Abstract
The G2-M transition in Aspergillus nidulans requires the NIMA kinase, the founding member of the Nek kinase family. Inactivation of NIMA results in a late G2 arrest, while overexpression of NIMA is sufficient to promote mitotic events independently of cell cycle phase. Endogenously tagged NIMA-GFP has dynamic mitotic localizations appearing first at the spindle pole body and then at nuclear pore complexes before transitioning to within nuclei and the mitotic spindle and back at the spindle pole bodies at mitotic exit, suggesting that it functions sequentially at these locations. Since NIMA is indispensable for mitotic entry, it has been difficult to determine the requirement of NIMA for subaspects of mitosis. We show here that when NIMA is partially inactivated, although mitosis can be initiated, a proportion of cells fail to successfully generate two daughter nuclei. We further define the mitotic defects to show that normal NIMA function is required for the formation of a bipolar spindle, nuclear pore complex disassembly, completion of chromatin segregation, and the normal structural rearrangements of the nuclear envelope required to generate two nuclei from one. In the remaining population of cells that enter mitosis with inadequate NIMA, two daughter nuclei are generated in a manner dependent on the spindle assembly checkpoint, indicating highly penetrant defects in mitotic progression without sufficient NIMA activity. This study shows that NIMA is required not only for mitotic entry but also sequentially for successful completion of stage-specific mitotic events.
INTRODUCTION
Identification of cell cycle specific mutations in model organisms has been instrumental in the discovery of proteins required for progression through all stages of the cell cycle. The pioneering work of Ron Morris (1) enabled the isolation of numerous highly conserved genes required for mitotic progression utilizing the model filamentous fungus Aspergillus nidulans (2). A. nidulans undergoes both sexual and asexual development to produce dormant ascospores and conidiospores, respectively. Because of their ease of production, conidiospores (conidia) are most often used as inoculum for A. nidulans cell cycle analysis. Conidia are uninucleated dormant cells that upon exposure to suitable growth conditions first undergo isotropic growth, during which the first mitosis is often completed. After a single site for polarized growth is established, germ tube extension occurs, during which the two nuclei transition the cell cycle and parasynchronously undergo mitosis to generate germlings with four nuclei in a common cytoplasm. Typically only after the third synchronous mitotic division does septation occur (3–5).
In A. nidulans, the never in mitosis A (NIMA) and cell division cycle 1 (CDK1) kinases are essential for mitotic entry but not for short-term germling growth (6). Like Cdk1 (7–13) NIMA was identified through a genetic screen to isolate temperature-sensitive (ts) cell cycle mutants, some of which caused an arrest in interphase (never in mitosis [nim]mutants) at the restrictive temperature (1). Two nimA alleles contain point mutations causing amino acid substitutions in the catalytic domain of the kinase (nimA5 [Y91N] and nimA7 [E41G]), while the nimA1 allele has a change in the C-terminal regulatory domain (L304P) just downstream of the catalytic domain (14). Cells carrying these temperature-sensitive nimA alleles, when incubated at the restrictive temperature, arrest with a single G2 nucleus, duplicated spindle pole bodies (SPBs), and cytoplasmic microtubule architecture (15). When restored to the permissive temperature, these cells synchronously enter mitosis, indicating that mitotic entry is contingent upon NIMA activation (16, 17). NIMA is the founding member of the Nek family of NIMA-related kinases conserved through all eukaryotes, which have diverse roles in mitosis as well as ciliogenesis (18, 19). In Schizosaccharomyces pombe, the NIMA ortholog Fin1 plays a central role in the G2-M transition by mitigating the inhibition of Cdk1 activity by the Pom1 kinase as well as by establishing a positive feedback of Polo kinase activity to cause a switch-like increase in Cdk1 activity (20, 21). However, in A. nidulans, although NIMA is required for entry into mitosis, it is not required for mitotic activation of Cdk1 kinase activity via tyrosine dephosphorylation (17). On the other hand, mitotic activation of Cdk1 is required for the final complete mitotic activation of NIMA during entry into mitosis, most likely via phosphorylation of NIMA by mitotic Cdk1 (22). Moreover, through the use of temperature-sensitive nimA mutants, NIMA function has been shown to be required for the nuclear localization of cyclin B, which is critical for Cdk1-cyclin B-mediated phosphorylation of mitotic substrates (23). It has been shown that the dramatic mitotic targeting of NIMA to SPBs (24) and nuclear pore complexes (NPCs) at the initiation of mitosis also requires mitotic activation of Cdk1 (25, 26).
Emphasizing its functional conservation, overexpression of NIMA causes mitotic chromatin condensation not only in A. nidulans but also, strikingly, in fission yeast, Xenopus, and human cells (27, 28). NIMA can also promote the phosphorylation of histone H3S10, a mark of mitotic chromatin (25). Using a forward genetic screen, mutations in two NPC proteins, SONA and SONB, were identified as suppressors of the nimA1 temperature-sensitive mutation, suggesting that NIMA may regulate these nuclear pore proteins (23, 29). Consistent with that expectation, NIMA is required and sufficient to promote NPC disassembly, one of the earliest mitotic events, not only in A. nidulans (26, 30–33) but also vertebrate systems (34), and NIMA and related human kinases can phosphorylate the NPC protein Nup98 in vitro (34).
Since NIMA function is essential for all aspects for mitosis, it has been difficult to assess whether NIMA is required for specific mitotic events subsequent to the start of mitosis. However, the analysis of cells that are mutated in the gene encoding the anaphase-promoting complex (APC) subunit BIME in addition to carrying a nimA5 mutant allele (nimA5 bimE7) suggested the possibility that specific mitotic events might require NIMA function. The absence of bimE function abrogates the G2-M-mediated arrest in nimA5 cells, promoting premature mitotic entry causing abnormal spindle formation and nuclear envelope (NE) invaginations potentially due to the initiation of mitosis in the absence of normal NIMA function (35).
Our studies presented here using cells with partial NIMA function provide further strong evidence that in addition to being required for mitotic entry, NIMA is also required to regulate spindle pole body functions, nuclear pore complex permeability, and NE dynamics for successful mitotic generation of daughter nuclei.
MATERIALS AND METHODS
Standard conditions were used for propagating and generating A. nidulans strains, as described in reference 36 with minor alterations. The genotypes of strains used in this study are provided in Table S1 in the supplemental material. Live-cell confocal imaging was performed using a 60×, 1.49-numerical aperture (NA) TIRF objective lens on a Nikon 484 Eclipse TE 2000-U (Nikon, Inc.) microscope equipped with an UltraView ERS spinning-disk 485 confocal system (PerkinElmer Inc.), and images were captured using a Hamamatsu ORCA-AG 486 camera. Temperature-controlled experiments were carried out using Bioptechs Delta T dishes and heating equipment from Bioptechs, Inc., PA, USA. The cells were grown at the required temperature in humidity chambers for 4.5 h. The objective and the stage were calibrated for the correct temperature, and then the imaging dish was transferred to the temperature-controlled setup on the microscope. For confocal imaging at room temperature, conidiospores were germinated in 35-mm glass-bottom petri dishes (MatTek Cultureware). Images and movies were processed and signal intensity inside the nucleus measured using ImageJ version 1.46m (http://rsbweb.nih.gov/ij/). All images are maximum-intensity projections unless indicated otherwise. Rotational analysis was done using the Projector 4D plugin on ImageJ. Statistical analyses were done using MS Excel, except for box plots, which were generated on Stata software. For the box plots, the ends of the whiskers extend to 1.5 times the height of the box or, if there is no value in that range, to the minimum and maximum values. If the data are distributed normally, approximately 95% of the data are expected to lie between the whiskers. For analysis of mitosis in the presence of unpolymerized tubulin, cells were germinated in the presence of 2.4 μg/ml benomyl at the required temperature for 4.5 h before imaging at the same temperature.
RESULTS
Using live-cell spinning disk 4D confocal microscopy, we examined the first mitosis in germinating uninucleated conidia in which the function of the mitotic NIMA kinase was partially inhibited to identify the earliest defects caused and avoid the possibility of looking at cumulative effects of previous mitotic failures. We employed the nimA7 mutation because this is a tight nimA allele encoding a mutation in the catalytic domain that facilitated temperature-controlled imaging at 35°C under conditions that allowed some cell growth and mitotic progression. We examined nimA7 strains that carried fluorescently tagged proteins locating to the kinetochore, nucleus, and NE by live-cell microscopy. These studies revealed that 46% (n = 54) of cells with partial NIMA function fail to complete the first mitosis successfully (strains MG190, MG229, MG227, and MG228). Of these 46%, 11% undergo mitotic failure due to an inability to form a bipolar spindle, while 35% exhibit defects downstream of bipolar spindle formation. Below we report in more detail how normal NIMA function is required for different aspects of mitosis.
Cells with reduced NIMA function have defects in forming a bipolar spindle.
In A. nidulans, the kinetochores are clustered near the spindle pole body at the nuclear periphery during interphase. At mitotic entry, nuclear spindle microtubules are initially formed from duplicated, unseparated SPBs in a monopolar fashion. Subsequently, the separation of the SPBs results in the bipolarization of the spindle. We generated nimA7 strains that carried green fluorescent protein (GFP)-Tub (TubA) (37) and the kinetochore marker Ndc80-mCherry Red (Ndc80-CR) (38) to monitor spindle formation and kinetochore segregation, respectively. In nimA7 cells grown at the permissive temperature of 22°C, the kinetochores are in a single focus during interphase, as expected. During mitosis, the kinetochores become spread along the spindle and then divide equally, regenerating SPB-associated foci within daughter nuclei upon successful completion of mitosis (Fig. 1A). When NIMA is partially inactivated by germinating the same strain at the semipermissive temperature of 35°C, we find that 11% (n = 54) of the cells form a monopolar spindle. These spindles appear to be arrested at the monopolar stage due to an inability to separate the duplicated SPBs, as indicated by the presence of clustered unseparated kinetochores (Fig. 1B). The monopolarity of the spindle was further confirmed in nimA7 cells carrying GFP-Tub and an SPB marker, the A. nidulans ortholog of Schizosaccharomyces pombe Sad1 (annotated as AN6868 at The Aspergillus Genome Database [AspGD]) (39), at the semipermissive temperature (data not shown), where 14% of SPBs (n = 28) were seen to remain as a single focus for the duration of mitosis.
FIG 1.
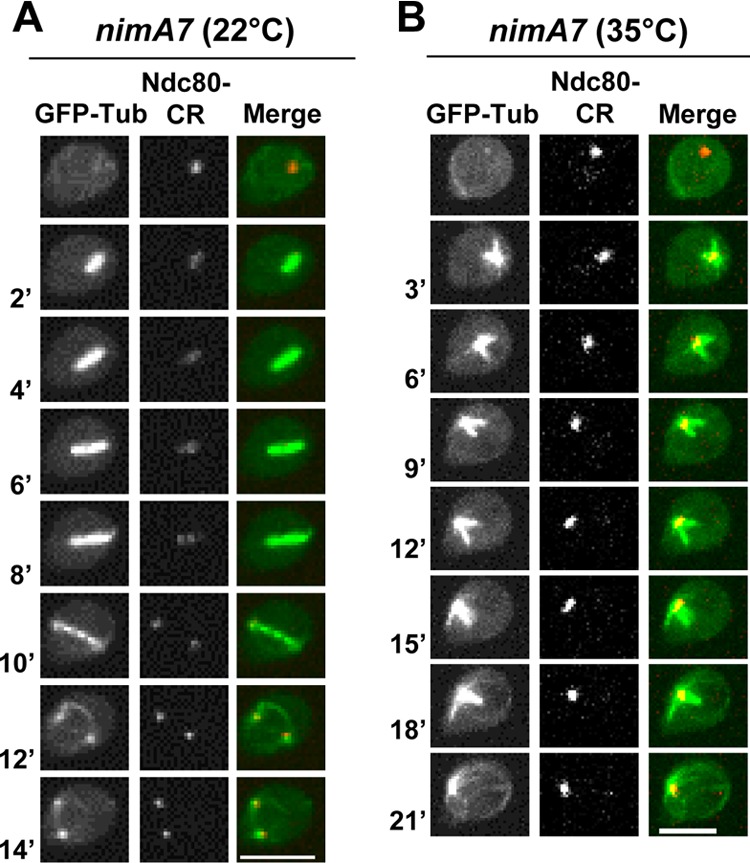
Monopolar spindle formation in cells with partial NIMA function. nimA7 cells carrying GFP-Tub and the kinetochore protein Ndc80-CR (strain MG190) were followed through mitosis at 22°C (A) or at 35°C (B). Bars, 5 μm.
Cells with reduced NIMA function show defects in anaphase completion, defective segregation of the nucleolus, and abnormal NE dynamics during mitosis.
In wild-type (WT) cells, mitotic entry is accompanied by chromatin condensation and spindle formation. During anaphase (Fig. 2A, 3′), the bipolar spindle segregates the chromatin and subsequent chromatin decondensation occurs during the formation of two G1 nuclei (Fig. 2A). In 35% of the nimA7 cells (n = 54), we find that a bipolar spindle is established; however, mitosis is not completed successfully, as chromatin fails to segregate completely upon spindle elongation (Fig. 2B, 8′, arrowhead) and collapses back into a single mass (Fig. 2B). In nimA7 cells that form a bipolar spindle carrying Ndc80-CR, GFP-Tub, and the spindle pole body marker GCP3-GFP (33, 40), the kinetochores could be seen to segregate, giving rise to two Ndc80 foci in an apparently normal fashion (Fig. 2C, arrowheads). Surprisingly, however, the segregation of Ndc80 was not maintained, and the kinetochores collapsed back to a single focus indicative of a defect in anaphase completion. Remarkably, the failure to segregate the kinetochores was accompanied by the separated SPBs also fusing back to give a single focus (Fig. 2C, 14′) at mitotic exit. Together with the examination of nimA7 strains carrying the SPB marker Sad1 or TinA (41) (data not shown), we find that 12% (n = 50) of nimA7 cells showed mitotic bipolar spindle formation and SPB separation followed by their reassociation into closely paired unresolvable foci, demonstrating that the phenotype is reproducible using different SPB markers. The population of nimA7 cells showing this phenotype is likely to be an underestimate, since clear observation of this phenotype is limited by the orientation of the mitotic apparatus. Collectively, these results show that partial inactivation of NIMA impairs successful anaphase completion.
FIG 2.
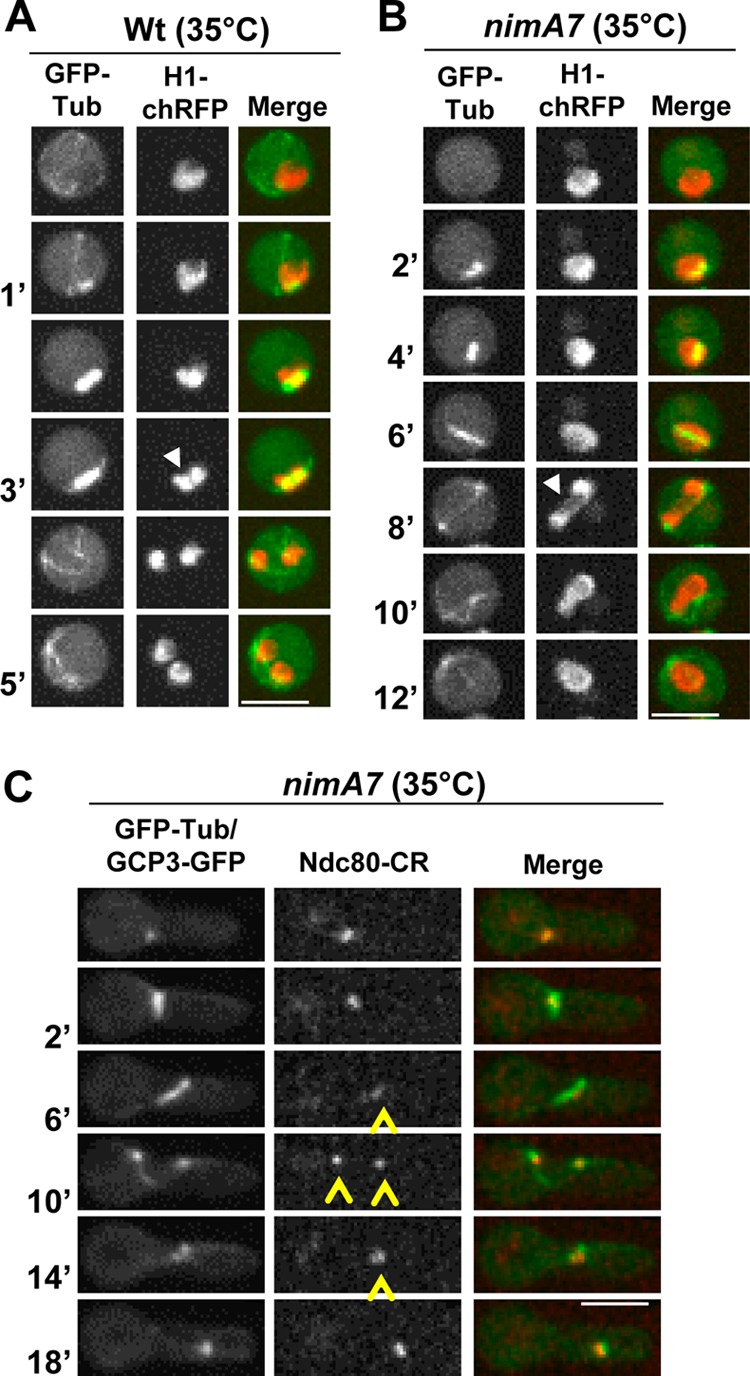
Failure of nuclear division in cells with partial NIMA function subsequent to bipolar spindle formation. (A and B) Histone H1-chRFP and GFP-Tub were used to follow mitosis in WT (strain MG300) (A) and nimA7 (strain MG298) (B) cells at 35°C. In panel A, the arrowhead points to chromatin segregation in anaphase. In panel B, the arrowhead points to chromatin that is attempting to segregate but eventually collapses into a single nucleus after failed anaphase. (C) nimA7 cell carrying GFP-Tub, GCP3-GFP and Ndc80-CR (strain MG190) followed through mitosis at the semipermissive temperature of 35°C. Arrowheads indicate kinetochore foci as they first become segregated (6 to 10 min) and then return back together (10 to 14 min). The GCP3-GFP foci at separated SPBs at 10 min also collapse back together by 14 min. Bars, 5 μm.
To monitor NE dynamics, a recently identified inner nuclear membrane (INM) marker that contains a C-terminal transmembrane domain with similarity to the S. pombe INM protein Bqt4 (S. A. Osmani and M. Chemudupati, unpublished data) (annotated at AspGD as AN0162 [http://www.aspergillusgenome.org/cgi-bin/locus.pl?dbid=ASPL0000057939]) was utilized. During WT mitosis, the NE, marked by CR-AN0162, restricts at two points, giving rise transiently to 3 compartments (Fig. 3A, arrows). The middle compartment contains the nucleolus, and the two compartments at either end encompass the segregated chromatin of the forming daughter nuclei (42). Interestingly, 30% (n = 27) of cells with reduced NIMA function show a defect in executing the NE double pinch (Fig. 3B). Moreover, a significant proportion of nimA7 cells (65%, n = 17 failed first mitoses) show the formation of an apparent intranuclear ring-like structure of NE (Fig. 3B, arrowhead), a phenomenon not observed after normal mitosis. We refer to this NE configuration as “theta” (θ), owing to its resemblance to the Greek letter. Intriguingly, we found, using a tagged nucleolar marker protein fibrillarin (Fib-CR) (42), that the nucleolus is present within the ring-like structure in the θ nuclei observed (Fig. 3D). Z sections through the nucleus (Fig. 3E) and three-dimensional (3D) rotations (see Movie S1 in the supplemental material) revealed that the nucleolus was positioned in an aberrant location within the θ nuclei, causing a protrusion which changed the normal oval shape of interphase nuclei. Notably this configuration was dynamic, with the nucleolar protrusion changing to become more or less prominent through time. In contrast to the normal nucleolar segregation observed in WT cells at the same temperature (see Fig. S1 in the supplemental material) (42), we also observed that some nimA7 nuclei apparently managed to segregate their nucleolus into two (Fig. 3C, 5′ to 6′), only to then have the two nucleoli collapse back into one (Fig. 3C, 7′). The data indicate that successful generation of daughter nuclei via mitotic NE double restrictions, coupled with nucleolar segregation, requires normal NIMA functions.
FIG 3.
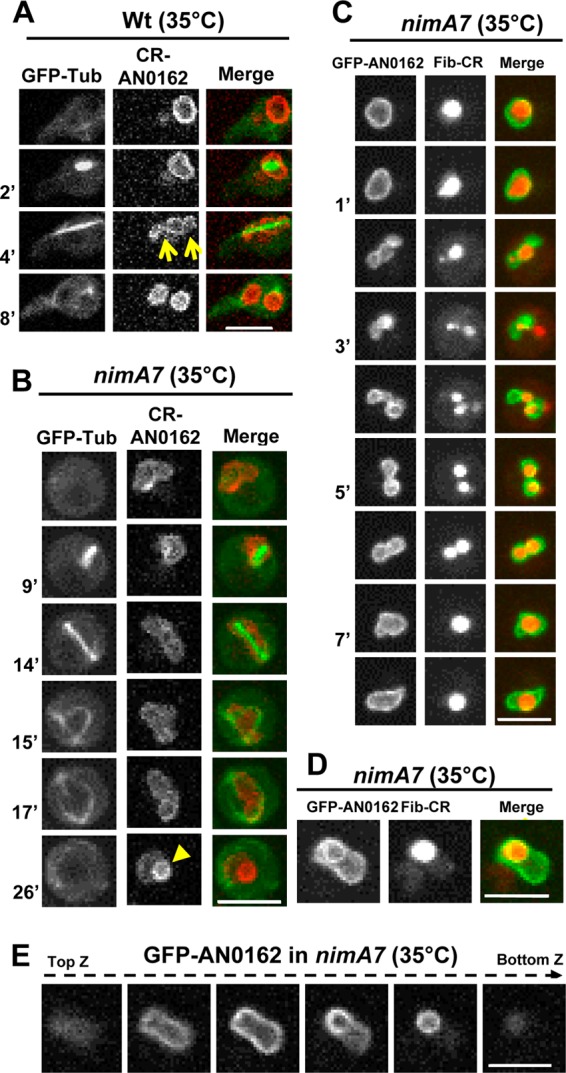
Defective NE dynamics and nucleolar segregation in cells with partial NIMA function. (A and B) Mitotic dynamics of the NE marker protein CR-AN0162 and GFP-Tub in WT (A) (strain MG224) and nimA7 (B) (strain MG227) cells. The arrows in panel A indicate the normal NE restriction points during telophase. The arrowhead in panel B at 26 min shows the formation of the “theta”-shaped nuclear structure (see the text for details). (C) Mitotic dynamics of the nucleolus marked by fibrillarin-CR and GFP-AN162 in a nimA7 nucleus (strain MG294) that fails to generate two daughter nuclei after first mitosis even though the nucleolus first appears to be segregated into two. (D) The nucleolar protein fibrillarin-CR (strain MG294) is apparently enveloped by GFP-AN0162 (arrow). (E) Z sections through a nimA7 nucleus marked with GFP-AN162 reveal an NE protrusion and the abnormal shape of the nucleus (see also Movie S1 in the supplemental material). Bars, 5 μm.
Cells with reduced NIMA function are unable to undergo normal nuclear pore complex disassembly during mitosis.
In A. nidulans, around half of the nuclear pore complex (NPC) proteins, representing the peripherally located components, disperse from NPCs during mitosis, abolishing nuclear transport (26, 30). Complete NIMA inactivation prevents initiation of mitosis and thus prevents downstream NPC disassembly, while ectopic expression of NIMA promotes partial nuclear pore complex disassembly, even out of cell cycle phase (26). Therefore, NIMA has been proposed to initiate mitosis at least in part by promoting NPC disassembly, which allows tubulin and mitotic regulators to access the nucleoplasm. The mitotic failure in cells with partial NIMA function might therefore be due to a defect in disassembling the NPCs. To test this, we generated strains having WT nimA or the nimA7 mutant allele, in which the location of the NPC protein Nup49-CR can be followed in addition to GFP-Tub. Nup49 is a peripheral nucleoporin that locates to the NPCs during interphase but disperses from them at mitosis (26, 43). Most surprisingly, in cells with reduced NIMA function, the majority of the Nup49-CR signal remained at the nuclear periphery during mitosis, when it should be dispersed as the mitotic spindle was formed (Fig. 4A, 3′). This dramatic defect, which was seen in all nimA7 cells at 35°C, indicates that in addition to being sufficient to promote NPC disassembly, NIMA function is actually required for NPC disassembly during mitosis after nuclei are triggered to undergo the G2-M transition. To further define the status of NPCs in mitotic nimA7 cells, we visualized nuclear transport by following NLS (nuclear localization sequence)-DsRed, a marker protein that is actively transported into interphase nuclei (44). During interphase, NLS-DsRed is exclusively nuclear (Fig. 4C). During mitosis, partial NPC disassembly results in the dispersal of NLS-DsRed until active transport is reestablished in daughter G1 nuclei (Fig. 4C). However, when NIMA is partially functional, NLS-DsRed does not disperse from nuclei upon mitotic entry, as marked by formation of the mitotic spindle, but continues to remain within nuclei (Fig. 4D, 2′ and 4′). This indicates that in mitotic nimA7 cells, both partial NPC disassembly and the opening of nuclear pores are defective.
FIG 4.
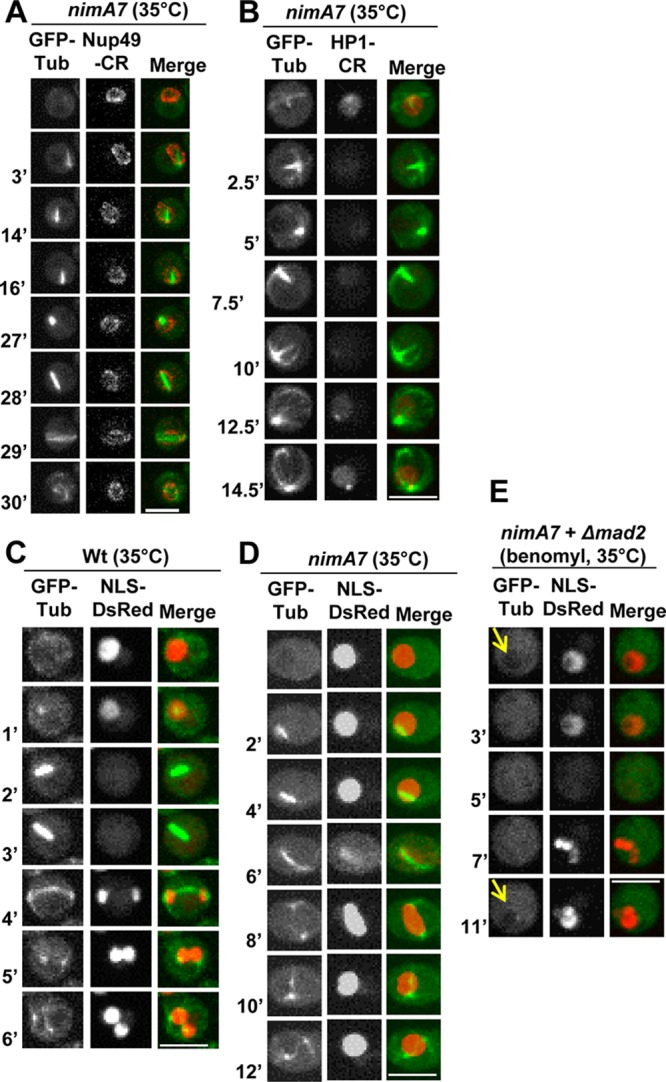
Cells with reduced NIMA function have defects in mitotic NPC disassembly. (A) Mitosis in a nimA7 cell carrying GFP-Tub and the peripheral nucleoporin Nup49-CR (strain MG321) at the semipermissive temperature. During spindle formation, unlike in WT cells, most Nup49 remains associated at the NE. (B) Mitosis in a nimA7 cell carrying the heterochromatin binding protein HP1-CR and GFP-Tub (strain MG378) at the semipermissive temperature. When spindles form, HP1-CR is released from nuclei. (C and D) Nuclear transport marked by NLS-DsRed and spindle formation (GFP-Tub) followed in mitotic WT cells (C) (strain MG273) and nimA7 cells (D) (strain MG229). (E) Behavior of NLS-DsRed in nimA7 Δmad2 cells (strain MG313) during mitosis in the presence of benomyl. Arrows indicate the nuclear shadow revealed by cytoplasmic GFP-Tub. Bars, 5 μm.
It has been proposed that NPC disassembly functions to allow the entry of tubulin into the nucleus, thus promoting spindle formation. However, since spindles were able to form in nimA7 cells even though NLS-DsRed fails to escape nuclei, we wanted to confirm whether tubulin is able to enter the nucleus during mitotic entry in these cells. To this end, we quantitated the change in tubulin signal inside the nucleus using single confocal sections through the middle of WT and nimA7 nuclei (Fig. 5). In WT interphase nuclei, unpolymerized fluorescently tagged tubulin is excluded from the nucleus, revealing a nuclear shadow in late G2 (Fig. 5A, arrow). Upon mitotic entry, this mitotic shadow fills in as tubulin equilibrates across the NE concurrent with the dispersal of NLS-DsRed from the nucleus (Fig. 5A to C). Interestingly in cells with partial NIMA function, nuclear GFP-Tub increases sharply at the G2-M transition; however, the nuclear signal of NLS-DsRed does not decrease precipitously as it does in the WT cells (Fig. 5D to F). This shows that when NIMA is partially inactive, tubulin is able to access the mitotic nucleoplasm and hence form the spindle even though NLS-DsRed is unable to escape from the mitotic nucleus in the normal fashion. This might suggest that tubulin is actively transported into these nuclei at mitosis. One alternative explanation, however, is that in the absence of normal NIMA function, the nuclear pores are insufficiently opened during mitosis, such that α-β tubulin dimers (GFP-TubA-BenA [127 kDa] [39]) can diffuse across the NE, while proteins of larger molecular size and/or shape, such as the tetrameric NLS-DsRed (190 kDa) (45), are prevented from escaping the nucleus. To test this idea, we examined the behavior of heterochromatic protein 1 (HP1) (also termed HepA in A. nidulans) (46), which is nuclear during interphase and disperses during mitosis. We find that HP1-CR, which is expected to be 53 kDa, does indeed diffuse from mitotic nuclei in nimA7 cells (Fig. 4B). This indicates that partial inactivation of NIMA allows only incomplete NPC disassembly, such that small proteins like tubulin and HP1 can diffuse across nuclear pores while larger protein complexes, such as NLS-DsRED, remain trapped within mitotic nuclei.
FIG 5.
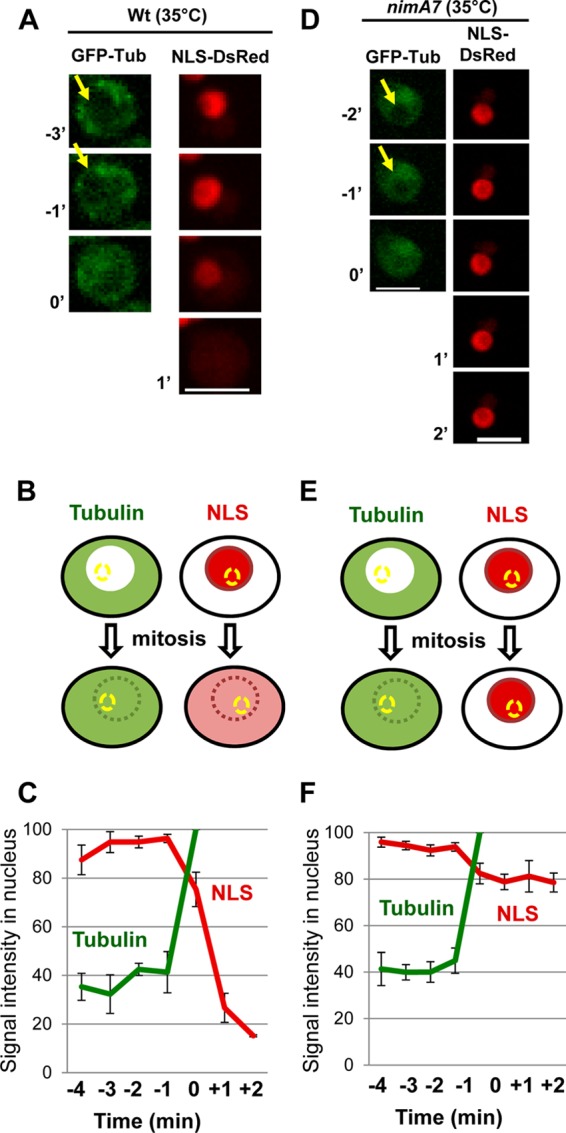
Tubulin can enter the mitotic nucleus in cells with reduced NIMA function. (A and D) Single Z-slice images of GFP-Tub and NLS-DsRed in WT and nimA7 cells at 35°C during the G2-M transition. Arrows mark the shadow of the nucleus revealed by the absence of GFP-tubulin. Bars, 5 μm. (B and E) Schematic representation of the location of tubulin and NLS-DsRed in interphase versus mitosis in WT cells (B) and nimA7 cells (E). Nuclear pore complex disassembly in mitosis is indicated by the dotted line enveloping the nucleus. The signal intensity in the yellow-dotted area was measured. (C and F) Changes in signal intensities of tubulin (green) and NLS-DsRed (red) with time during the G2-M transition of WT (C) (n = 4, strain MG273) or nimA7 (F) (n = 6, strain MG229) cells.
Intriguingly, in nimA7 cells that form a bipolar spindle, although NLS-DsRed does not disperse at mitotic entry, it does escape the nucleus transiently at a point corresponding to spindle elongation, suggesting that perhaps the mechanical force of spindle elongation compromises NE integrity (Fig. 4D, 6′). To test whether the release of NLS-DsRed is caused by mechanical forces acting on the NE driven by the elongating spindle, we germinated nimA7 cells with mad2 deleted and carrying GFP-Tub and NLS-DsRed in the presence of benomyl, a microtubule-depolymerizing drug. The absence of Mad2 prevents the engagement of the spindle assembly checkpoint (SAC)-mediated mitotic arrest in the presence of unpolymerized tubulin (32, 43), allowing nuclei to enter and then exit mitosis without a mitotic spindle. As in WT cells, during G2, GFP-Tub is excluded from the nucleus, revealing a nuclear shadow in the nimA7 Δmad2 cells treated with benomyl (Fig. 4E, arrow). Entry into mitosis was marked by the diffusion of soluble GFP-Tub into the nucleus, as expected (37), but NLS-DsRed remained nuclear at this time. In contrast, in WT cells with normal NIMA function, NLS-DsRed is known to be released upon mitotic entry under identical conditions (43). Importantly, we found that NLS-DsRed still dispersed briefly in the absence of spindle formation in mitotic nimA7 cells (Fig. 4E, 5′) before then being reimported into the G1 nucleus. In G1, GFP-Tub is once again excluded from the nucleus (Fig. 4E, 11', arrow). The data show that in the absence of forces exerted by spindle microtubules, NLS-DsRed is able to disperse briefly in cells with partial NIMA function, indicating that the mitotic permeability properties of the NE are being modified in a manner independent of the mitotic spindle (see Discussion).
Insufficient NIMA function causes a delay in mitotic progression.
The mitotic defects seen in cells with partial NIMA function suggest that nimA7 cells may be delayed in mitotic progression, as indicated by comparing the times of mitosis for WT and nimA7 cells in Fig. 1 and 2. To quantitate this, we calculated the average time that spindle microtubules are detectable in WT and nimA7 cells from time lapse analysis of mitosis at the semipermissive temperature for nimA7. Cells with partial NIMA function show the presence of spindle microtubules for more than double the time on average that WT cells do and also display more variability in time in mitosis (see Fig. S2A and B in the supplemental material). There was no correlation between the time spent in mitosis and whether the mitosis was successful or not (data not shown). In order to determine whether the delay in mitosis seen in cells with partial NIMA function is specific to this allele of nimA, we followed mitosis in strains with the two other available temperature-sensitive nimA alleles, nimA1 and nimA5. We find that spindle microtubules are similarly present for a prolonged time period in nimA1 and nimA5 cells compared to WT cells (see Fig. S2C in the supplemental material), demonstrating that partial inactivation of NIMA results in prolonged mitosis independent of the allele used.
Cells with partial NIMA function are dependent on the spindle assembly checkpoint for their survival.
The spindle assembly checkpoint functions to monitor mitotic errors and impose a delay in mitosis so that these errors can be corrected before anaphase is triggered. Given that partial NIMA function results in multiple mitotic defects, as well as a delay in mitosis, it is possible that the completion of mitosis in nimA7 cells might be dependent on the SAC. To test this, we generated strains carrying nimA7 that have a deletion of a gene essential for SAC function, mad2 (32). To examine whether the deletion of mad2 affects the growth of nimA7 cells, we compared the colony growth of the nimA7 Δmad2 mutant and the single mutants at the semipermissive temperature. The double mutants lacking mad2 with partial NIMA function are unable to form any visible colonies at this temperature (Fig. 6A), highlighting the requirement of the SAC for the survival of nimA7 cells with partial NIMA functions. Using live-cell microscopy and NLS-DsRed and GFP-Tub as markers, we find that nimA7 Δmad2 cells show a highly penetrant inability to complete their first mitosis successfully, where nuclei enter mitosis but fail to generate daughter nuclei (Fig. 6B and C). Because GFP-Tub is excluded from G2 nuclei but diffuses into the nucleus during mitosis and is reexported back out of the nucleus during G1, nuclear tubulin can be used as a marker of mitosis. We find that mitotic nimA7 Δmad2 cells have tubulin within the nucleus for an average of 5 min, compared to a longer 12 min in nimA7 cells (Fig. 6B and D). However, surprisingly, we see that even after tubulin is exported from the nucleus, astral microtubules continue to extend from the cytoplasmic side of nimA7 Δmad2 nuclei (Fig. 6B). Taken together, these data show that completion of mitoses in cells with partial NIMA function is dependent on SAC function.
FIG 6.
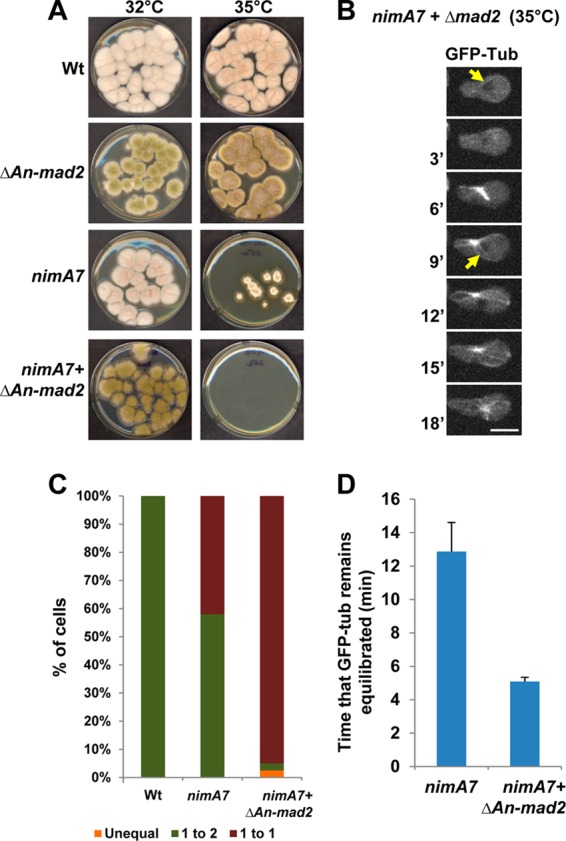
Cells with reduced NIMA function depend on the spindle assembly checkpoint (SAC) for survival. (A) Colony growth of strains of the indicated genotypes at 35°C after 72 h. Strains: WT, R153; nimA7, CDS790; ΔAn-mad2, CDS629; nimA7+ΔAn-mad2, MG381. (B) Mitosis as followed using NLS-DsRed and GFP-Tub in a nimA7 cell that also lacks mad2 (strain MG313). Arrows point to the exclusion of GFP-Tub from the nucleus in G2 and then G1. Bar, 5 μm. (C) Quantitation of the number of cells that complete the first mitosis successfully versus those which do not in WT (n = 15), nimA7 (n = 26), and nimA7 Δmad2 (n = 40) strains. Unequal division refers to a mitosis resulting in daughter nuclei of different sizes. (D) Quantitation of the time that GFP-Tub is seen equilibrated across the NE (n = 11 for each).
To ascertain whether the two nuclei generated in nimA7 cells after an apparently successful first mitosis are normal, we followed them into their second mitosis (see Fig. S3 in the supplemental material). Surprisingly, we find that in such cells, daughter nuclei can behave differently from each other. As shown in the example in Fig. S3B in the supplemental material, although both nuclei enter mitosis forming spindles, one nucleus (n2) attempts anaphase, while the other (n1) does not. The nucleus that does not attempt anaphase also does not disperse NLS-DsRed and likely forms a monopolar spindle, whereas nucleus n2 disperses NLS-DsRed briefly (at 6′). However, since nucleus n1 is actively transporting, much of the NLS-DsRed dispersed from n2 is available for import into nucleus n1. Thus, the two nuclei undergo asynchronous mitoses in an autonomous manner, in sharp contrast to the parasynchronous mitoses seen in all WT cells.
The generation of two nuclei after first mitosis in 54% of nimA7 cells depends on SAC function, indicating that the SAC is functional. However, the remaining 46% of the first mitoses fail to generate daughter nuclei, although nuclei exit mitosis, suggesting that perhaps SAC function is not completely normal in these nimA7 cells. To test whether SAC-mediated mitotic arrest can be appropriately engaged in the absence of normal NIMA function, we followed mitosis in nimA7 cells at the semipermissive temperature in the presence of unpolymerized GFP-tubulin. Mitotic initiation can be followed by the entry of GFP-Tub into the nucleus, which in WT cells is followed immediately by the dispersal of NLS-DsRed from the nucleus (Fig. 7A and C; see Fig. S4A in the supplemental material). However, in 50% of the nimA7 cells (n = 30), entry of GFP-Tub into the nucleus was not accompanied by dispersal of NLS-DsRed (data not shown), while in the other half of the population, NLS-DsRed dispersed after an average delay of 18 min (Fig. 7B and C; see Fig. S4A in the supplemental material). This suggests that the delay in mitosis imposed by the SAC allows some nimA7 cells to accumulate enough NIMA function so as to eventually affect NLS dispersal. To compare the time of mitotic SAC arrest between WT and nimA7 cells, mitotic exit could be monitored by the reimport of NLS-DsRed, as this is an M-G1 transition-regulated event. The actual time of mitotic SAC arrest could therefore be measured as the time between entry of GFP-Tub into nuclei (start of mitosis) and the nuclear reimport of NLS-DsRed (mitotic exit) (Fig. 7). In WT cells, mitotic entry in the presence of the microtubule-depolymerizing drug benomyl results in a SAC-mediated mitotic arrest, as previously documented, for an average of 40.6 ± 10 min (Fig. 7A and D; see Fig. S4B in the supplemental material) (43). Importantly, mitosis in cells with partial NIMA function in the absence of spindles was similarly prolonged to an average of 46 ± 13 min (Fig. 7B and D; see Fig. S4B in the supplemental material) in a manner dependent on the SAC (Fig. 4E). The data indicate that the SAC is engaged normally during first mitosis in the absence of spindles in nimA7 cells at the semipermissive temperature.
FIG 7.
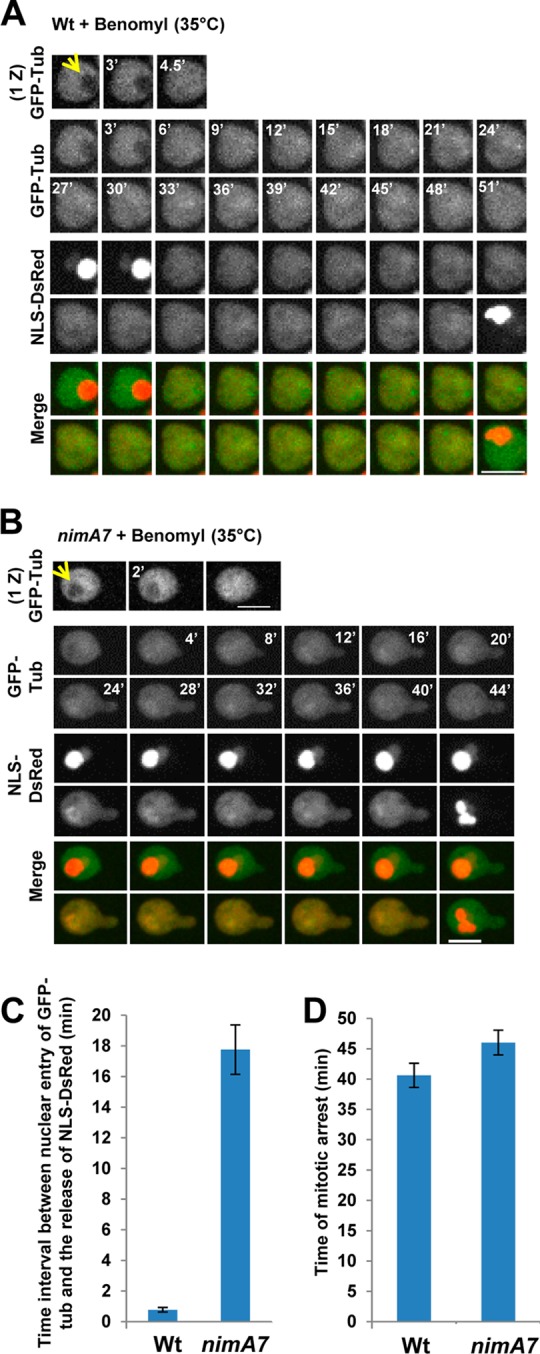
Cells with partial NIMA function are capable of engaging the SAC-mediated mitotic arrest in response to unpolymerized GFP-Tub. (A) Mitosis in a WT cell (strain MG273) in the presence of benomyl, a microtubule-depolymerizing drug. Mitotic entry is indicated by the filling in of the GFP-Tub nuclear shadow (arrow) followed immediately by the dispersal of NLS-DsRed throughout the cytoplasm. (B) Mitosis in a nimA7 cell (strain MG229) in the presence of benomyl at the semipermissive temperature (35°C). The dispersal of NLS-DsRed is seen after a time lag following the entry of GFP-Tub into the nucleus. Bars, 5 μm. (C) Quantitation of the time interval between the entry of GFP-Tub into the nucleus and the dispersal of NLS-DsRed in WT (n = 25) versus nimA7 (n = 41) cells at 35°C. The difference in time interval between nuclear entry of GFP-Tub and the release of NLS-DsRed between WT and nimA7 cells is highly significant (P = 0.00 by unpaired t test). (D) The length of SAC-mediated mitotic arrest in the absence of spindles is quantitated as the time interval between the entry of GFP-Tub into the nucleus and the reimport of NLS-DsRed at mitotic exit for WT (n = 25) cells and nimA7 (n = 41) cells at the semipermissive temperature. The difference in mitotic arrest time between WT and nimA7 cells is not significant (P > 0.01 using unpaired t test).
DISCUSSION
Initiation of mitosis in the absence of sufficient NIMA function.
When NIMA is partially inhibited by incubating strains having the nimA7 temperature-sensitive allele at semipermissive temperatures, nuclear division can be initiated. Because NIMA is not required for activation of mitotic Cdk1 kinase activity, this is presumably because low levels of NIMA7 activity are sufficient to trigger mitotic entry, since CDK1 is expected to be mitotically activated. However, in the absence of normal NIMA function, penetrant defects in specific mitotic events ensue. Therefore, the analysis of nimA7 cells at the semipermissive temperature enabled the uncoupling of NIMA's requirement for mitotic initiation from its functions during mitotic progression and proved to be a powerful approach to uncover the requirement for NIMA in regulating specific mitotic events (Fig. 8).
FIG 8.
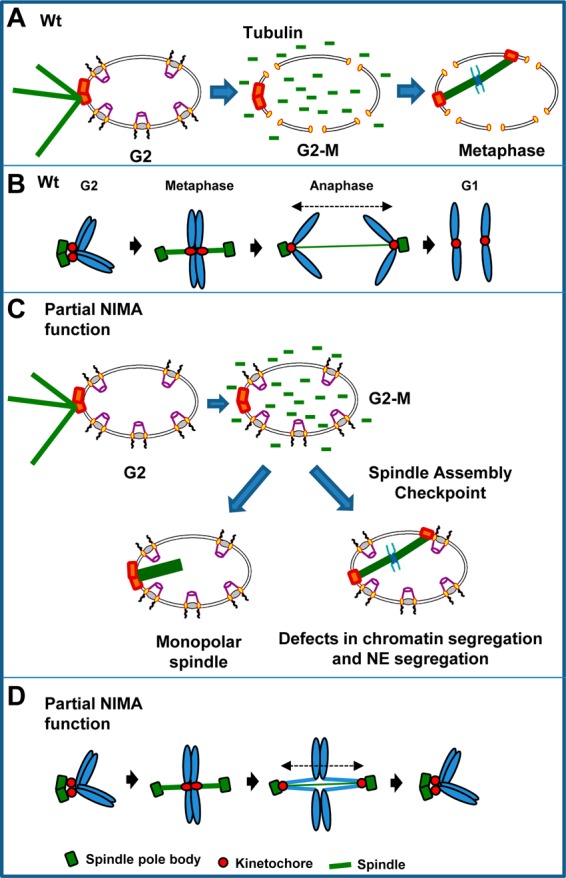
Schematic representation of the mitotic defects seen in cells with partial NIMA function. (A) In WT cells at the semipermissive temperature, mitotic entry is accompanied by the partial disassembly of the nuclear pore complexes, which allows the entry of unpolymerized tubulin into the nucleus and the formation of the metaphase spindle. (B) Schematic representation of the behavior of the spindle pole body, kinetochores, and chromatin in WT cells. (C) In cells with partial NIMA function (nimA7 cells, 35°C), the disassembly of nuclear pore complexes at the G2-M transition is incomplete. However, this incomplete NPC disassembly is sufficient to allow tubulin into the nucleus and spindle formation. In the absence of sufficient NIMA, about half of the cells fail to complete mitosis successfully either due to an inability to establish a bipolar spindle or due to defects in anaphase completion and NE dynamics. The successful mitoses in nimA7 cells are dependent on the function of the spindle assembly checkpoint. (D) Schematic representation of the behavior of the spindle pole body, kinetochores, and the chromatin in some cells with partial NIMA function. (The parts of the cartoons in panels A and C depicting the NE and nuclear pore complexes were adapted from reference 63 with permission.)
NIMA-mediated regulation of spindle pole body separation.
Partial inactivation of NIMA leads to monopolar spindle formation implicating NIMA in the separation of the duplicated spindle pole bodies during mitotic entry (Fig. 8A and C). This provides strong evidence for the functional significance of the localization of NIMA-GFP to SPBs as the first step in promoting the G2-M transition (24). The formation of monopolar spindles is likely not due to a defect in SPB duplication, since SPB duplication has been shown to occur in the absence of NIMA function and bipolar spindles form immediately upon return of nimA mutants to a permissive temperature (15, 47, 48). The human NIMA-related kinase Nek2 regulates the removal of centrosomal linker proteins C-Nap1, rootletin, and β-catenin through phosphorylation, while the Nek9-Nek6/Nek7 phosphorylation cascade regulates the function of the BIMC kinesin Eg5 during centrosome separation (19, 49–52). Along similar lines, a temperature-sensitive mutant of the S. pombe NIMA ortholog fin1 was identified in a screen for spindle architecture defects where the microtubule nucleation of one of the spindle pole bodies was affected (53). Moreover, Fin1 localizes to the spindle pole body from G2 to mitotic exit and regulates SPB function via phosphorylation during the G2-M transition (20, 54). Our functional analysis of NIMA, revealing its role in regulating microtubule-organizing centers during mitosis, further demonstrates that this function is highly conserved.
NIMA-mediated regulation of mitotic NPC disassembly and NE permeability.
Dispersal of peripheral nucleoporins (Nups) can be induced by NIMA overexpression in a cell cycle-independent manner (26). Our data showing that the peripheral Nup49 does not disperse at mitosis in cells with partial NIMA function provide strong evidence that NIMA is not only sufficient but also required for partial mitotic disassembly of NPCs in cells that have transitioned from G2 into mitosis. Furthermore, our data revealed that in the absence of normal NIMA function, nuclear pores are opened incompletely compared to those of WT cells during mitotic progression (Fig. 8A and C). This allowed HP1-CR and GFP-tubulin to traverse the mitotic NPCs at mitotic entry, but not the larger NLS-DsRED tetramer. Because mitotic nuclear pore complex disassembly happens in a hierarchical manner (55), it is possible that partial NIMA function is sufficient to promote only the early steps of NPC disassembly. Alternatively, there might be a more general defect in the extent of dispersal of all peripheral Nups. In either scenario, our findings indicate that the distinctive location of NIMA at nuclear pore complexes during mitosis (24, 26) is reflective of its function to open NPCs to promote mitosis, a function recently shown to be conserved in mammalian cells (34).
Interestingly, although in nimA7 cells NLS-DsRed does not disperse at mitotic entry it does disperse, albeit momentarily, coincident with spindle elongation, even though there is no detectable dispersal of Nup49. This suggested that perhaps mechanical forces driven by spindle elongation might be responsible for NE tearing and transient release of NLS-DsRed. Intriguingly, however, we found that NLS-DsRed disperses in nimA7 mitotic cells even when microtubules are depolymerized. This indicates that subsequent to the partial disassembly of the NPCs at the G2-M transition, permeability across the NE is further increased at a later time point in mitosis. In support of this idea, the dispersal of proteasomal components, which are expected to be in large complexes, has been observed to occur later than NPC disassembly in WT A. nidulans cells but can be made to occur earlier when the mitotic nuclear pores are more dramatically disassembled and further opened in the absence of nup37 and elys (Yi Xiong and Berl Oakley, personal communication). Furthermore, NE breakdown in anaphase has been demonstrated in the fission yeast Schizosaccharomyces japonicas in a manner independent of the mitotic spindle, similar to the case for our nimA7 cells (56). This suggests the existence of a second mechanism, in addition to partial NPC disassembly, in A. nidulans regulating the permeability properties of the NE late in mitosis that is potentially important for normal mitotic progression.
Functions for NIMA in successful segregation of SPBs, kinetochores, and chromosomes.
Some cells with partial NIMA function fail to divide their nuclei after establishment of a bipolar spindle, with the kinetochores and chromatin transiently segregating but then collapsing back together. This potentially reflects defects in sister chromatin separation, which is known to involve cohesin and topoisomerase II (57, 58), and potentially defects in chromosome condensation and the regulation of condensin (59). Such defects would help explain how kinetochores can apparently be successfully segregated but still be attached via unresolved sister chromatid cohesion and/or catenation (Fig. 8D). Then, upon mitotic spindle dissolution, sister chromatids might collapse back together, effectively reassociating sister kinetochores (Fig. 8D). Importantly, it has been shown that chromosomes can act like springs to counteract the motive force of the mitotic spindle, which would also support this model (60). Because separated SPBs also collapse back together in some nimA7 cells, this suggests that some connection is maintained between the segregated kinetochores and the separated SPBs before kinetochores are pulled back together via unresolved sister chromatids (Fig. 8D).
It is possible that some defects during mitosis caused by lack of NIMA function might reflect secondary effects of insufficient NIMA activity toward SPBs and/or NPCs earlier during mitosis. Although we do not discount this possibility, because NIMA has dynamic mitotic location on the mitotic apparatus after it has located to SPBs and NPCs, it is also possible, if not likely, that later mitotic defects do reflect deficiencies in NIMA function at these later mitotic locations.
Full NIMA activity is required for normal NE dynamics and G1 nuclear structure.
During A. nidulans mitosis, the NE undergoes restriction and abscission at two points to generate two daughter nuclei coupled with segregation of the nucleolus (42). NIMA has been implicated in regulation of these complex mitotic NE dynamics. Expression of a truncated version of TINC, a yeast two-hybrid-interacting protein of NIMA, results in defects in the normal dynamics of the NE, causing large masses of chromatin to be enveloped within a single NE as well as premature loss of NIMA in mitotic cells (61). In our study, we found that in cells with partial NIMA, the NE fails to complete its normal double restrictions and abscissions, causing failure to successfully generate two nuclei during mitotic exit. This further implicates NIMA in the regulation of NE dynamics during mitotic exit. We also observed marked defects in interphase nuclear structure after mitotic failure in nimA7 nuclei. In these nuclei, the nucleolus was pushed out from the nucleus, forming a protrusion of the normal ovoid nuclei. This indicates that the normal spatial constraints acting upon the nucleolus to position it within interphase nuclei are defective in postmitotic cells formed with insufficient NIMA function. Interestingly, recent work also points to a potential function for NIMA with SONC, a chromatin-associated protein encoded by a gene identified as an extragenic suppressor of the nimA1 mutant, in regulating global chromatin organization during mitosis (62). Taking this together with our functional analysis of NIMA during mitosis, it is becoming clearer that NIMA is important for execution of several key aspects of mitotic nuclear restructuring that can affect subsequent interphase nuclear architecture when impaired.
The spindle assembly checkpoint and NIMA.
Colony growth and successful mitoses at 35°C of nimA7 cells are completely dependent on the function of the SAC, indicating that the SAC is functional in cells with partial NIMA function. However, in cells with partial NIMA function that undergo mitotic failure, their SAC-mediated mitotic delay is far shorter than the mitotic arrest seen in WT cells treated with microtubule poisons that prevents spindle formation (43). This suggests that the SAC in nimA7 cells may be defective. Importantly, however, we find that nimA7 cells undergo cell cycle delay the same as WT cells during drug-induced mitotic SAC arrest. Therefore, the core SAC machinery is functional in the absence of normal levels of NIMA function. The inability of some nimA7 cells to complete mitosis successfully might therefore stem from defects downstream of SAC satisfaction or possibly be due to ineffective engagement of the checkpoint in the absence of normal NIMA function due to the unique mitotic defects cause by partial NIMA function.
In conclusion, by utilizing a temperature-sensitive allele of NIMA at a semipermissive temperature, we have been able to query what mitotic events require NIMA activity after mitosis has been initiated. Our findings indicate that NIMA plays complex roles during mitosis by functioning as a spatially regulated rheostat-like regulator as well as an on/off switch at the initiation of mitosis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jian-Qiu Wu (Molecular Genetics, The Ohio State University) for assistance with the live-cell confocal microscopy, Colin P. De Souza and Mahesh Chemudupati (Molecular Genetics, The Ohio State University) for strains, and Sridhar Vedachalam (Cornell University) for technical assistance in generating the box plots. We are also grateful to the past and present members of the Osmani lab for their feedback and support.
This study was funded by NIH grant GM042564 to S.A.O. and a Pelotonia Graduate Fellowship to M.G.
Footnotes
Published ahead of print 1 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00231-13.
REFERENCES
- 1.Morris NR. 1975. Mitotic mutants of Aspergillus nidulans. Genet. Res. 26:237–254. 10.1017/S0016672300016049 [DOI] [PubMed] [Google Scholar]
- 2.Morris NR, Enos AP. 1992. Mitotic gold in a mold: Aspergillus genetics and the biology of mitosis. Trends Genet. 8:32–37. 10.1016/0168-9525(92)90022-V [DOI] [PubMed] [Google Scholar]
- 3.Rosenberger RF, Kessel M. 1967. Synchrony of nuclear replication in individual hyphae of Aspergillus nidulans. J. Bacteriol. 94:1464–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clutterbuck AJ. 1970. Synchronous nuclear division and septation in Aspergillus nidulans. J. Gen. Microbiol. 60:133–135. 10.1099/00221287-60-1-133 [DOI] [PubMed] [Google Scholar]
- 5.Fiddy C, Trinci AP. 1976. Mitosis, septation, branching and the duplication cycle in Aspergillus nidulans. J. Gen. Microbiol. 97:169–184. 10.1099/00221287-97-2-169 [DOI] [PubMed] [Google Scholar]
- 6.Osmani SA, Ye XS. 1996. Cell cycle regulation in Aspergillus by two protein kinases. Biochem. J. 317:633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nurse P, Thuriaux P, Nasmyth K. 1976. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146:167–178. 10.1007/BF00268085 [DOI] [PubMed] [Google Scholar]
- 8.Hartwell LH, Culotti J, Pringle JR, Reid BJ. 1974. Genetic control of the cell division cycle in yeast. Science 183:46–51. 10.1126/science.183.4120.46 [DOI] [PubMed] [Google Scholar]
- 9.Hartwell LH, Mortimer RK, Culotti J, Culotti M. 1973. Genetic control of the cell division cycle in yeast. V. Genetic analysis of cdc mutants. Genetics 74:267–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartwell LH. 1971. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 69:265–276 [DOI] [PubMed] [Google Scholar]
- 11.Culotti J, Hartwell LH. 1971. Genetic control of the cell division cycle in yeast. III. Seven genes controlling nuclear division. Exp. Cell Res. 67:389–401 [DOI] [PubMed] [Google Scholar]
- 12.Hartwell LH. 1971. Genetic control of the cell division cycle in yeast. II. Genes controlling DNA replication and its initiation. J. Mol. Biol. 59:183–194 [DOI] [PubMed] [Google Scholar]
- 13.Hartwell LH, Culotti J, Reid B. 1970. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc. Natl. Acad. Sci. U. S. A. 66:352–359. 10.1073/pnas.66.2.352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pu RT, Osmani SA. 1995. Mitotic destruction of the cell cycle regulated NIMA protein kinase of Aspergillus nidulans is required for mitotic exit. EMBO J. 14:995–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osmani SA, May GS, Morris NR. 1987. Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J. Cell Biol. 104:1495–1504. 10.1083/jcb.104.6.1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Connell MJ, Osmani AH, Morris NR, Osmani SA. 1992. An extra copy of nimEcyclinB elevates pre-MPF levels and partially suppresses mutation of nimTcdc25 in Aspergillus nidulans. EMBO J. 11:2139–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osmani AH, McGuire SL, Osmani SA. 1991. Parallel activation of the NIMA and p34cdc2 cell cycle-regulated protein kinases is required to initiate mitosis in A. nidulans. Cell 67:283–291. 10.1016/0092-8674(91)90180-7 [DOI] [PubMed] [Google Scholar]
- 18.Quarmby LM, Mahjoub MR. 2005. Caught Nek-ing: cilia and centrioles. J. Cell Sci. 118:5161–5169. 10.1242/jcs.02681 [DOI] [PubMed] [Google Scholar]
- 19.Fry AM, O'Regan L, Sabir SR, Bayliss R. 2012. Cell cycle regulation by the NEK family of protein kinases. J. Cell Sci. 125:4423–4433. 10.1242/jcs.111195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grallert A, Chan KY, Alonso-Nunez ML, Madrid M, Biswas A, Alvarez-Tabares I, Connolly Y, Tanaka K, Robertson A, Ortiz JM, Smith DL, Hagan IM. 2013. Removal of centrosomal PP1 by NIMA kinase unlocks the MPF feedback loop to promote mitotic commitment in S. pombe. Curr. Biol. 23:213–222. 10.1016/j.cub.2012.12.039 [DOI] [PubMed] [Google Scholar]
- 21.Grallert A, Connolly Y, Smith DL, Simanis V, Hagan IM. 2012. The S. pombe cytokinesis NDR kinase Sid2 activates Fin1 NIMA kinase to control mitotic commitment through Pom1/Wee1. Nat. Cell Biol. 14:738–745. 10.1038/ncb2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye XS, Xu G, Pu RT, Fincher RR, McGuire SL, Osmani AH, Osmani SA. 1995. The NIMA protein kinase is hyperphosphorylated and activated downstream of p34cdc2/cyclin B: coordination of two mitosis promoting kinases. EMBO J. 14:986–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu L, Osmani SA, Mirabito PM. 1998. A role for NIMA in the nuclear localization of cyclin B in Aspergillus nidulans. J. Cell Biol. 141:1575–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen KF, Osmani SA. 23 October 2013. Regulation of mitosis by the NIMA kinase involves TINA and its newly discovered partner An-WDR8 at spindle pole bodies. Mol. Biol. Cell. 10.1091/mbc.E13-07-0422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Souza CP, Osmani AH, Wu LP, Spotts JL, Osmani SA. 2000. Mitotic histone H3 phosphorylation by the NIMA kinase in Aspergillus nidulans. Cell 102:293–302. 10.1016/S0092-8674(00)00035-0 [DOI] [PubMed] [Google Scholar]
- 26.De Souza CP, Osmani AH, Hashmi SB, Osmani SA. 2004. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 14:1973–1984. 10.1016/j.cub.2004.10.050 [DOI] [PubMed] [Google Scholar]
- 27.O'Connell MJ, Norbury C, Nurse P. 1994. Premature chromatin condensation upon accumulation of NIMA. EMBO J. 13:4926–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu KP, Hunter T. 1995. Evidence for a NIMA-like mitotic pathway in vertebrate cells. Cell 81:413–424. 10.1016/0092-8674(95)90394-1 [DOI] [PubMed] [Google Scholar]
- 29.De Souza CP, Horn KP, Masker K, Osmani SA. 2003. The SONB(NUP98) nucleoporin interacts with the NIMA kinase in Aspergillus nidulans. Genetics 165:1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osmani AH, Davies J, Liu HL, Nile A, Osmani SA. 2006. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell 17:4946–4961. 10.1091/mbc.E06-07-0657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Souza CP, Osmani SA. 2007. Mitosis, not just open or closed. Eukaryot. Cell 6:1521–1527. 10.1128/EC.00178-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Souza CP, Hashmi SB, Nayak T, Oakley B, Osmani SA. 2009. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol. Biol. Cell 20:2146–2159. 10.1091/mbc.E08-08-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu HL, De Souza CP, Osmani AH, Osmani SA. 2009. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex. Mol. Biol. Cell 20:616–630. 10.1091/mbc.E08-06-0628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, Kutay U. 2011. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell 144:539–550. 10.1016/j.cell.2011.01.012 [DOI] [PubMed] [Google Scholar]
- 35.Osmani AH, O'Donnell K, Pu RT, Osmani SA. 1991. Activation of the nimA protein kinase plays a unique role during mitosis that cannot be bypassed by absence of the bimE checkpoint. EMBO J. 10:2669–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141–238. 10.1016/S0065-2660(08)60408-3 [DOI] [PubMed] [Google Scholar]
- 37.Ovechkina Y, Maddox P, Oakley CE, Xiang X, Osmani SA, Salmon ED, Oakley BR. 2003. Spindle formation in Aspergillus is coupled to tubulin movement into the nucleus. Mol. Biol. Cell 14:2192–2200. 10.1091/mbc.E02-10-0641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang L, Ukil L, Osmani A, Nahm F, Davies J, De Souza CP, Dou X, Perez-Balaguer A, Osmani SA. 2004. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3:1359–1362. 10.1128/EC.3.5.1359-1362.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arnaud MB, Cerqueira GC, Inglis DO, Skrzypek MS, Binkley J, Chibucos MC, Crabtree J, Howarth C, Orvis J, Shah P, Wymore F, Binkley G, Miyasato SR, Simison M, Sherlock G, Wortman JR. 2012. The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 40:D653–D659. 10.1093/nar/gkr875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong Y, Oakley BR. 2009. In vivo analysis of the functions of gamma-tubulin-complex proteins. J. Cell Sci. 122:4218–4227. 10.1242/jcs.059196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osmani AH, Davies J, Oakley CE, Oakley BR, Osmani SA. 2003. TINA interacts with the NIMA kinase in Aspergillus nidulans and negatively regulates astral microtubules during metaphase arrest. Mol. Biol. Cell 14:3169–3179. 10.1091/mbc.E02-11-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ukil L, De Souza CP, Liu HL, Osmani SA. 2009. Nucleolar separation from chromosomes during Aspergillus nidulans mitosis can occur without spindle forces. Mol. Biol. Cell 20:2132–2145. 10.1091/mbc.E08-10-1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Souza CP, Hashmi SB, Yang X, Osmani SA. 2011. Regulated inactivation of the spindle assembly checkpoint without functional mitotic spindles. EMBO J. 30:2648–2661. 10.1038/emboj.2011.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suelmann R, Sievers N, Fischer R. 1997. Nuclear traffic in fungal hyphae: in vivo study of nuclear migration and positioning in Aspergillus nidulans. Mol. Microbiol. 25:757–769. 10.1046/j.1365-2958.1997.5131873.x [DOI] [PubMed] [Google Scholar]
- 45.Toews MW, Warmbold J, Konzack S, Rischitor P, Veith D, Vienken K, Vinuesa C, Wei H, Fischer R. 2004. Establishment of mRFP1 as a fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in vitro (GATEWAY). Curr. Genet. 45:383–389. 10.1007/s00294-004-0495-7 [DOI] [PubMed] [Google Scholar]
- 46.Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J. 2010. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol. 76:1376–1386. 10.1111/j.1365-2958.2010.07051.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osmani SA, Pu RT, Morris NR. 1988. Mitotic induction and maintenance by overexpression of a G2-specific gene that encodes a potential protein kinase. Cell 53:237–244. 10.1016/0092-8674(88)90385-6 [DOI] [PubMed] [Google Scholar]
- 48.Oakley BR, Morris NR. 1983. A mutation in Aspergillus nidulans that blocks the transition from interphase to prophase. J. Cell Biol. 96:1155–1158. 10.1083/jcb.96.4.1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA. 1998. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 141:1563–1574. 10.1083/jcb.141.7.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA. 2005. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell Biol. 171:27–33. 10.1083/jcb.200504107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bahmanyar S, Kaplan DD, Deluca JG, Giddings TH, Jr, O'Toole ET, Winey M, Salmon ED, Casey PJ, Nelson WJ, Barth AI. 2008. β-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 22:91–105. 10.1101/gad.1596308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bertran MT, Sdelci S, Regue L, Avruch J, Caelles C, Roig J. 2011. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J. 30:2634–2647. 10.1038/emboj.2011.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grallert A, Hagan IM. 2002. Schizosaccharomyces pombe NIMA-related kinase, Fin1, regulates spindle formation and an affinity of Polo for the SPB. EMBO J. 21:3096–3107. 10.1093/emboj/cdf294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krien MJ, West RR, John UP, Koniaras K, McIntosh JR, O'Connell MJ. 2002. The fission yeast NIMA kinase Fin1p is required for spindle function and nuclear envelope integrity. EMBO J. 21:1713–1722. 10.1093/emboj/21.7.1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, Ellenberg J. 2008. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 180:857–865. 10.1083/jcb.200707026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yam C, He Y, Zhang D, Chiam KH, Oliferenko S. 2011. Divergent strategies for controlling the nuclear membrane satisfy geometric constraints during nuclear division. Curr. Biol. 21:1314–1319. 10.1016/j.cub.2011.06.052 [DOI] [PubMed] [Google Scholar]
- 57.Andrews CA, Vas AC, Meier B, Gimenez-Abian JF, Diaz-Martinez LA, Green J, Erickson SL, Vanderwaal KE, Hsu WS, Clarke DJ. 2006. A mitotic topoisomerase II checkpoint in budding yeast is required for genome stability but acts independently of Pds1/securin. Genes Dev. 20:1162–1174. 10.1101/gad.1367206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Remeseiro S, Losada A. 2013. Cohesin, a chromatin engagement ring. Curr. Opin. Cell Biol. 25:63–71. 10.1016/j.ceb.2012.10.013 [DOI] [PubMed] [Google Scholar]
- 59.Cuylen S, Haering CH. 2011. Deciphering condensin action during chromosome segregation. Trends Cell Biol. 21:552–559. 10.1016/j.tcb.2011.06.003 [DOI] [PubMed] [Google Scholar]
- 60.Stephens AD, Haggerty RA, Vasquez PA, Vicci L, Snider CE, Shi F, Quammen C, Mullins C, Haase J, Taylor RM, 2nd, Verdaasdonk JS, Falvo MR, Jin Y, Forest MG, Bloom K. 2013. Pericentric chromatin loops function as a nonlinear spring in mitotic force balance. J. Cell Biol. 200:757–772. 10.1083/jcb.201208163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davies JR, Osmani AH, De Souza CP, Bachewich C, Osmani SA. 2004. Potential link between the NIMA mitotic kinase and nuclear membrane fission during mitotic exit in Aspergillus nidulans. Eukaryot. Cell 3:1433–1444. 10.1128/EC.3.6.1433-1444.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Larson JR, Facemyer EM, Shen K-F, Ukil L, Osmani SA. 8 November 2013. Insights into dynamic mitotic chromatin organization through the NIMA kinase suppressor SonC, a chromatin-associated protein involved in the DNA damage response. Genetics. 10.1534/genetics.113.156745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Souza CP, Osmani SA. 2009. Double duty for nuclear proteins—the price of more open forms of mitosis. Trends Genet. 25:545–554. 10.1016/j.tig.2009.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


