Abstract
Pyrimidines are important nucleic acid precursors which are constantly synthesized, degraded, and rebuilt in the cell. Four degradation pathways, two of which are found in eukaryotes, have been described. One of them, the URC pathway, has been initially discovered in our laboratory in the yeast Lachancea kluyveri. Here, we present the global changes in gene expression in L. kluyveri in response to different nitrogen sources, including uracil, uridine, dihydrouracil, and ammonia. The expression pattern of the known URC genes, URC1-6, helped to identify nine putative novel URC genes with a similar expression pattern. The microarray analysis provided evidence that both the URC and PYD genes are under nitrogen catabolite repression in L. kluyveri and are induced by uracil or dihydrouracil, respectively. We determined the function of URC8, which was found to catalyze the reduction of malonate semialdehyde to 3-hydroxypropionate, the final degradation product of the pathway. The other eight genes studied were all putative permeases. Our analysis of double deletion strains showed that the L. kluyveri Fui1p protein transported uridine, just like its homolog in Saccharomyces cerevisiae, but we demonstrated that is was not the only uridine transporter in L. kluyveri. We also showed that the L. kluyveri homologs of DUR3 and FUR4 do not have the same function that they have in S. cerevisiae, where they transport urea and uracil, respectively. In L. kluyveri, both of these deletion strains grew normally on uracil and urea.
INTRODUCTION
Pyrimidines, together with purines, are important precursors for many macromolecules in the cell, such as nucleic acids, and their internal pools are strictly regulated to provide the cell with a balanced supply of nucleotides. The main metabolic pathways affecting the size of the intracellular nucleotide pools are de novo, salvage, and catabolic pathways. In addition, degradation of pyrimidines and other macromolecules is a way for many microorganisms to obtain nitrogen when more preferable, easily accessible sources are scarce (1).
As a model to study pyrimidine catabolism, we have previously developed the yeast Lachancea kluyveri (2, 3) (formerly Saccharomyces kluyveri [4]), which, unlike its distant relative Saccharomyces cerevisiae, is able to degrade and utilize the nitrogen from most known intermediates in nucleotide metabolism (5).
Four different pyrimidine degradation pathways have been described so far, of which the reductive pathway is the most common, being found among most eukaryotes and some bacteria. The URC pathway is found in several fungi and bacteria, while the oxidative and Rut pathways have been found in only a few bacteria (6–9). The reductive pathway starts with the reduction of uracil to dihydrouracil by a dihydropyrimidine dehydrogenase (EC 1.3.1.2), an enzyme which has not been found in any fungi. The pyrimidine ring is then opened by dihydropyrimidinase (EC 3.5.2.2) to create β-ureidopropionate, which becomes hydrolyzed to β-alanine by a β-alanine synthase (EC 3.5.1.6). The β-alanine can then be further degraded by a beta-alanine aminotransferase (EC 2.6.1.2), yielding malonate semialdehyde (10). In L. kluyveri the enzymes required for the second, third, and fourth steps, encoded by PYD2, PYD3, and PYD4, respectively, have been identified and characterized (11–15), while S. cerevisiae is not able to perform any of these steps. Many other yeasts, however, also have the ability to degrade one or more of the reductive pathway intermediates (16). Despite the complete lack of dihydropyrimidine dehydrogenase-encoding genes among fungi, many yeasts are able to degrade uracil (16).
We have previously discovered in L. kluyveri a novel pyrimidine catabolic pathway, the URC pathway, found in many yeasts, fungi, and bacteria, that represents the only known way for yeast to degrade uracil (17). Six loci, URC1-6, have been found to be necessary for the degradation of uracil (Fig. 1). URC2 encodes a Zn2Cys6 zinc finger transcription factor, URC3 and URC5 together encode a urea amidolyase (EC 6.3.4.6 and EC 3.5.1.54), and URC6 encodes a uracil phosphoribosyltransferase (EC 2.4.2.9). URC1 and URC4 are conserved genes, encoding proteins of unknown function. The outcomes of the pathway are 3-hydroxypropionate, which is excreted, and urea, which is further hydrolyzed to yield ammonia and carbon dioxide (17). Ribosyl-urea or phosphoribosyl-urea has been proposed to be an intermediate of the pathway (18), but other intermediates, as well as functions of some of the involved enzymes, remain largely unknown.
FIG 1.
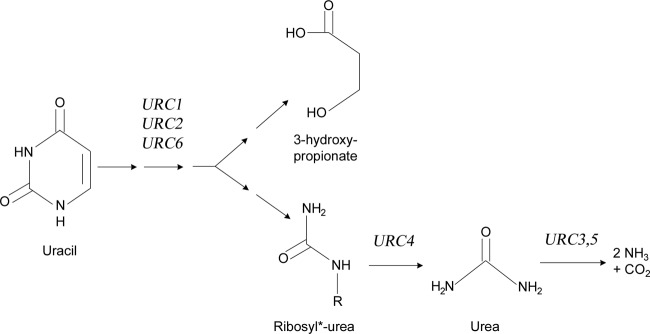
Overview of the known parts of the URC pathway. Uracil is transformed by the action of the URC1 and URC6 gene products and possibly one or several other enzymes. This results in the formation of 3-hydroxypropionate, which is excreted, and (phospho)ribosyl urea, which is somehow degraded to urea (17, 18). Urc4p has been proposed to catalyze the degradation of the (phospho)ribosyl urea. The nitrogen is then released in the form of ammonia by the action of Urc3,5 enzyme. URC2 encodes a transcription factor which is likely required for a functional URC pathway (17).
It is apparent that the four pyrimidine degradation pathways do not share many intermediates and enzymatic activities, but 3-hydroxypropionate is the final product in both the URC and Rut pathways. In Escherichia coli, 3-hydroxypropionate is produced by the action of RutE or YdfG through the reduction of malonate semialdehyde (19).
In this report, we performed a global expression analysis in L. kluyveri and screened for additional URC genes. Microarrays were used to study changes in gene expression of all 5,321 predicted genes in response to different pyrimidines and other nitrogen-containing metabolites present as the sole nitrogen source. Here, we can show that the known URC genes are all upregulated by uracil, uridine, and, to some extent, dihydrouracil but are downregulated by ammonia, which means they are under nitrogen catabolite repression (1). Nine putative URC genes were further studied through deletion analysis, and subsequently the function of a new URC gene, URC8, encoding a short-chain dehydrogenase, was determined through an enzyme assay.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The L. kluyveri strains used and developed in this study can be found in Table 1. The yeast strains were grown in rich yeast extract-peptone-dextrose (YPD) media unless stated differently. For selection, 100 μg/ml G418 was used.
TABLE 1.
L. kluyveri strains used and developed during this research project
| No. | Strain | Genotypea | Originating strain |
|---|---|---|---|
| 1 | Y057 | Diploid, prototroph | NRRL Y-12651 |
| 2 | Y156 | MATα ura3 | GRY1175 (J. Strathern) |
| 3 | Y1392 | MATa prototroph | Y057 |
| 4 | Y1161 | MATα ura3 urc2::KANMX3 | Y156 |
| 5 | Y1616 | MATa urc8::KANMX3 | Y1392 |
| 6 | Y1694 | MATa Lkdur3::KANMX3 | Y1392 |
| 7 | Y1700 | MATα ura3 Lkfui1::KANMX3 | Y156 |
| 8 | Y1701 | MATα ura3 Lkfur4::KANMX3 | Y156 |
| 9 | Y1702 | MATα ura3 Lkdal5::KANMX3 | Y156 |
| 10 | Y1703 | MATα ura3 Lkdal4::KANMX3 | Y156 |
| 11 | Y1705 | MATα ura3 Lkdur3::KANMX3 | Y156 |
| 12 | Y1704 | MATα ura3 SAKL0A07480::KANMX3 | Y156 |
| 13 | Y1773 | MATα ura3 Lkfui1::KANMX3 Lkfui1 hom::URA3 | Y156 |
| 14 | Y1774 | MATα ura3 Lkfui1::KANMX3 Lkdal4::URA3 | Y156 |
| 15 | Y1775 | MATα ura3 Lkfui1::KANMX3 Lkfur4::URA3 | Y156 |
| 16 | Y1776 | MATα ura3 Lkfui1::KANMX3 Lkdal5::URA3 | Y156 |
| 17 | Y1777 | MATα ura3 Lkfui1::KANMX3 Lkdal5 hom::URA3 | Y156 |
| 18 | Y1778 | MATα ura3 Lkfui1::KANMX3 SAKL0A07480::URA3 | Y156 |
| 19 | Y1779 | MATα ura3 Lkfui1::KANMX3 Lkdur3::URA3 | Y156 |
| 20 | Y1780 | MATα ura3 Lkfui1 hom::KANMX3 | Y156 |
| 21 | Y1781 | MATα ura3 Lkdal5 hom::KANMX3 | Y156 |
| 22 | Y1782 | MATα ura3 Lkdur3::KANMX3 SAKL0A07480::URA3 | Y156 |
| 23 | Y1783 | MATα ura3 Lkfui1 hom::KANMX3 Lkdal4::URA3 | Y156 |
| 24 | Y1784 | MATα ura3 Lkfui1 hom::KANMX3 Lkfur4::URA3 | Y156 |
| 25 | Y1785 | MATα ura3 Lkfui1 hom::KANMX3 Lkdal5::URA3 | Y156 |
| 26 | Y1786 | MATα ura3 Lkfui1 hom::KANMX3 Lkdal5 hom::URA3 | Y156 |
| 27 | Y1787 | MATα ura3 Lkfui1 hom::KANMX3 SAKL0A07480::URA3 | Y156 |
| 28 | Y1788 | MATα ura3 Lkfui1 hom::KANMX3 Lkdur3::URA3 | Y156 |
| 29 | Y1789 | MATα ura3 Lkdal5::KANMX3 Lkdal5 hom::URA3 | Y156 |
| 30 | Y1790 | MATα ura3 Lkdal5::KANMX3 Lkdal4::URA3 | Y156 |
| 31 | Y1791 | MATα ura3 Lkdal5::KANMX3 Lkfur4::URA3 | Y156 |
| 32 | Y1792 | MATα ura3 Lkdal5::KANMX3 Lkdur3::URA3 | Y156 |
| 33 | Y1793 | MATα ura3 Lkdal5::KANMX3 SAKL0A07480::URA3 | Y156 |
| 34 | Y1794 | MATα ura3 Lkdal5 hom::KANMX3 Lkdal4::URA3 | Y156 |
| 35 | Y1795 | MATα ura3 Lkdal5 hom::KANMX3 Lkfur4::URA3 | Y156 |
| 36 | Y1796 | MATα ura3 Lkdal5 hom::KANMX3 Lkdur3::URA3 | Y156 |
| 37 | Y1797 | MATα ura3 Lkdal5 hom::KANMX3 SAKL0A07480::URA3 | Y156 |
| 38 | Y1798 | MATα ura3 Lkdal4::KANMX3 Lkfur4::URA3 | Y156 |
| 39 | Y1799 | MATα ura3 Lkdal4::KANMX3 Lkdur3::URA3 | Y156 |
| 40 | Y1800 | MATα ura3 Lkdal4::KANMX3 SAKL0A07480::URA3 | Y156 |
| 41 | Y1801 | MATα ura3 Lkfur4::KANMX3 Lkdur3::URA3 | Y156 |
| 42 | Y1802 | MATα ura3 Lkfur4::KANMX3 SAKL0A07480::URA3 | Y156 |
hom, homolog.
For the microarray experiment and the growth test, N-minimal medium was made from defined minimal medium complemented with various nitrogen sources at 0.1% (wt/vol), including uracil, dihydrouracil, uridine, urea, allantoin, proline, or ammonium sulfate, as described previously (13).
For selection of transformants with the URA3 gene marker, SC URA dropout medium was used (1% succinic acid, 0.6% NaOH, 2% glucose, 0.19% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, 0.1926% synthetic complete mixture [Kaiser], and dropout mixture lacking uracil [Formedium]).
Escherichia coli, which was used for protein production, was grown in LB medium (1% NaCl, 0.5% yeast extract, 1% peptone, pH 7).
Cell cultivation for microarray experiment.
All cultivations of L. kluyveri were performed at 25°C, and E. coli was grown at 37°C. An overnight culture of the diploid sequenced strain Y057 (NRRL strain Y12651) in YPD was used to inoculate flasks with N-minimal medium with uracil, dihydrouracil, uridine, proline, or ammonia as the nitrogen source. Four parallel cultures (biological replicates) were made for each condition. The cells were grown to an optical density at 600 nm (OD600) of between 0.3 and 0.5 and then harvested by transferring the culture to small centrifugation tubes filled with crushed ice. The cells were collected by centrifugation at 4°C at 3,000 rpm for 10 min.
RNA extraction and cDNA synthesis and labeling.
The yeast cells were immediately resuspended in 1 ml ice-cold TRIzol reagent (Invitrogen) and then disrupted using a mini-beadbeater (6 m/s; five times for 25 s each) using 0.5-μm glass beads. The samples were kept on ice, and total RNA was extracted according to the provided protocol, except that the RNA was precipitated at −20°C for 30 min. Cleanup was performed by running the RNA on RNeasy spin columns (Qiagen) according to the manufacturer's specifications. The RNA was treated on column with RNase-free DNase (Qiagen) according to the manufacturer's protocol, and the integrity of extracted total RNA was analyzed by capillary electrophoresis (2100 Bioanalyzer; Agilent).
For cDNA synthesis, SuperScript III reverse transcriptase (RT; Invitrogen) was used, with 5 μg of total RNA as the template and the oligo(dT)12-18 primers (Invitrogen). The cDNA was labeled using a Pronto! plus labeling kit (Promega) according to the manufacturer's protocol. Test samples were labeled with Cy3-dCTP (Amersham), and the reference was labeled with Cy5-dCTP. The protocols provided by the manufacturer were followed.
Microarray hybridization and analysis.
Equimolar amounts of labeled test and reference cDNA were hybridized onto L. kluyveri microarray slides using the MAUI hybridization system. The microarrays, which were made on Corning epoxy slides, were provided by Mark Johnston, St. Louis, MO. Each spot contains a 65-base-long oligonucleotide representing one gene or open reading frame out of the total 5,353 included and is present in triplicates on each slide. In total, L. kluyveri has 5,321 predicted genes, but on the array some are represented by two different oligonucleotides; hence, 5,353 oligonucleotide-containing spots. The oligonucleotide sequences were based on the genome sequence determined by the Washington University School of Medicine (20), which was the only one available for L. kluyveri when the arrays were printed. Details of the platform can be found in the GEO database (platform no. GPL17318; http://www.ncbi.nlm.nih.gov/geo/). The slides were scanned and resulting images were analyzed in the GenePix Pro 4.1 software package (Molecular Devices, LLC).
Data analysis was conducted in R using the Bioconductor package. The raw data were normalized by within-print-tip-group local regression (LOESS) location normalization. An average of the signal from the three replicates on each array was then compared to the corresponding average of the control from the same array, and the log2 value of the sample/reference ratio was calculated. A standard t test was applied to assess the significance of the changes in expression, and the obtained P values were corrected for false discovery rates (FDR) due to the large sample size. The median of the log2 ratios from the four biological replicates was used instead of the mean, since, in several cases, one out of the four replicates had large deviations from the other three. More details on the microarray experiment can be found in the GEO database (accession no. GSE48135).
Seven genes regulated in a manner similar to that of the known URC1-6 genes, i.e., upregulated by uracil and uridine and repressed by ammonia, were selected for further phenotypic studies. In addition, two other genes, both paralogs of genes selected from the microarray analysis, were included in the study. All nine genes can be found in Table 2.
TABLE 2.
L. kluyveri genes selected for knockout analysis and the name and function of their homologs in S. cerevisiaea
|
L. kluyveri name |
S. cerevisiae name |
Function in S. cerevisiae | ||
|---|---|---|---|---|
| Common | Systematic | Common | Systematic | |
| LkFUI1 | SAKL0H03476g | FUI1 | YBL042C | Uridine transporter |
| LkFUI1 hom | SAKL0H03498g | None | Not found in S. cerevisiae; unknown | |
| LkDAL5 | SAKL0C00352g | DAL5 | YJR152W | Ureidosuccinate/allantoate transporter |
| LkDAL5 hom | SAKL0G16456g | None | Not found in S. cerevisiae; unknown | |
| LkDAL4 | SAKL0G04532g | DAL4 | YIR028W | Allantoin permease |
| LkFUR4 | SAKL0C11594g | FUR4 | YBR021W | Uracil transporter |
| LkDUR3 | SAKL0A10010g | DUR3 | YHL016C | Urea/polyamine permease |
| SAKL0A07480 | Noneb | Transport of basic amino acids | ||
| URC8 | SAKL0H04730g | YMR226C | 3-Hydroxy acid reductase | |
All of these genes, except the LkFUI1 homolog (hom) and LkDAL5 homolog, were highly expressed on uracil but not on ammonia.
Weakly similar to VBA2 (YBR293W).
Cassettes for gene replacement.
Gene replacement cassettes were made for each of the nine genes and were used to create the knockout strains. The cassettes consisted of a marker gene flanked by regions of 470 to 520 bases homologous to the upstream and downstream sequence of the target gene (17). All sequences used, both coding and noncoding, were retrieved from the Génolevures database (http://www.genolevures.org/) (21). The constructs for the cassettes were amplified separately by PCR, purified, and then assembled in a special primer-free assembly PCR, where about 48 ng marker gene and 20 ng flanking region were assembled with the help of overhangs on the flanking regions designed to be complementary to the marker gene. A second reaction with primers was run to amplify the correct product, using part of the assembly reaction as the template (∼5 μl for a 30-μl reaction). All primers used are specified in Table S1 in the supplemental material. For all PCRs, the Phusion polymerase (Finnzymes/Thermo Scientific) was used and its recommended protocol was followed, using 65°C as annealing temperature, or, in difficult cases, a touchdown protocol from 70 or 72 to 65°C. The assembly reaction was run for 15 cycles, while the second reaction was run, as normal, for 30 cycles.
Creation of knockout strains.
Transformation of the gene replacement cassettes to create the knockout strains was performed by electroporation into competent L. kluyveri cells as described previously (13), with some minor adjustments. The cells were grown to an OD600 of 1 to 1.5, and the amount of cells used for each transformation corresponded to 2 to 3 ml of cells. For all transformations where minimal media was used for selection, an extra step of two washes with 2 ml of sterile water or minimal media was included after the incubation in YPD before spreading the cells on the selective media to avoid transfer of the rich media to the selective minimal media.
Single deletions of all transporter-like genes were made by transformation of the cassettes with KANMX3 as the marker into the ura3 strain Y156. For the reductase gene, URC8, the same procedure was used but employing the prototrophic strain Y1392 instead of Y156, since no double deletion was planned for this gene and the URA3 marker was not needed. Transformants were selected on YPD with G418. Double knockouts were then created for all putative transporter genes by transforming the single knockout strains with deletion cassettes, now using URA3 as the marker gene, which allowed selection on SD media.
The correct insertion of the deletion cassette was confirmed by colony PCR in all deletion strains created. Cells were dissolved in 5 μl 0.02 M NaOH, incubated for 10 min at 95°C, and then spun down. Three μl of the supernatant was used as the template in a regular PCR using DreamTaq DNA polymerase (Fermentas).
All of the resulting strains, both single and double knockouts, were tested for growth on minimal media with various sources of nitrogen (described above). The sequenced strain Y057 and the parent strain were used as positive controls, and the Δurc2 strain Y1161, which has a known URC phenotype, was used as the negative control. The strains were grown overnight in YPD and then harvested and washed in sterile water. They were diluted to ODs of 0.1 and 0.01 and spotted onto solid sole-nitrogen-source media. Analysis of the growth was done after 3 and 6 days.
The knockout strains created in this study are represented under the numbers 6 to 42 in Table 1.
Urc8p overexpression and assay.
The URC8 gene from L. kluyveri was cloned and inserted into the pET151/D-TOPO vector (Invitrogen), which adds an N-terminal His tag to the protein product. The construct (P1027) was transformed into Escherichia coli strain BL21 (Invitrogen) for expression. A culture was grown at 37°C to an OD600 of 0.6, and then expression was induced with 0.1 mM IPTG (isopropyl β-d-thiogalactopyranoside) followed by overnight incubation at 16°C. The cells were harvested and homogenized by French press two times at 16,000 and 18,000 lb/in2, and the protein was purified using nickel-nitrilotriacetic acid (Ni-NTA) chromatography. The storage buffer was subsequently changed to 10 μM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 0.1 M KP buffer, pH 7.4 using PD-10 columns (GE Healthcare).
The L. kluyveri β-alanine aminotransferase, Pyd4p, was purified according to a previous description (15) and then used for production of malonate semialdehyde under the following conditions: 83 mM KP buffer, pH 7.4, 80.8 mM β-alanine, 2.6 mM α-ketoglutarate, 0.3 μg/μl Pyd4p, 30°C, 5 min. NADPH and Urc8p were added (0.13 μg/μl and 0.42 ng/μl, respectively) to assay the Urc8p activity. Under these conditions, neither the Pyd4p concentration nor the produced malonate semialdehyde should be limiting. The reaction was monitored by the decrease in absorbance at 340 nm, where NADPH has its absorption maximum.
Microarray data accession numbers.
The microarray data determined in the course of this work have been deposited in the GEO database under accession no. GSE48135 and the microarray platform is found under accession no. GPL17318.
RESULTS
Global expression analysis.
Global expression analysis was performed on L. kluyveri by specifically designed microarrays (GEO accession no. GSE48135; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE48135), based on the first available genome sequence (20), in order to study the pyrimidine catabolism and its relationship to general nitrogen metabolism. To cover the different aspects of pyrimidine and nitrogen metabolism, we chose four growth conditions, where uracil, uridine, dihydrouracil, or ammonia was used are the only source of nitrogen. For each experiment, cells grown with proline as the nitrogen source were used as the control, since proline, like uracil, uridine, and dihydrouracil, is a poor nitrogen source and because its metabolism is not related to that of the other sources used. The top 20 upregulated genes under each condition are shown in Table 3. The highest number of upregulated genes was seen under the uridine condition, with 250 genes being at least 2-fold upregulated. On dihydrouracil at least 122 genes were 2-fold or more upregulated, on uracil 91 genes, and on ammonia 57 genes. The complete lists of genes which are at least 2-fold upregulated are found in Table S2 in the supplemental material.
TABLE 3.
Top 20 upregulated genes under the four conditions analyzed: uracil, dihydrouracil, uridine, and ammonia media
| Condition and no. | ID | Systematic name | Common name | S. cerevisiae homolog | Log2 fold induction | P value |
|---|---|---|---|---|---|---|
| Uracil | ||||||
| 1 | Contig145.1.A:821 | SAKL0H14498g | URC1 | None | 9.703 | 0.00141 |
| 2 | Contig31.2.A:628 | SAKL0A09988g | URC4 | None | 8.410 | 0.00104 |
| 3 | Skluy_39.4.2:2 | SAKL0C00352g | DAL5 | 5.951 | 0.00107 | |
| 4 | Skluy_32.2.16:16 | SAKL0H03476g | FUI1 | 5.728 | 0.00069 | |
| 5 | Skluy_1.3.12:12 | SAKL0E11792g | URC6 | FUR1 | 5.465 | 0.00061 |
| 6 | Skluy_36.1.5:5 | SAKL0H10560g | URC3,5 | DUR1,2 | 5.264 | 0.00110 |
| 7 | Skluy_116.1.12:12 | SAKL0D01386g | ATR1 | 4.976 | 0.00073 | |
| 8 | Skluy_11.1.2:2 | SAKL0H23210g | YGR015C | 4.668 | 0.00175 | |
| 9 | Skluy_104.1.4:4 | SAKL0C02178g | Nonea | 4.626 | 0.00458 | |
| 10 | Skluy_104.1.5:5 | SAKL0C02200g | PRM1 | 4.550 | 0.00828 | |
| 11 | Skluy_31.2.8:8 | SAKL0A10010g | DUR3 | 4.483 | 0.00116 | |
| 12 | Skluy_54.3.14:14 | SAKL0G04532g | DAL4 | 4.280 | 0.00155 | |
| 13 | Skluy_182.1.2:2 | SAKL0C11594g | FUR4 | 3.890 | 0.00140 | |
| 14 | Skluy_76.1.28:28 | SAKL0H04730g | URC8 | YMR226C | 3.819 | 0.00206 |
| 15 | Contig29.2.E:116 | SAKL0H20526g | None | 3.804 | 0.00515 | |
| 16 | Skluy_13.2.3:3 | SAKL0B05588g | PYD2 | None | 3.721 | 0.00337 |
| 17 | Skluy_61.2.32:32 | SAKL0B10054g | FUN26 | 3.702 | 0.00140 | |
| 18 | Contig147.1.A:1027 | SAKL0C11748g | PYD3 | None | 3.590 | 0.00207 |
| 19 | Skluy_102.2.15:15 | SAKL0G14762g | UGA4 | 3.475 | 0.00259 | |
| 20 | Skluy_0.1.1:1 | SAKL0H05412g | TPO4 | 3.400 | 0.00731 | |
| Dihydrouracil | ||||||
| 1 | Contig147.1.A:1027 | SAKL0C11748g | PYD3 | None | 9.015 | 0.00129 |
| 2 | Skluy_13.2.3:3 | SAKL0B05588g | PYD2 | None | 6.908 | 0.00326 |
| 3 | Skluy_177.1.3:3 | SAKL0B10120g | DAL5 | 6.305 | 0.00135 | |
| 4 | Skluy_30.1.33:33 | SAKL0B12562g | UGA1 | 6.217 | 0.00097 | |
| 5 | Skluy_182.1.2:2 | SAKL0C11594g | FUR4 | 5.903 | 0.00075 | |
| 6 | Skluy_76.1.28:28 | SAKL0H04730g | URC8 | YMR226C | 4.984 | 0.00126 |
| 7 | Skluy_1.3.12:12 | SAKL0E11792g | URC6 | FUR1 | 4.622 | 0.00083 |
| 8 | Contig145.1.A:821 | SAKL0H14498g | URC1 | None | 4.158 | 0.00122 |
| 9 | Contig31.2.A:628 | SAKL0A09988g | URC4 | None | 4.048 | 0.00190 |
| 10 | Skluy_30.1.35:35 | SAKL0B12606g | HPA2 | 4.018 | 0.00103 | |
| 11 | Skluy_23.5.2:2 | SAKL0G07480g | GIT1 | 3.825 | 0.01224 | |
| 12 | Contig73.1.B:480 | SAKL0A08514g | None | 3.758 | 0.00135 | |
| 13 | Skluy_104.1.4:4 | SAKL0C02178g | None | 3.581 | 0.00405 | |
| 14 | Skluy_45.3.31:31 | SAKL0E15290g | FEN2 | 3.469 | 0.00278 | |
| 15 | Skluy_104.1.5:5 | SAKL0C02200g | PRM1 | 3.244 | 0.00460 | |
| 16 | Skluy_20.1.9:9 | SAKL0F16632g | ESBP6 | 3.113 | 0.00411 | |
| 17 | Skluy_3.3.12:12 | SAKL0E05016g | ARG8 | 3.001 | 0.00127 | |
| 18 | Contig6.3.B:1543 | SAKL0H11176g | None | 2.960 | 0.00191 | |
| 19 | Contig20.1.B:21 | SAKL0F16654g | None | 2.834 | 0.00092 | |
| 20 | Skluy_3.3.13:13 | SAKL0E04994g | HER2 | 2.715 | 0.00242 | |
| Uridine | ||||||
| 1 | Contig145.1.A:821 | SAKL0H14498g | URC1 | None | 10.131 | 0.00138 |
| 2 | Skluy_33.4.3:3 | SAKL0D00814g | PHO84 | 9.393 | 0.00161 | |
| 3 | Skluy_23.5.2:2 | SAKL0G07480g | GIT1 | 9.293 | 0.00146 | |
| 4 | Contig31.2.A:628 | SAKL0A09988g | URC4 | None | 8.012 | 0.00137 |
| 5 | Contig1.2.A:494 | SAKL0E12144g | None | 7.726 | 0.00143 | |
| 6 | Skluy_32.2.16:16 | SAKL0H03476g | FUI1 | 6.070 | 0.00159 | |
| 7 | Skluy_39.4.2:2 | SAKL0C00352g | DAL5 | 5.786 | 0.00167 | |
| 8 | Skluy_53.2.1:1 | SAKL0H13222g | ENA2 | 5.271 | 0.00143 | |
| 9 | Skluy_6.8.1:1 | SAKL0H13222g | ENA5 | 4.828 | 0.00160 | |
| 10 | Skluy_98.1.15:15 | SAKL0A00726g | VBA5 | 4.819 | 0.00128 | |
| 11 | Skluy_53.1.1:1 | SAKL0H13222g | ENA5 | 4.756 | 0.00160 | |
| 12 | Contig30.1.B:723 | SAKL0B12232g | NOHBY208 | 4.754 | 0.00330 | |
| 13 | Skluy_4.6.2:2 | SAKL0E07964g | XBP1 | 4.752 | 0.00229 | |
| 14 | Skluy_61.2.32:32 | SAKL0B10054g | FUN26 | 4.551 | 0.00125 | |
| 15 | Skluy_1.3.12:12 | SAKL0E11792g | URC6 | FUR1 | 4.502 | 0.00134 |
| 16 | Contig11.2.B:654 | SAKL0H24310g | None | 4.343 | 0.00122 | |
| 17 | Skluy_36.1.5:5 | SAKL0H10560g | URC3,5 | DUR1,2 | 4.326 | 0.00129 |
| 18 | Skluy_46.2.9:9 | SAKL0B03212g | ICL2 | 4.315 | 0.00397 | |
| 19 | Skluy_76.1.28:28 | SAKL0H04730g | URC8 | YMR226C | 4.161 | 0.00286 |
| 20 | Skluy_0.4.5:5 | SAKL0H08580g | YPL110C | 4.123 | 0.00126 | |
| Ammonia | ||||||
| 1 | Skluy_88.1.2:2 | SAKL0D02948g | AGP1 | 3.515 | 0.019478 | |
| 2 | Skluy_140.1.3:3 | SAKL0D09152g | CIT1 | 2.660 | 0.009231 | |
| 3 | Skluy_2.1.29:29 | SAKL0F04356g | TIR3 | 2.417 | 0.026387 | |
| 4 | Skluy_95.2.7:7 | SAKL0B02090g | HSP12 | 2.190 | 0.05253 | |
| 5 | Skluy_80.1.22:22 | SAKL0F13310g | ITR2 | 2.123 | 0.03577 | |
| 6 | Skluy_27.1.38:38 | Sequence on chromosome F, no matching genes in Génolevures | 2.089 | 0.027403 | ||
| 7 | Skluy_22.1.1:1 | Sequence on chromosome G, no matching genes in Génolevures | 2.054 | 0.042043 | ||
| 8 | Skluy_34.1.4:4 | SAKL0D04510g | JEN1 | 2.024 | 0.013497 | |
| 9 | Contig113.1.A:1140 | SAKL0B01672g | NOHBY648 | 1.961 | 0.007016 | |
| 10 | Skluy_22.1.46:46 | SAKL0G10164g | PDC1 | 1.924 | 0.021277 | |
| 11 | Skluy_112.3.8:8 | SAKL0C11528g | GAL7 | 1.906 | 0.040539 | |
| 12 | Skluy_375.1.1:1 | Matches 10 retrotransposon Gag genes | YHR214C-C | 1.882 | 0.201569 | |
| 13 | Contig573.1.A:203 | SAKL0F13310g | 1.831 | 0.050309 | ||
| 14 | Skluy_105.1.10:10 | SAKL0A09394g | OPT1 | 1.712 | 0.022063 | |
| 15 | Skluy_40.1.1:1 | SAKL0G16038g | CTR3 | 1.689 | 0.022914 | |
| 16 | Skluy_12.2.10:10 | SAKL0G19140g | MDH1 | 1.671 | 0.013244 | |
| 17 | Skluy_2.1.30:30 | SAKL0F04334g | PDC1 | 1.616 | 0.02575 | |
| 18 | Skluy_25.3.2:2 | SAKL0G14014g | HIP1 | 1.540 | 0.029876 | |
| 19 | Skluy_15.3.12:12 | SAKL0D11946g | SUC2 | 1.497 | 0.03344 | |
| 20 | Skluy_161.1.5:5 | SAKL0G08052g | YHR087W | 1.490 | 0.036698 |
Weakly similar to VBA2 (YBR293W).
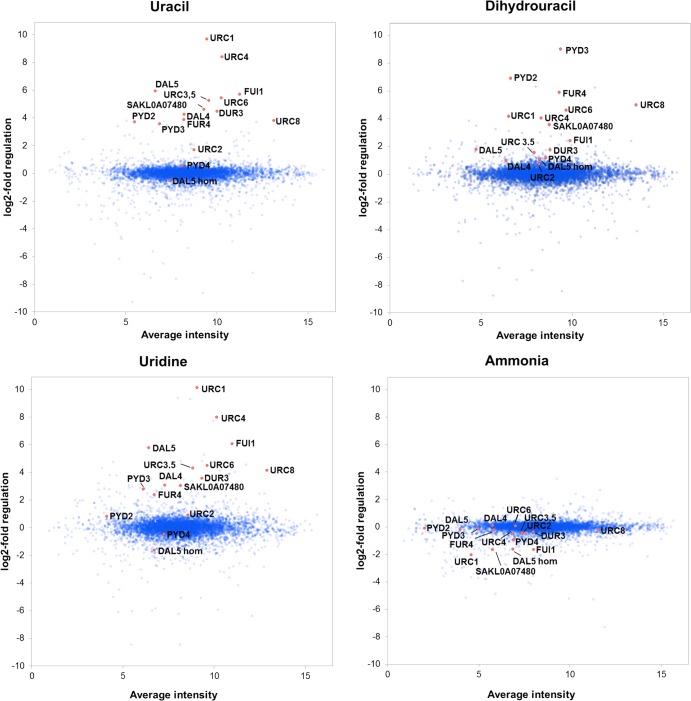
Through this analysis, we show that all of the five already known URC genes (17) were upregulated in the presence of uracil and uridine, while no induction was observed on ammonia. For URC2, which encodes a transcription factor, the expression was comparatively constitutive, with only a 3-fold induction on uracil. An important observation was that URC1 is the most upregulated gene on both uracil and uridine, being 800 to 1,000 times induced, closely followed by URC4, which was increased 250 to 350 times. Surprisingly, we found that most of the URC genes were also induced by dihydrouracil, even though its metabolism is not related to that of uracil and uridine in yeast, in contrast to mammals and other organisms which possess the full reductive pathway. In addition, PYD2 and PYD3 of the reductive pathway also were upregulated on uracil, but the induction by dihydrouracil was stronger. A similar but much weaker effect was seen for the last gene of the pathway, PYD4. The genes of the reductive pathway also were downregulated by ammonia. These results show that both the PYD and the URC genes in L. kluyveri are subjected to the nitrogen catabolite repression. The change in expression is represented in Table 4 and graphically visualized in Fig. 2.
TABLE 4.
Log2 fold regulation obtained from the microarray experiment of L. kluyveri genes related to pyrimidine metabolism in response to different sole nitrogen sourcesa
| L. kluyveri name | S. cerevisiae homolog | Sequence ID | Log2 fold regulation in response to: |
|||
|---|---|---|---|---|---|---|
| Uracil | Dihydrouracil | Uridine | Ammonia | |||
| URC1 | None | Contig145.1.A:821 | 9.703 | 4.158 | 10.131 | −2.016 |
| URC2 | YDR520C | Skluy_39.6.2:2 | 1.715 | 0.726 | 0.939 | −0.418 |
| URC3,5 | DUR1,2 | Skluy_36.1.5:5 | 5.264 | 1.554 | 4.326 | −0.406 |
| URC4 | None | Contig31.2.A:628 | 8.410 | 4.048 | 8.012 | −0.501 |
| URC6 | FUR1 | Skluy_1.3.12:12 | 5.465 | 4.622 | 4.502 | 0.080 |
| PYD2 | None | Skluy_13.2.3:3 | 3.721 | 6.908 | 0.839 | −0.078 |
| PYD3 | None | Contig147.1.A:1027 | 3.590 | 9.015 | 2.798 | −0.106 |
| PYD4 | UGA1 | Skluy_29.2.17:17 | 0.585 | 1.213 | −0.441 | −0.941 |
| URC8 | YMR226C | Skluy_76.1.28:28 | 3.819 | 4.984 | 4.161 | −0.201 |
| DAL4 | DAL4 | Skluy_54.3.14:14 | 4.280 | 0.986 | 3.090 | 0.107 |
| DAL5 | DAL5 | Skluy_39.4.2:2 | 5.951 | 1.801 | 5.786 | −0.121 |
| DAL5 homolog | DAL5 | Skluy_40.3.7:7 | −0.513 | 1.105 | −1.614 | −1.609 |
| FUI1 | FUI1 | Skluy_32.2.16:16 | 5.728 | 2.406 | 6.070 | −1.632 |
| FUI1 homolog | FUI1 | SAKL0H03498gb | NA | NA | NA | NA |
| FUR4 | FUR4 | Skluy_182.1.2:2 | 3.890 | 5.903 | 2.406 | −0.211 |
| SAKL0A07480 | Nonec | Skluy_104.1.4:4 | 4.626 | 3.581 | 3.061 | −1.632 |
| DUR3 | DUR3 | Skluy_31.2.8:8 | 4.483 | 1.749 | 3.580 | −0.351 |
Cells grown on single-nitrogen-source media were used, and the expression under the four conditions was compared to that in cells grown on media with proline as the nitrogen source. The log2 fold regulation was calculated by logarithmically normalizing the sample/reference ratio of the values recovered from the scanned arrays. NA, not applicable.
The oligonucleotide matching this gene could not be identified; therefore, its systematic name found in the Génolevures database was used.
Weakly similar to VBA2 (YBR293W).
FIG 2.
Expression analysis of L. kluyveri grown on different nitrogen sources, as analyzed by microarrays. The oligonucleotides representing genes known to be involved in the URC and reductive pyrimidine catabolic pathways are marked in red. All of the genes selected for further analysis, which also are marked in the figure, were previously unknown but have expression patterns similar to those of the known URC genes or are homologous to a gene with a similar expression pattern. The exception is the FUI1 homolog SAKL0H03498g, which could not be identified among the oligonucleotides represented on the array. The y axis shows the sample/reference signal log2 ratio, and the x axis is the average intensity of the total signal for each oligonucleotide (average of the technical and biological replicates).
Deletion studies revealed novel URC genes.
From the microarray results, seven potential new URC genes were selected for further studies (Table 2). The selection was made on the basis (i) that their expression profile was similar to that of the known URC genes, i.e., upregulation on uracil and uridine and no change or downregulation on ammonia (Table 4), and (ii) that the function in L. kluyveri was unknown. Two additional genes (SAKL0H03498g and SAKL0G16456g), whose function in L. kluyveri were also unknown, were included as well. These two genes had different expression patterns but were included since they are paralogs of the FUI1 and DAL5 genes selected from the microarray study. An extensive collection of knockout strains, including 9 single and 26 double knockout strains (Table 1), was prepared through gene replacement for the selected genes. All of the created strains were found to be viable and were tested for growth on media with different nitrogen sources (Table 5) using the parental strain as a positive control and the Δurc2 strain as the negative control.
TABLE 5.
Growth phenotype analysis of single and double deletion strains
| Deleted gene(s)a | Growthb on: |
|||||
|---|---|---|---|---|---|---|
| Ammonia | Uracil | Uridine | DHU | Allantoin | Urea | |
| Single deletion | ||||||
| LkFUI1 | +++ | +++ | + | +++ | +++ | +++ |
| LkFUI1 hom | +++ | +++ | +++ | +++ | +++ | +++ |
| LkDAL5 | +++ | +++ | +++ | +++ | +++ | +++ |
| LkDAL5 hom | +++ | +++ | +++ | +++ | +++ | +++ |
| LkDAL4 | +++ | +++ | +++ | +++ | +++ | +++ |
| LkFUR4 | +++ | +++ | +++ | +++ | +++ | +++ |
| LkDUR3 | +++ | +++ | +++ | +++ | +++ | +++ |
| SAKL0A07480 | +++ | +++ | +++ | +++ | +++ | +++ |
| URC8 | +++ | +/++ | + | +/− | +++ | +++ |
| Double deletion | ||||||
| ΔLkfur4 ΔLkdal4 | +++ | +++ | +++ | +++ | +++ | +++ |
| ΔLkfui1 ΔLkfur4 | +++ | +++ | − | ++ | +++ | +++ |
| ΔLkfui1 Δfui1 hom | +++ | +++ | − | +++ | +++ | +++ |
| ΔLkfui1 ΔLkdur3 | +++ | +++ | + | ++ | +++ | +++ |
| ΔLkfui1 ΔLkdal4 | +++ | +++ | + | +++ | +++ | +++ |
| ΔLkfui1 ΔLkdal5 | +++ | +++ | + | +++ | +++ | +++ |
| ΔLkfui1 dal5 hom | +++ | +++ | + | +++ | +++ | +++ |
| ΔLkfui1 ΔSAKL0A07480 | +++ | +++ | ++ | +++ | +++ | +++ |
| ΔLkdur3 ΔSAKL0A07480 | +++ | ++ | ++ | +++ | +++ | +++ |
| ΔLkdur3 ΔLkdal5 | +++ | ++ | ++ | +++ | +++ | +++ |
| ΔLkdur3 ΔLkdal4 | +++ | ++ | ++ | +++ | +++ | +++ |
| Δfui1 hom ΔLkdur3 | +++ | +++ | +++ | ++ | +++ | +++ |
| Δfui1 hom ΔLkdal4 | +++ | +++ | +++ | +++ | +++ | +++ |
| Δfui1 hom ΔLkdal5 | +++ | +++ | +++ | +++ | +++ | +++ |
| Δfui1 hom ΔLkfur4 | +++ | +++ | +++ | +++ | +++ | +++ |
| Δfui1 hom ΔSAKL0A07480 | +++ | +++ | +++ | +++ | +++ | +++ |
| Δdal5 hom ΔLkdal5 | +++ | +++ | +++ | +++ | +++ | +++ |
| ΔLkdal4 ΔLkdal5 | +++ | +++ | +++ | +++ | +++ | +++ |
| ΔLkfur4 ΔLkdal5 | +++ | +++ | +++ | +++ | +++ | +++ |
| ΔSAKL0A07480 ΔLkdal5 | +++ | +++ | +++ | +++ | +++ | +++ |
| Δdal5 hom ΔLkdal4 | +++ | +++ | +++ | +++ | +++ | +++ |
| Δdal5 hom ΔLkfur4 | +++ | +++ | +++ | +++ | +++ | +++ |
| Δdal5 hom ΔLkdur3 | +++ | +++ | +++ | +++ | +++ | +++ |
| Δdal5 hom ΔSAKL0A07480 | +++ | +++ | +++ | +++ | +++ | +++ |
| ΔLkfur4 ΔSAKL0A07480 | +++ | +++ | +++ | +++ | +++ | +++ |
| ΔLkdal4 ΔSAKL0A07480 | +++ | +++ | +++ | +++ | +++ | +++ |
hom, homolog.
Shading highlights the results which differed from those for the wild-type strain. +++, wild-type-like growth; ++, growth, but less than that of the wild type; +, very limited and slow growth; −, no growth/equal to negative control; NA, not assayed. DHU, dihydrouracil. Growth was inspected after 3 and 6 days.
A majority of the genes selected for deletion analysis encode putative permeases. So far, no permeases have been characterized in L. kluyveri. Uptake of nutrients is crucial for the cell, so often there is more than one transporter for each nutrient, although their affinity for that nutrient may differ. Therefore, it is not very surprising that most of the permease gene deletions did not show any deviating phenotype; however, a few strains grew slower than their parental strain. The strain with the deletion of the L. kluyveri FUI1 (SAKL0H03476g) gene showed a decrease in growth on uridine. Several of the double-knockout strains where this gene was missing had a changed phenotype on uridine, and in a few cases a slight decrease in growth was seen on dihydrouracil as well. Two strains, in which LkFUI1 was disrupted together with the LkFUI1 paralog or LkFUR4, only had a little background growth, similar to the negative control, thereby showing a stronger growth phenotype than the single deletion of LkFUI1.
A slight but still clear decrease in growth on both uracil and uridine was observed for several double knockout combinations involving the LkDUR3 gene. No effect, however, was seen on urea, although this is the substrate of DUR3 in S. cerevisiae (22).
A newly characterized URC gene encodes a C3-modifying enzyme.
The only non-permease-like gene selected for further analysis was SAKL0H04730g, which was later named URC8 (see Fig. S3 in the supplemental material). The deletion strain showed significantly reduced growth on uracil and uridine, indicating involvement of this gene in the URC pathway. In addition, it also showed less growth on dihydrouracil, which is degraded by the reductive pathway and would be expected to be unrelated to the URC pathway. The Urc8p homologs in S. cerevisiae (YMR226C) and E. coli (YdfG) have been characterized previously (23). They belong to the NADP+/NADPH-dependent short-chain dehydrogenase/reductase family (23, 24), and YdfG is known to reduce malonate semialdehyde to 3-hydroxypropionate in the Rut pathway in E. coli (19), which is also the outcome of the URC pathway (17). We hypothesize that the role of URC8 is similar to that of YdfG.
Overexpression and purification of Urc8p.
We successfully subcloned the URC8 gene of L. kluyveri and produced the protein in E. coli, using a pET/TOPO vector in order to add a His tag to facilitate purification. The purified protein was analyzed by SDS-PAGE (Fig. 3), and a band just below 35 kDa was observed, which should be compared to the predicted size of the monomer of the native protein, 29.2 kDa, and the 4 kDa added by the vector. After confirmation of the protein size, the concentration was determined by Bradford assay and the yield was estimated to be about 10 mg per liter of culture. The fractions containing the highest protein concentration were pooled, desalted, and diluted for activity assays.
FIG 3.
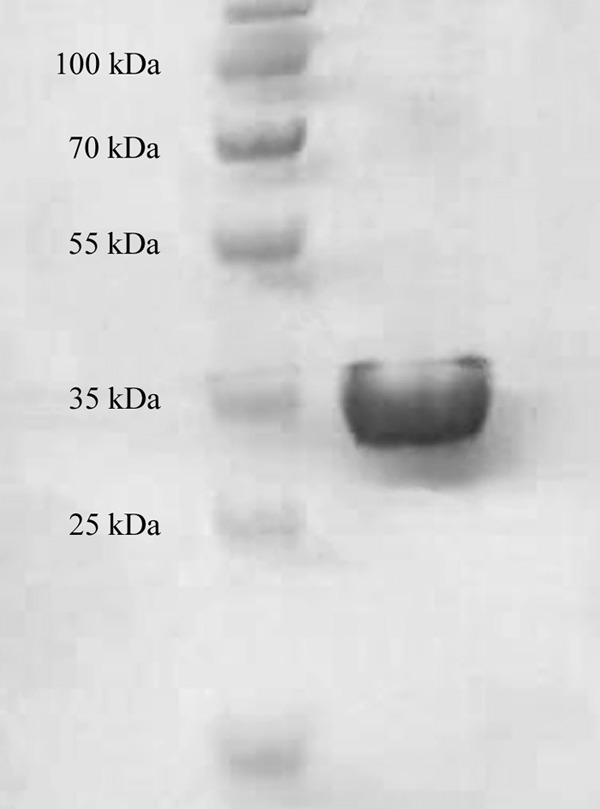
Purified His-tagged Urc8 protein from L. kluyveri run on SDS-PAGE.
Urc8p catalyzes the reduction of malonate semialdehyde to 3-hydroxypropionate.
To test our hypothesis that Urc8p catalyzes the same reaction as YdfG in the Rut pathway (19), i.e., converts malonate semialdehyde to 3-hydroxypropionate, we used the purified protein in a coupled assay with the β-alanine amino-transferase from L. kluyveri, Pyd4p, to make the proposed substrate. β-Alanine amino-transferase catalyzes the fourth step of the common reductive pyrimidine degradation pathway, where β-alanine is converted to malonate semialdehyde (15). The purified Pyd4p enzyme was incubated with β-alanine and α-ketoglutarate (as an acceptor of the ammonia moiety) at 30°C for 5 min to produce malonate semialdehyde before addition of NADPH and the Urc8 protein. The conditions were optimized, so the concentrations of Pyd4p and its substrates should not be limiting.
The reaction was monitored by the absorbance change of NADPH at 340 nm. A linear decrease from 0.825 to 0.494 was observed during a 10-min period (Fig. 4), indicating that NADPH is the reducing agent in the reaction. A negligible decrease in absorbance was seen without Urc8p added or when NADH was used instead of NADPH. This confirms that Urc8p catalyzes the reduction of malonate semialdehyde to 3-hydroxypropionate, which is a waste product of the URC pathway (Fig. 4).
FIG 4.
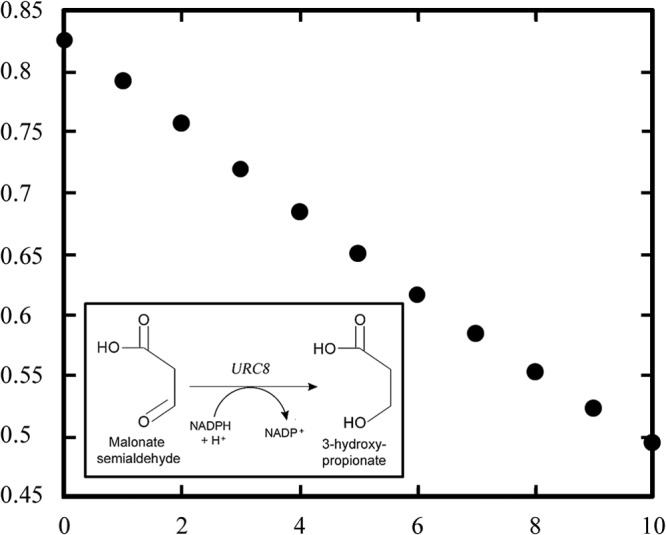
Assay for the new URC enzyme, Urc8p. For the Urc8p assay, the absorbance was monitored at 340 nm for 10 min, corresponding to the decrease of NADPH. In the inset in the lower left corner, the Urc8p reaction, where malonic semialdehyde is converted into 3-hydroxypropionate with the use of NADPH as the reductant, is presented. No change in absorbance was observed when NADH was used instead of NADPH in a parallel sample.
DISCUSSION
We designed a microarray to deepen our knowledge on the URC pathway. L. kluyveri had already been studied by microarray analysis (25, 26), but only with cells grown in rich medium and not minimal media with different nitrogen sources. Our transcription analysis of L. kluyveri clearly shows that all genes known to be involved in the pyrimidine catabolism are also under nitrogen catabolite repression (Table 4). The nitrogen catabolite repression has been well studied in S. cerevisiae and in some filamentous fungi (1, 27), and we confirmed here that this regulatory circuit also exists in L. kluyveri, making the nitrogen metabolism more energy efficient by using the best source available. All URC genes were indeed induced when uracil or uridine, but not ammonia, was used as the source of nitrogen (Table 4 and Fig. 2). However, the fact that a quite substantial induction of most URC genes is also observed in the dihydrouracil condition is more surprising, considering that the catabolism of uracil and dihydrouracil occurs by two separate pathways in fungi, although they are both degraded by the same reductive pathway in most other organisms. The fact that the URC genes were strongly upregulated in response to uracil and uridine implies that these compounds also can be utilized as a source of nitrogen in nature. This would give L. kluyveri an advantage in niches where preferred nitrogen sources, such as ammonia and glutamate, are absent but uracil or uridine is present.
In contrast to mammals, insects, plants, and many bacteria, which have a complete reductive pathway in which degradation of uracil proceeds through reduction to dihydrouracil, yeasts that are able to degrade pyrimidines possess two separate pathways, one for uracil and one for dihydrouracil catabolism (17). The dihydrouracil is degraded by the same reductive pathway as that in other organisms, but the enzyme performing the first step, namely, the reduction of uracil to dihydrouracil, has not been found in any yeast. Therefore, yeast must use the URC pathway for degradation of uracil and uridine. Surprisingly, we found that the genes involved in the reductive pathway, namely, PYD2 (13) and PYD3 (14), were significantly upregulated by uracil, although the induction by dihydrouracil was higher. Similarly, the URC genes were found to be upregulated not only by uracil but also by dihydrouracil. In short, all URC and PYD genes were strongly regulated by their expected inducers. The observed induction by uracil and dihydrouracil could be a leftover process from the complete reductive pathway in early unicellular eukaryotes. It could also be explained by the fact that the two molecules only differ by one double bond in the pyrimidine ring and might be similar enough to activate the specific regulator proteins for each other's pathways. A similar overlap could be seen in the transport, where the same transporter carries both uracil and dihydrouracil (28). Lastly, one could also speculate that the two molecules get slowly interconverted in the yeast cell.
The high induction by uracil observed for URC1 and URC4 (about 830 and 350 times, respectively) can be compared to a previous study of uracil induction in Schizosaccharomyces pombe; however, that study was performed in the presence of ammonia. Here, the URC1 homolog urg1 was upregulated 100-fold. As in L. kluyveri, the S. pombe homologs of URC1, URC4, and URC6 were the three most strongly regulated genes in response to uracil, although the URC6 homolog urg2 was stronger than the URC4 homolog urg3 (29).
After our microarray analysis, we developed a large collection of strains with single and double gene disruptions for the eight putative transporters included in this study (Table 1). The single deletion strains were all viable, meaning that none of the genes was essential. Likewise, all of the double deletions created were found to be viable.
All deletion strains of FUI1 showed restricted growth on the minimal plate containing uridine as a sole nitrogen source (Table 5). Together, these results suggest that FUI1, which is a high-affinity uridine permease in S. cerevisiae (30), also is responsible for uridine transport in L. kluyveri. However, in L. kluyveri, there are additional permeases whose functions overlap this function. The double deletions where FUI1 was disrupted together with FUR4 or the LkFUI1 paralog had a null phenotype, indicating that these two genes function as weak uridine transporters. In S. cerevisiae, FUR4 is known to be the only uracil transporter (31), which is clearly not the case with the L. kluyveri homolog, since no effect at all was seen on uracil for any of its deletions.
The existence of several transporters for uridine explains why strains with the single deletion of FUI1 still grow slightly on uridine. The function of the LkFui1p paralog (SAKL0H03498g) has not been characterized before, but it is 70% identical to LkFui1p and 69% identical to S. cerevisiae Fui1p. We speculate that the function of the three proteins is similar. The main FUR4 function in S. cerevisiae is transport of uracil. Interestingly, no deviating phenotype was observed for any other double deletion strain where FUR4 was disrupted; instead, they all grew like the parental strain, even on uracil (Table 5). However, we cannot rule out uracil transport as a function of FUR4 in L. kluyveri, since other transporters might have overlapping functions, but it is clear that the uracil transport in L. kluyveri is different from that in S. cerevisiae, where FUR4 is the only uracil transporter (31, 32).
DUR3 also seems to have some role in the transport of both uracil and uridine, since several of its double knockout combinations showed some deviation in phenotype on these media, although no change was seen for the single knockout strain (Table 5). The other genes deleted in these double combinations apart from FUI1, namely, SAKL0A07480, DAL4, and DAL5, might also play a small role in uracil and uridine transport. The slight phenotype observed for the dur3 double knockouts, with fui1 and the Lkfui1 paralog, indicates that it also is involved in the transport of dihydrouracil. In S. cerevisiae, DUR3 is known to be a permease for polyamines and urea (22, 33), but no deviating phenotype was observed on urea for any dur3 strain in our growth tests, so it seems that the function of DUR3 in L. kluyveri is different.
The new URC gene presented in this paper, URC8, had a similar expression level under dihydrouracil, uridine, and uracil conditions (Table 4), and the corresponding knockout grew weakly on uracil, uridine, and dihydrouracil. We successfully overproduced and purified the encoded protein, Urc8p, and showed that it can catalyze the reduction of a 3-carbon molecule just like its homologs in S. cerevisiae and E. coli, YMR226c and YdfG. The confirmed reaction, i.e., reduction of malonate semialdehyde to 3-hydroxypropionate, is the same reaction catalyzed by RutE and YdfG in E. coli (19). However, the Urc8p homologs YMR226c and YdfG are known multisubstrate enzymes, meaning that Urc8p is likely to be one too. We speculate that the now confirmed reaction is the one which is required for the URC pathway, since 3-hydroxypropionate has already been confirmed as a waste product of this pathway (17).
Now that the function of URC8 is clear, its regulation by uracil and dihydrouracil also makes sense, as this gene apparently provides a link between the reductive and URC pathways (Fig. 5). Malonate semialdehyde, the substrate of Urc8p, is formed in both pathways and then processed by Urc8p. In the URC pathway, it is likely formed directly or spontaneously from the three-carbon part when the pyrimidine ring is opened and in the reductive pathway from beta-alanine by the action of Pyd4 (10). The reactions needed for extracting the two nitrogens from the pyrimidine ring are still unrelated in the two pathways, as Urc8p is only involved in taking care of the carbon waste product. The Urc8p homolog in E. coli, YdfG, degrades the malonate semialdehyde formed in the Rut pathway, producing 3-hydroxypropionate, thereby overlapping in function with RutE, which is a malonate semialdehyde reductase not homologous to YdfG or Urc8p. The known catalytic amino acids (34) are conserved in the E. coli and S. cerevisiae homologs of Urc8p. They are present also in the L. kluyveri protein (see Fig. S2 in the supplemental material), suggesting a similar function. In addition, the E. coli homologs produce the same product as that formed in the URC pathway. These facts together led us to hypothesize that Urc8p performed the same reaction as YdfG. In E. coli, the reaction is proposed to have a detoxifying role, helping to speed up the removal of toxic intermediates formed during the degradation (19).
FIG 5.
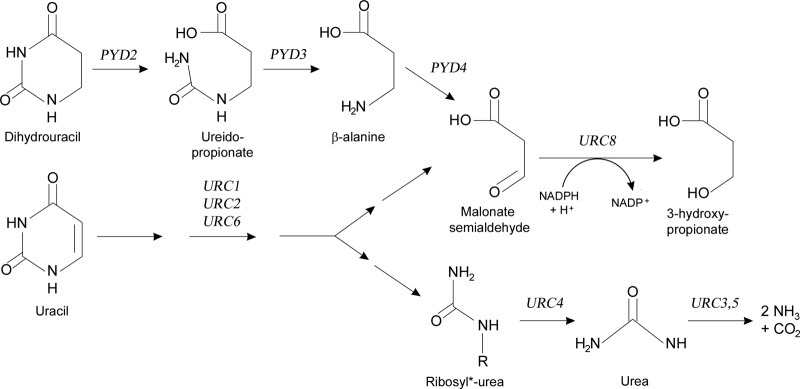
Overview of pyrimidine degradation in L. kluyveri, showing a link between the reductive and the URC pathways. At the top is the reductive pathway, which degrades dihydrouracil to β-alanine via ureidopropionate using the PYD2 and PYD3 gene products. In yeast, the gene encoding the dihydropyrimidine dehydrogenase, which catalyzes the reduction of uracil to dihydrouracil, does not exist, but other species, like mammals, plants, and insects, possess the complete reductive pathway. In yeast, uracil is degraded by the URC pathway (bottom). Malonate semialdehyde is produced both in the reductive pathway when Pyd4p deaminates β-alanine and in the URC pathway after the pyrimidine ring opening, presumably by the action of Urc1p. The malonate semialdehyde is degraded by Urc8p to the end product 3-hydroxypropionate.
The fact that the Δurc8 strain is still able to grow on both uracil and dihydrouracil, but much slower than the parent strain, also points to a detoxifying role of URC8. This is the first report on functional homology among the four pyrimidine degradation pathways.
In conclusion, we have now provided proof of nitrogen catabolite repression in L. kluyveri and provided a deeper knowledge of the regulation of genes involved in general nitrogen metabolism and pyrimidine degradation from the microarrays. A new URC gene, URC8, was found necessary for full growth of L. kluyveri on uracil, and its function was determined. Our growth test analysis of the large single and double knockout collection for putative transporters brought new insight into the complex transport of pyrimidines and other metabolically related compounds. LkFui1p was found to be involved in the uptake of uridine, but unlike its homolog in S. cerevisiae, it is not the only uridine transporter in L. kluyveri.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mark Johnston and Christopher S. Sawyer at the Genome Technology Access Center (GTAC; Washington University, St. Louis, MO) for designing and printing the L. kluyveri DNA microarrays. GTAC is supported by NCI Cancer Center support grant P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA grant UL1RR024992 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and the NIH Roadmap for Medical Research.
Many thanks to Jeanette Valchich and Johan Staaf at SCIBLU Genomics at Lund University for patience with helping us perform the cDNA labeling, hybridization, and analysis of the microarray slides. We are grateful to Jessica Abbott at the Department of Biology at Lund University, who helped us to extract the microarray data from R files. We also thank Hans-Erik Åkerlund from the Department of Biochemistry and Structural Biology at Lund University for technical assistance, Mingming Lai for making the URC8 knockout strain, and You Lv for purifying the Urc8p and Pyd4p enzymes.
This work was supported by the Swedish Research Council, the Nilsson-Ehle foundation at The Royal Physiographic Society in Lund, the Jörgen Lindström foundation, and the Sven and Lilly Lawski Foundation for Scientific Studies.
Footnotes
Published ahead of print 1 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00202-13.
REFERENCES
- 1.ter Schure EG, van Riel NAW, Verrips CT. 2000. The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 24:67–83 [DOI] [PubMed] [Google Scholar]
- 2.Beck H, Dobritzsch D, Piskur J. 2008. Saccharomyces kluyveri as a model organism to study pyrimidine degradation. FEMS Yeast Res. 8:1209–1213. 10.1111/j.1567-1364.2008.00442.x [DOI] [PubMed] [Google Scholar]
- 3.Gojkovic Z, Paracchini S, Piskur J. 1998. A new model organism for studying the catabolism of pyrimidines and purines. Adv. Exp. Med. Biol. 431:475–479. 10.1007/978-1-4615-5381-6_94 [DOI] [PubMed] [Google Scholar]
- 4.Kurtzman CP. 2003. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 4:233–245. 10.1016/S1567-1356(03)00175-2 [DOI] [PubMed] [Google Scholar]
- 5.Larue TA, Spencer JFT. 1968. Utilization of purines and pyrimidines by yeast. Can. J. Microbiol. 14:79–86. 10.1139/m68-012 [DOI] [PubMed] [Google Scholar]
- 6.Loh KD, Gyaneshwar P, Papadimitriou EM, Fong R, Kim KS, Parales R, Zhou ZR, Inwood W, Kustu S. 2006. A previously undescribed pathway for pyrimidine catabolism. Proc. Natl. Acad. Sci. U. S. A. 103:5114–5119. 10.1073/pnas.0600521103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayaishi O, Kornberg A. 1952. Metabolism of cytosine, thymine, uracil, and barbituric acid by bacterial enzymes. J. Biol. Chem. 197:717–732 [PubMed] [Google Scholar]
- 8.Soong CL, Ogawa J, Sakuradani E, Shimizu S. 2002. Barbiturase, a novel zinc-containing amidohydrolase involved in oxidative pyrimidine metabolism. J. Biol. Chem. 277:7051–7058. 10.1074/jbc.M110784200 [DOI] [PubMed] [Google Scholar]
- 9.Soong CL, Ogawa J, Shimizu S. 2001. Novel amidohydrolytic reactions in oxidative pyrimidine metabolism: analysis of the barbiturase reaction and discovery of a novel enzyme, ureidomalonase. Biochem. Biophys. Res. Commun. 286:222–226. 10.1006/bbrc.2001.5356 [DOI] [PubMed] [Google Scholar]
- 10.Andersen G, Andersen B, Dobritzsch D, Schnackerz KD, Piskur J. 2007. A gene duplication led to specialized gamma-aminobutyrate and beta-alanine aminotransferase in yeast. FEBS J. 274:1804–1817. 10.1111/j.1742-4658.2007.05729.x [DOI] [PubMed] [Google Scholar]
- 11.Dobritzsch D, Andersen B, Piskur J. 2005. Crystallization and X-ray diffraction analysis of dihydropyrimidinase from Saccharomyces kluyveri. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61:359–362. 10.1107/S174430910500610X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobritzsch D, Gojkovic Z, Andersen B, Piskur J. 2003. Crystallization and preliminary X-ray analysis of beta-alanine synthase from the yeast Saccharomyces kluyveri. Acta Crystallogr. D Biol. Crystallogr. 59:1267–1269. 10.1107/S0907444903009120 [DOI] [PubMed] [Google Scholar]
- 13.Gojkovic Z, Jahnke K, Schnackerz KD, Piskur J. 2000. PYD2 encodes 5,6-dihydropyrimidine amidohydrolase, which participates in a novel fungal catabolic pathway. J. Mol. Biol. 295:1073–1087. 10.1006/jmbi.1999.3393 [DOI] [PubMed] [Google Scholar]
- 14.Gojkovic Z, Sandrini MPB, Piskur J. 2001. Eukaryotic beta-alanine synthases are functionally related but have a high degree of structural diversity. Genetics 158:999–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schnackerz KD, Andersen G, Dobritzsch D, Piskur J. 2008. Degradation of pyrimidines in Saccharomyces kluyveri: transamination of beta-alanine. Nucleosides Nucleotides Nucleic Acids 27:794–799. 10.1080/15257770802145983 [DOI] [PubMed] [Google Scholar]
- 16.Andersen G, Merico A, Bjornberg O, Andersen B, Schnackerz KD, Dobritzsch D, Piskur J, Compagno C. 2006. Catabolism of pyrimidines in yeast: a tool to understand degradation of anticancer drugs. Nucleosides Nucleotides Nucleic Acids 25:991–996. 10.1080/15257770600889386 [DOI] [PubMed] [Google Scholar]
- 17.Andersen G, Bjornberg O, Polakova S, Pynyaha Y, Rasmussen A, Moller K, Hofer A, Moritz T, Sandrini MPB, Merico AM, Compagno C, Akerlund HE, Gojkovic Z, Piskur J. 2008. A second pathway to degrade pyrimidine nucleic acid precursors in eukaryotes. J. Mol. Biol. 380:656–666. 10.1016/j.jmb.2008.05.029 [DOI] [PubMed] [Google Scholar]
- 18.Bjornberg O, Vodnala M, Domkin V, Hofer A, Rasmussen A, Andersen G, Piskur J. 2010. Ribosylurea accumulates in yeast urc4 mutants. Nucleosides Nucleotides Nucleic Acids 29:433–437. 10.1080/15257771003741265 [DOI] [PubMed] [Google Scholar]
- 19.Kim KS, Pelton JG, Inwood WB, Andersen U, Kustu S, Wemmer DE. 2010. The Rut pathway for pyrimidine degradation: novel chemistry and toxicity problems. J. Bacteriol. 192:4089–4102. 10.1128/JB.00201-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, Waterston R, Cohen BA, Johnston M. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71–76. 10.1126/science.1084337 [DOI] [PubMed] [Google Scholar]
- 21.Genolevures C, Souciet JL, Dujon B, Gaillardin C, Johnston M, Baret PV, Cliften P, Sherman DJ, Weissenbach J, Westhof E, Wincker P, Jubin C, Poulain J, Barbe V, Segurens B, Artiguenave F, Anthouard V, Vacherie B, Val ME, Fulton RS, Minx P, Wilson R, Durrens P, Jean G, Marck C, Martin T, Nikolski M, Rolland T, Seret ML, Casaregola S, Despons L, Fairhead C, Fischer G, Lafontaine I, Leh V, Lemaire M, de Montigny J, Neuveglise C, Thierry A, Blanc-Lenfle I, Bleykasten C, Diffels J, Fritsch E, Frangeul L, Goeffon A, Jauniaux N, Kachouri-Lafond R, Payen C, Potier S, Pribylova L, Ozanne C, Richard GF, Sacerdot C, Straub ML, Talla E. 2009. Comparative genomics of protoploid Saccharomycetaceae. Genome Res. 19:1696–1709. 10.1101/gr.091546.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elberry HM, Majumdar ML, Cunningham TS, Sumrada RA, Cooper TG. 1993. Regulation of the urea active transporter gene (DUR3) in Saccharomyces cerevisiae. J. Bacteriol. 175:4688–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujisawa H, Nagata S, Misono H. 2003. Characterization of short-chain dehydrogenase/reductase homologues of Escherichia coli (YdfG) and Saccharomyces cerevisiae (YMR226C). Biochim. Biophys. Acta Proteins Proteomics 1645:89–94. 10.1016/S1570-9639(02)00533-2 [DOI] [PubMed] [Google Scholar]
- 24.Yang Y, Zhu D, Piegat TJ, Hua L. 2007. Enzymatic ketone reduction: mapping the substrate profile of a short-chain alcohol dehydrogenase (YMR226c) from Saccharomyces cerevisiae. Tetrahedron Asymmetry 18:1799–1803. 10.1016/j.tetasy.2007.08.008 [DOI] [Google Scholar]
- 25.Agier N, Romano OM, Touzain F, Cosentino Lagomarsino M, Fischer G. 2013. The spatiotemporal program of replication in the genome of Lachancea kluyveri. Genome Biol. Evol. 5:370–388. 10.1093/gbe/evt014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsankov AM, Thompson DA, Socha A, Regev A, Rando OJ. 2010. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol. 8:e1000414. 10.1371/journal.pbio.1000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marzluf GA. 1997. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 61:17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gojković Z. 1999. Degradation of pyrimidines in yeast Saccharomyces kluyveri: genetic and molecular characterization of a novel fungal catabolic pathway. Ph.D. dissertation University of Copenhagen, Copenhagen, Denmark [Google Scholar]
- 29.Watt S, Mata J, Lopez-Maury L, Marguerat S, Burns G, Bahler J. 2008. urg1: a uracil-regulatable promoter system for fission yeast with short induction and repression times. PLoS One 3:e1428. 10.1371/journal.pone.0001428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vickers MF, Yao SYM, Baldwin SA, Young JD, Cass CE. 2000. Nucleoside transporter proteins of Saccharomyces cerevisiae. Demonstration of a transporter (FUI1) with high uridine selectivity in plasma membranes and a transporter (FUN26) with broad nucleoside selectivity in intracellular membranes. J. Biol. Chem. 275:25931–25938. 10.1074/jbc.M000239200 [DOI] [PubMed] [Google Scholar]
- 31.Jund R, Chevallier MR, Lacroute F. 1977. Uracil transport in Saccharomyces cerevisiae. J. Membr. Biol. 36:233–235. 10.1007/BF01868153 [DOI] [PubMed] [Google Scholar]
- 32.Grenson M. 1969. The utilization of exogenous pyrimidines and the recycling of uridine-5′-phosphate derivatives in Saccharomyces cerevisiae, as studied by means of mutants affected in pyrimidine uptake and metabolism. Eur. J. Biochem. 11:249–260. 10.1111/j.1432-1033.1969.tb00767.x [DOI] [PubMed] [Google Scholar]
- 33.Uemura T, Kashiwagi K, Igarashi K. 2007. Polyamine uptake by DUR3 and SAM3 in Saccharomyces cerevisiae. J. Biol. Chem. 282:7733–7741. 10.1074/jbc.M611105200 [DOI] [PubMed] [Google Scholar]
- 34.Filling C, Berndt KD, Benach J, Knapp S, Prozorovski T, Nordling E, Ladenstein R, Jornvall H, Oppermann U. 2002. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 277:25677–25684. 10.1074/jbc.M202160200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.