of the 7-bladed beta-propeller structure (43) across all homologues (see Fig. S2 in the supplemental material). The Cia1 phylogeny is poorly resolved but does not appear to indicate any lateral gene transfer between eukaryotic lineages (see Fig. S13 in the supplemental material).
(vi) MMS19.
MMS19 is the most recently discovered component of the CIA machinery (9, 12). This chaperone forms a complex with Nar1 and Cia1 components. This protein is patchily distributed among eukaryotes, suggesting that it has been lost numerous times along various lineages (Table 1). However, a high degree of divergence between homologous sequences was observed. We thus cannot exclude the possibility that other eukaryotic homologues have diverged beyond recognition and that MMS19 is universally present but unrecognizable at the sequence level in many eukaryotes (see Materials and Methods). In particular, the large degree of sequence divergence observed specifically in homologues from Blastocystis spp., alveolates, and excavates suggests that the protein may have a different function in these lineages. Although the phylogeny is poorly resolved, the presence of MMS19 homologues in most organisms of all major groups, as well as the absence of obvious LGT between eukaryotic lineages, suggests that this component already existed in LECA (see Fig. S14 in the supplemental material). As no homologues of this protein have been identified in prokaryotes, this component likely evolved after the divergence of eukaryotes from their prokaryotic ancestor, although here again, the high rate of evolution of this gene might preclude the identification of prokaryotic homologues.
(vii) Cia2.
The Cia2 protein family has recently been identified as acting at the end of the CIA pathway (9, 12). In animals, two paralogous members of this family, Cia2A and Cia2B, are present. Cia2B associates with Cia1 and MMS19 to form the so-called CIA-targeting complex, which binds directly to the coordinating cysteines of Fe/S clusters of most nuclear/cytosolic apoproteins that are matured by the complex (44). This suggests that Cia2B has a role in connecting donor and acceptor Fe/S proteins (9). In contrast, Cia2A seems to be specifically involved in cellular iron homeostasis through the maturation of iron regulatory protein 1 and the stabilization of iron regulatory protein 2 (45).
Phylogenetic analyses showed that virtually all eukaryotes, including Blastocystis, possess at least one homologue of the Cia2 family (see Fig. S15 in the supplemental material). The phylogenetic tree is poorly supported overall but seems to show a basal split between two monophyletic groups. The first one includes the human Cia2B homologue and contains sequences from all eukaryotic lineages (with very few exceptions), suggesting that this protein was already present in LECA. The second group, which includes the human Cia2A, displays a much narrower taxonomic distribution, with orthologues present only in some lineages of animals, Amoebozoa, ciliates, Trypanosoma, and red algae. Given the very weak support for this clade and its sparse taxonomic distribution, the existence of a second paralogue in these lineages can be explained by three different hypotheses: (i) the two paralogues were already present in LECA and have been independently lost many times along the eukaryotic tree, (ii) the duplication event occurred in one of these eukaryotic lineages and the gene has subsequently been laterally transferred to the other lineages represented in the Cia2A clade, or (iii) several duplication events occurred independently and the second copy of each lineage artifactually groups with the others in the tree.
Proteins of the Cia2 families are composed of a unique DUF59 domain and represent the only proteins carrying this domain in eukaryotic organisms (with the exception of the HCF101 protein, discussed below). Cia2 possess homologues in both archaea and bacteria. Surprisingly, the closest prokaryotic homologues can be found among bacteria and correspond to a DUF59-containing protein called SufT. The function of SufT is unknown, but members of this family are commonly found in operons for the iron/sulfur cluster biosynthesis SUF system. This is particularly noteworthy since, to our knowledge, it is the first reported evidence for an evolutionary link between the SUF and the CIA pathways.
Other bacterial and archaeal homologues are found in operons associated with phenylacetic acid degradation pathways and are represented by the PaaD protein (46, 47). PaaD is essential in vivo and is thought to be involved in the maturation of the other Fe/S cluster-baring members of the operon (48). Finally, the only other eukaryotic protein carrying a DUF59 domain is the plastid HCF101 (high chlorophyll fluorescence 101) protein. HCF101 is essential for the accumulation of two [4Fe-4S]-containing chloroplast proteins (49). Interestingly, this protein is a fusion of a DUF59 domain, a member of the P-loop NTPase superfamily (a superfamily that also contains the Nbp35 and Cfd1 proteins of the CIA pathway), and a DUF971 domain.
On the origin and evolution of the CIA machinery.
Our phylogenetic analyses suggest that all of the eight known components of the CIA system originated before the diversification of extant eukaryotic lineages and that LECA had a complete and functional CIA pathway, likely resembling the one found in most animals and fungi. While some components have been universally conserved during eukaryotic evolution (i.e., Nbp35, Nar1, Cia1, and Cia2), a few of the CIA proteins, such as Cfd1 and MMS19, display a very patchy distribution (Table 1). According to the corresponding phylogenetic trees, this distribution is more likely to be explained by multiple loss events than by lateral gene transfers (see above). This presence/absence pattern also has no obvious correlation with organismal lifestyle. However, we can speculate that it may correlate with the complement of Fe/S cluster-containing proteins and the type of Fe/S clusters needed and assembled through the CIA pathway in these organisms. In contrast, the presence/absence pattern of Tah18 and Dre2 seems to have a clearer correlation with organismal lifestyle. With the exception of Cryptosporidium species, all of the protists that live in hypoxic environments and contain mitochondrion-related organelles (i.e., Entamoeba, Mastigamoeba, Trichomonas, Giardia, and Blastocystis) lack Dre2 and a canonical Tah18 orthologue (Table 1). This is unexpected because the NADPH-Tah18-Dre2 electron transfer chain was shown (in yeast) to be required early in the biosynthetic pathway for the incorporation of the Fe/S cluster into all proteins carrying stable Fe/S clusters (i.e., not scaffold proteins), including the CIA Fe/S proteins Nbp35 and Nar1 (10). Consequently, it seems like another protein (or pair of proteins) carries out the Tah18-Dre2 functions when the latter are absent. For example, the bona fide Tah18 orthologue, which seems to have been lost in some protists, could have been replaced by another member of the superfamily showing the same FMN-FAD-NAD domain organization (e.g., as found in Blastocystis and Giardia). In contrast, Dre2 could have been replaced by a nonhomologous protein, since no paralogue of this component can be found in the aforementioned protists. This is consistent with the fact that eukaryotic diflavin reductases can be coupled with various electron-accepting proteins (e.g., cytochrome P450, methionine synthase, nitric oxide synthase 1). It is thus possible that Tah18 is transferring electrons to an analogue of Dre2 (i.e., a nonhomologous but functionally similar protein). It is interesting to correlate the loss of Dre2 with the presence in some of these anaerobes of recently acquired unique cytosolic Fe/S biosynthetic proteins; NifS/NifU in the cases of Entamoeba and Mastigamoeba and a SufCB fusion protein in the case of Blastocystis. It is possible that the NIF and SUF components have a Dre2-like role in the electron transfer chain and therefore interface with the remainder of the CIA system in these organisms.
In addition, the Tah18-Dre2 complex putatively interfaces with the mitochondrial ISC system in yeast and is implicated in processing its product, compound X (1). Compound X is thought to be a sulfur-containing compound of an as yet unknown nature that serves as the sulfur donor for the CIA pathway. It is produced by the mitochondrion-localized ISC assembly machinery and exported from the mitochondrial matrix to the cytosol. In Giardia mitosomes, the only proteins carrying Fe/S clusters are the components of the ISC Fe/S cluster assembly machinery itself. Consequently, one of the main roles of mitosomes could be to export compound X to other cellular compartments (50). The potentially essential role of compound X for the proper functioning of the CIA pathway is another line of evidence that a Tah18-Dre2 analogue must exist in this organism.
The various patterns of the presence/absence of components across eukaryotes suggest that while there are a number of core essential components of the CIA system (e.g., Nbp35, Cia1, Cia2, and Nar1), the remaining set of proteins appears to be more evolutionarily plastic. Since this pathway has been extensively studied in only a few opisthokont lineages, further investigations focusing on microbial eukaryotes will likely provide us with insights into other novel components involved in the CIA system that may functionally replace some of the less conserved known CIA proteins. In any case, a broader study of the CIA system and its target apoproteins across eukaryotic diversity should illuminate both universal ancestral and lineage-specific functions of this essential pathway.
Supplementary Material
ACKNOWLEDGMENTS
This work, E.G., and D.G. were supported by operating grant MOP-62809 from the Canadian Institutes of Health Research awarded to A.J.R. A.D.T. was supported by a Marie Curie international outgoing fellowship, and L.E. was supported by a postdoctoral fellowship from the Tula Foundation. A.J.R. acknowledges support from the Canadian Institute for Advanced Research Program in Integrated Microbial Biodiversity.
We thank Roland Lill for providing us with the antibodies against the Nbp35, Nar1, and Tah18 proteins. We also thank Mary Ann Trevors for her assistance on the electron microscopy and Stephen Whitefield for his guidance on using the Cellular & Molecular Digital Imaging facility. Finally, we are thankful to the three anonymous reviewers for valuable comments.
Footnotes
Published ahead of print 15 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00158-13.
REFERENCES
- 1.Lill R. 2009. Function and biogenesis of iron-sulphur proteins. Nature 460:831–838. 10.1038/nature08301 [DOI] [PubMed] [Google Scholar]
- 2.Balk J, Lobreaux S. 2005. Biogenesis of iron-sulfur proteins in plants. Trends Plant Sci. 10:324–331. 10.1016/j.tplants.2005.05.002 [DOI] [PubMed] [Google Scholar]
- 3.Lill R, Muhlenhoff U. 2008. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77:669–700. 10.1146/annurev.biochem.76.052705.162653 [DOI] [PubMed] [Google Scholar]
- 4.Balk J, Aguilar Netz DJ, Tepper K, Pierik AJ, Lill R. 2005. The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron-sulfur protein assembly. Mol. Cell. Biol. 25:10833–10841. 10.1128/MCB.25.24.10833-10841.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balk J, Pierik AJ, Netz DJ, Muhlenhoff U, Lill R. 2004. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 23:2105–2115. 10.1038/sj.emboj.7600216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hausmann A, Aguilar Netz DJ, Balk J, Pierik AJ, Muhlenhoff U, Lill R. 2005. The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery. Proc. Natl. Acad. Sci. U. S. A. 102:3266–3271. 10.1073/pnas.0406447102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roy A, Solodovnikova N, Nicholson T, Antholine W, Walden WE. 2003. A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J. 22:4826–4835. 10.1093/emboj/cdg455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urzica E, Pierik AJ, Muhlenhoff U, Lill R. 2009. Crucial role of conserved cysteine residues in the assembly of two iron-sulfur clusters on the CIA protein Nar1. Biochemistry 48:4946–4958. 10.1021/bi900312x [DOI] [PubMed] [Google Scholar]
- 9.Stehling O, Vashisht AA, Mascarenhas J, Jonsson ZO, Sharma T, Netz DJ, Pierik AJ, Wohlschlegel JA, Lill R. 2012. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science 337:195–199. 10.1126/science.1219723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Netz DJ, Stumpfig M, Dore C, Muhlenhoff U, Pierik AJ, Lill R. 2010. Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat. Chem. Biol. 6:758–765. 10.1038/nchembio.432 [DOI] [PubMed] [Google Scholar]
- 11.Netz DJ, Pierik AJ, Stumpfig M, Bill E, Sharma AK, Pallesen LJ, Walden WE, Lill R. 2012. A bridging [4Fe-4S] cluster and nucleotide binding are essential for function of the Cfd1-Nbp35 complex as a scaffold in iron-sulfur protein maturation. J. Biol. Chem. 287:12365–12378. 10.1074/jbc.M111.328914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gari K, Leon Ortiz AM, Borel V, Flynn H, Skehel JM, Boulton SJ. 2012. MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science 337:243–245. 10.1126/science.1219664 [DOI] [PubMed] [Google Scholar]
- 13.Papatriantafyllou M. 2012. DNA metabolism: MMS19: CIA agent for DNA-linked affairs. Nat. Rev. Mol. Cell Biol. 13:538–539. 10.1038/nrm3411 [DOI] [PubMed] [Google Scholar]
- 14.Maralikova B, Ali V, Nakada-Tsukui K, Nozaki T, van der Giezen M, Henze K, Tovar J. 2010. Bacterial-type oxygen detoxification and iron-sulfur cluster assembly in amoebal relict mitochondria. Cell. Microbiol. 12:331–342. 10.1111/j.1462-5822.2009.01397.x [DOI] [PubMed] [Google Scholar]
- 15.Mi-ichi F, Abu Yousuf M, Nakada-Tsukui K, Nozaki T. 2009. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc. Natl. Acad. Sci. U. S. A. 106:21731–21736. 10.1073/pnas.0907106106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nyvltova E, Sutak R, Harant K, Sedinova M, Hrdy I, Paces J, Vlcek C, Tachezy J. 2013. NIF-type iron-sulfur cluster assembly system is duplicated and distributed in the mitochondria and cytosol of Mastigamoeba balamuthi. Proc. Natl. Acad. Sci. U. S. A. 110:7371–7376. 10.1073/pnas.1219590110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsaousis AD, Ollagnier de Choudens S, Gentekaki E, Long S, Gaston D, Stechmann A, Vinella D, Py B, Fontecave M, Barras F, Lukes J, Roger AJ. 2012. Evolution of Fe/S cluster biogenesis in the anaerobic parasite Blastocystis. Proc. Natl. Acad. Sci. U. S. A. 109:10426–10431. 10.1073/pnas.1116067109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Muller WE, Wetter T, Suhai S. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159. 10.1101/gr.1917404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nasirudeen AM, Tan KS. 2004. Isolation and characterization of the mitochondrion-like organelle from Blastocystis hominis. J. Microbiol. Methods 58:101–109. 10.1016/j.mimet.2004.03.008 [DOI] [PubMed] [Google Scholar]
- 20.Lantsman Y, Tan KS, Morada M, Yarlett N. 2008. Biochemical characterization of a mitochondrial-like organelle from Blastocystis sp. subtype 7. Microbiology 154:2757–2766. 10.1099/mic.0.2008/017897-0 [DOI] [PubMed] [Google Scholar]
- 21.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222. 10.1093/nar/gkp985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denoeud F, Roussel M, Noel B, Wawrzyniak I, Da Silva C, Diogon M, Viscogliosi E, Brochier-Armanet C, Couloux A, Poulain J, Segurens B, Anthouard V, Texier C, Blot N, Poirier P, Ng GC, Tan KS, Artiguenave F, Jaillon O, Aury JM, Delbac F, Wincker P, Vivares CP, El Alaoui H. 2011. Genome sequence of the stramenopile Blastocystis, a human anaerobic parasite. Genome Biol. 12:R29. 10.1186/gb-2011-12-3-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schultz J, Milpetz F, Bork P, Ponting CP. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95:5857–5864. 10.1073/pnas.95.11.5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Criscuolo A, Gribaldo S. 2010. BMGE (block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10:210. 10.1186/1471-2148-10-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 28.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25:1307–1320. 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- 29.Shimodaira H, Hasegawa M. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246–1247. 10.1093/bioinformatics/17.12.1246 [DOI] [PubMed] [Google Scholar]
- 30.Shi X, Gu H, Susko E, Field C. 2005. The comparison of the confidence regions in phylogeny. Mol. Biol. Evol. 22:2285–2296. 10.1093/molbev/msi226 [DOI] [PubMed] [Google Scholar]
- 31.Ali V, Nozaki T. 2013. Iron-sulphur clusters, their biosynthesis, and biological functions in protozoan parasites. Adv. Parasitol. 83:1–92. 10.1016/B978-0-12-407705-8.00001-X [DOI] [PubMed] [Google Scholar]
- 32.Stechmann A, Hamblin K, Perez-Brocal V, Gaston D, Richmond GS, van der Giezen M, Clark CG, Roger AJ. 2008. Organelles in Blastocystis that blur the distinction between mitochondria and hydrogenosomes. Curr. Biol. 18:580–585. 10.1016/j.cub.2008.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Lyver ER, Nakamaru-Ogiso E, Yoon H, Amutha B, Lee DW, Bi E, Ohnishi T, Daldal F, Pain D, Dancis A. 2008. Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol. Cell. Biol. 28:5569–5582. 10.1128/MCB.00642-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soler N, Craescu CT, Gallay J, Frapart YM, Mansuy D, Raynal B, Baldacci G, Pastore A, Huang ME, Vernis L. 2012. A S-adenosylmethionine methyltransferase-like domain within the essential, Fe-S-containing yeast protein Dre2. FEBS J. 279:2108–2119. 10.1111/j.1742-4658.2012.08597.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vernis L, Facca C, Delagoutte E, Soler N, Chanet R, Guiard B, Faye G, Baldacci G. 2009. A newly identified essential complex, Dre2-Tah18, controls mitochondria integrity and cell death after oxidative stress in yeast. PLoS One 4:e4376. 10.1371/journal.pone.0004376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leipe DD, Wolf YI, Koonin EV, Aravind L. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317:41–72. 10.1006/jmbi.2001.5378 [DOI] [PubMed] [Google Scholar]
- 37.Basu S, Leonard JC, Desai N, Mavridou DA, Tang KH, Goddard AD, Ginger ML, Lukes J, Allen JW. 2013. Divergence of Erv1-associated mitochondrial import and export pathways in trypanosomes and anaerobic protists. Eukaryot. Cell 12:343–355. 10.1128/EC.00304-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Netz DJ, Pierik AJ, Stumpfig M, Muhlenhoff U, Lill R. 2007. The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol. Nat. Chem. Biol. 3:278–286. 10.1038/nchembio872 [DOI] [PubMed] [Google Scholar]
- 39.Bych K, Netz DJ, Vigani G, Bill E, Lill R, Pierik AJ, Balk J. 2008. The essential cytosolic iron-sulfur protein Nbp35 acts without Cfd1 partner in the green lineage. J. Biol. Chem. 283:35797–35804. 10.1074/jbc.M807303200 [DOI] [PubMed] [Google Scholar]
- 40.Adl SM, Simpson AG, Lane CE, Lukes J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, Heiss A, Hoppenrath M, Lara E, Le Gall L, Lynn DH, McManus H, Mitchell EA, Mozley-Stanridge SE, Parfrey LW, Pawlowski J, Rueckert S, Shadwick L, Schoch CL, Smirnov A, Spiegel FW. 2012. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59:429–493. 10.1111/j.1550-7408.2012.00644.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyd JM, Pierik AJ, Netz DJ, Lill R, Downs DM. 2008. Bacterial ApbC can bind and effectively transfer iron-sulfur clusters. Biochemistry 47:8195–8202. 10.1021/bi800551y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyd JM, Drevland RM, Downs DM, Graham DE. 2009. Archaeal ApbC/Nbp35 homologs function as iron-sulfur cluster carrier proteins. J. Bacteriol. 191:1490–1497. 10.1128/JB.01469-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srinivasan V, Netz DJ, Webert H, Mascarenhas J, Pierik AJ, Michel H, Lill R. 2007. Structure of the yeast WD40 domain protein Cia1, a component acting late in iron-sulfur protein biogenesis. Structure 15:1246–1257. 10.1016/j.str.2007.08.009 [DOI] [PubMed] [Google Scholar]
- 44.van Wietmarschen N, Moradian A, Morin GB, Lansdorp PM, Uringa EJ. 2012. The mammalian proteins MMS19, MIP18, and ANT2 are involved in cytoplasmic iron-sulfur cluster protein assembly. J. Biol. Chem. 287:43351–43358. 10.1074/jbc.M112.431270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stehling O, Mascarenhas J, Vashisht AA, Sheftel AD, Niggemeyer B, Rosser R, Pierik AJ, Wohlschlegel JA, Lill R. 2013. Human CIA2A-FAM96A and CIA2B-FAM96B integrate iron homeostasis and maturation of different subsets of cytosolic-nuclear iron-sulfur proteins. Cell Metab. 18:187–198. 10.1016/j.cmet.2013.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferrandez A, Minambres B, Garcia B, Olivera ER, Luengo JM, Garcia JL, Diaz E. 1998. Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J. Biol. Chem. 273:25974–25986 [DOI] [PubMed] [Google Scholar]
- 47.Olivera ER, Minambres B, Garcia B, Muniz C, Moreno MA, Ferrandez A, Diaz E, Garcia JL, Luengo JM. 1998. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc. Natl. Acad. Sci. U. S. A. 95:6419–6424. 10.1073/pnas.95.11.6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grishin AM, Ajamian E, Tao L, Zhang L, Menard R, Cygler M. 2011. Structural and functional studies of the Escherichia coli phenylacetyl-CoA monooxygenase complex. J. Biol. Chem. 286:10735–10743. 10.1074/jbc.M110.194423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwenkert S, Netz DJ, Frazzon J, Pierik AJ, Bill E, Gross J, Lill R, Meurer J. 2010. Chloroplast HCF101 is a scaffold protein for [4Fe-4S] cluster assembly. Biochem. J. 425:207–214. 10.1042/BJ20091290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jedelsky PL, Dolezal P, Rada P, Pyrih J, Smid O, Hrdy I, Sedinova M, Marcincikova M, Voleman L, Perry AJ, Cambo Beltran N, Lithgow T, Tachezy J. 2011. The minimal proteome in the reduced mitochondrion of the parasitic protist Giardia intestinalis. PLoS One 6:e17285. 10.1371/journal.pone.0017285 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.