Abstract
Mitogen-activated protein kinase (MAPK) modules are evolutionarily conserved signaling cascades that function in response to the environment and play crucial roles in intracellular signal transduction in eukaryotes. The involvement of a MAP kinase in regulating cytokinesis in yeast, animals, and plants has been reported, but the requirement for a MAP kinase for cytokinesis in the early-branching protozoa is not documented. Here, we show that a MAP kinase homolog (TbMAPK6) from Trypanosoma brucei plays distinct roles in cytokinesis in two life cycle forms of T. brucei. TbMAPK6 is distributed throughout the cytosol in the procyclic form but is localized in both the cytosol and the nucleus in the bloodstream form. RNA interference (RNAi) of TbMAPK6 results in moderate growth inhibition in the procyclic form but severe growth defects and rapid cell death in the bloodstream form. Moreover, TbMAPK6 appears to be implicated in furrow ingression and cytokinesis completion in the procyclic form but is essential for cytokinesis initiation in the bloodstream form. Despite the distinct defects in cytokinesis in the two forms, RNAi of TbMAPK6 also caused defective basal body duplication/segregation in a small cell population in both life cycle forms. Altogether, our results demonstrate the involvement of the TbMAPK6-mediated pathway in regulating cytokinesis in trypanosomes and suggest distinct roles of TbMAPK6 in cytokinesis between different life cycle stages of T. brucei.
INTRODUCTION
Cytokinesis, the final step of cell division, requires the cooperative actions of many regulatory proteins at the initiation site of cytokinesis and on the cleavage furrow (1). Although the molecular mechanisms underlying the initiation, progression, and completion of cytokinesis in eukaryotes remain poorly understood, the core regulatory pathways are likely well conserved from fungi to humans (2, 3). The cleavage plane in fungi and animals is defined by the central spindle, and the cleavage furrow in these organisms is composed of actin and myosin II, forming the so-called actomyosin contractile ring, which plays a crucial role in pinching the cell in two (3–5).
Cytokinesis in Trypanosoma brucei, a unicellular microbial eukaryote and the causative agent of human sleeping sickness in Africa, appears to differ from that in fungi and animals (5, 6). It has been well documented that cytokinesis in trypanosomes is initiated from the anterior tip of the new flagellum attachment zone (FAZ), and the cleavage furrow ingresses unidirectionally along the longitudinal axis from the anterior toward the posterior end of the cell. Therefore, the cytokinesis cleavage plane in a dividing trypanosome cell likely is defined by the new flagellum and the new FAZ (6–8). Strikingly, no evidence supports the formation of an actomyosin contractile ring at the cleavage furrow in trypanosomes (9), implying that the cleavage furrow in trypanosomes likely is composed of novel components.
Increasing evidence has shown that there are some drastic differences in cell cycle regulation between the different life cycle forms of T. brucei. For example, inhibition of mitosis in the procyclic form does not block cytokinesis, whereas mitotic defects in the bloodstream form arrest cytokinesis but allow additional rounds of DNA replication (10, 11). Moreover, the regulatory pathway of cytokinesis also appears to differ between the two forms because silencing of some cytokinesis proteins results in distinct cytokinesis defects in the two life cycle forms. For example, the trypanosome homolog of Mob1, a downstream factor of the Polo-like kinase in the yeast septum initiation network pathway that controls mitotic exit and cytokinesis (12), is required for the completion of cytokinesis in the bloodstream form but is essential for proper cleavage furrow positioning in the procyclic form (13). These observations suggest the existence of distinct mechanisms in cytokinesis in the two life cycle forms of T. brucei.
Mitogen-activated protein kinase (MAPK) is known to play essential roles in intracellular signal transduction in eukaryotic cells by responding to environmental changes (14). Among the many MAPK cascades in plants, the NRK1 cascade regulates cytokinesis (15). This MAPK cascade stimulates the turnover of the microtubule array termed phragmoplast, a structure analogous to the midbody in animals, resulting in expansion of the phragmoplast, which is essential for formation of the cell division plate between the two daughter cells (15). All the components in the NRK1 pathway are concentrated at the midzone of the phragmoplast during cytokinesis. NRK1 phosphorylates microtubule-associated protein 65 (MAP65), which directly regulates disassembly of the phragmoplast microtubules, and depolymerization of the phragmoplast microtubules appears to exert feedback regulation of the activity of the NRK1 cascade (15). The requirement for a MAPK for cytokinesis in other systems has also been reported but was less well characterized than the plant NRK1 cascade. The fission yeast MAPK Pmk1, which regulates cell integrity (16), plays an additional role in cytokinesis by phosphorylating the RNA-binding protein Nrd1 for the latter to stabilize the mRNA of myosin, a key component of the actomyosin contractile ring (17). In HeLa cells, the p44/p42 MAP kinases ERK1/2 are localized to the midbody and are required for abscission, but the downstream target(s) of ERK1/2 on the midbody is not known, and therefore, the mechanistic role of ERK1/2 in cytokinesis remains elusive (18).
The T. brucei genome encodes 13 putative MAP kinase homologs (19), and five of them, KFR1, TbMAPK2, TbECK1, TbMAPK4, and TbMAPK5, have been characterized, each of which participates in cell proliferation or differentiation. KFR1 is likely involved in proliferation of the bloodstream form and is regulated by gamma interferon, an extracellular molecule (20). TbMAPK2 is required for differentiation of the bloodstream form to the procyclic form and for subsequent cell proliferation (21). TbECK1 has characteristics of both a MAPK and a cyclin-dependent kinase and is essential in all life cycle forms investigated (22). TbMAPK4 appears to be involved in the response to temperature stress (23), and TbMAPK5 is required for infection of the mammalian host and for differentiation of the slender form to the stumpy form (24). None of these MAPK homologs appear to play a role in cytokinesis in any life cycle forms of T. brucei.
In this paper, we report the functional characterization of a MAP kinase homolog, TbMAPK6 (Tb927.10.5140), in both the procyclic and bloodstream forms of T. brucei. TbMAPK6 is a close homolog of the Leishmania MAPK LmxMPK2 (19), and its function in T. brucei is not known. LmxMPK2 appears to be nonessential for cell viability because null mutants of LmxMPK2 can be readily generated (25). RNA interference (RNAi) of TbMAPK6 leads to distinct cytokinesis defects in the two forms. In the procyclic form, cells depleted of TbMAPK6 are able to initiate cytokinesis but appear to be arrested midway to the completion of cytokinesis. In the bloodstream form, however, TbMAPK6 RNAi arrests the cells before cytokinesis initiation. Moreover, silencing of TbMAPK6 only slows down cell proliferation in the procyclic form but results in severe growth inhibition and rapid cell death in the bloodstream form. Our results identify distinct phenotypes of TbMAPK6 RNAi in different life cycle stages of T. brucei and suggest TbMAPK6 as a potential drug target.
MATERIALS AND METHODS
Trypanosome cell culture and RNA interference.
The procyclic form 427 cell line was cultivated in SDM-79 medium supplemented with 10% fetal bovine serum (Atlanta Biologicals, Inc.) at 27°C, whereas the procyclic 29-13 cell line (26) was cultured at 27°C in SDM-79 medium supplemented with 10% fetal bovine serum, 15 μg/ml G418, and 50 μg/ml hygromycin to maintain the tetracycline repressor and T7 RNA polymerase constructs.
The bloodstream form 221 cell line was cultured in HMI-9 medium supplemented with 10% fetal bovine serum (Atlanta Biologicals, Inc.) at 37°C with 5% CO2. The bloodstream form SM (single marker) cell line (26) was cultivated in HMI-9 medium containing 10% fetal bovine serum and 2.5 μg/ml G418 in a 37°C incubator supplied with 5% CO2.
RNAi was carried out as described previously (27). A 503-bp fragment corresponding to the N-terminal coding region of TbMAPK6 was cloned into the pZJM vector and the stem-loop vector (28), and the resulting plasmids were linearized and electroporated into the procyclic form 29-13 cell line and the bloodstream form SM cell line. Twenty-four hours after electroporation, cells were selected under 2.5 μg/ml phleomycin. Successful transfectants were further cloned by limiting dilution in a 96-well plate. To induce RNAi, the clonal cell line was incubated with 1.0 μg/ml tetracycline, and cell growth was monitored daily with a hemocytometer. Three clonal cell lines were induced for potential phenotypes, and all three cell lines exhibited almost identical defects. Therefore, only one clonal cell line was chosen for in-depth characterization.
Northern blotting.
Total RNA was purified from T. brucei cells before and after RNAi induction for 3 days with the TRIzol reagent (Invitrogen). Northern blotting was performed as previously described (29). Briefly, 30 μg total RNA was denatured, separated on an agarose gel, and transferred onto a nitrocellulose membrane in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Northern hybridization was carried out overnight at 42°C in 50% formamide, 6× SSC, 0.5% SDS, 5× Denhardt's solution with 0.1 mg/ml salmon sperm DNA. DNA probes were labeled with [α-32P]dCTP. The membrane was washed three times for 30 min each time in 2× SSC-0.1% SDS, 1× SSC-0.1% SDS, and 0.5× SSC-0.1% SDS, respectively, and then exposed to the X-ray film. The same membrane was stripped and rehybridized with a tubulin gene fragment as the loading control.
Quantitative RT-PCR.
Total RNA was purified from T. brucei cells before and after RNAi induction for 2 days with the TRIzol reagent and treated with DNase I to remove any contaminated DNA. First-strand cDNA was then synthesized with Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega), and real-time (RT) PCR was carried out using the SYBR green PCR master mix (Bio-Rad) on the CFX Real-Time PCR System (Bio-Rad). A 100-bp DNA fragment from the 3′ portion of TbMAPK6 coding region was chosen as the RT-PCR target sequence. Actin transcripts were used as the control. Three replicates of PCR were run simultaneously in the Real-Time PCR machine, and the results represent the average of the three RT-PCRs.
Epitope tagging of endogenous TbMAPK6 in the procyclic and bloodstream forms.
A 601-bp DNA fragment corresponding to the C-terminal coding region of TbMAPK6 was cloned into the pC-3HA-PAC vector, which was derived from the pC-3HA-BSD vector (30) by replacing the blasticidin S deaminase (BSD) gene with the puromycin N-acetyltransferase (PAC) gene, and the resulting construct was electroporated into the procyclic form 427 cell line and the bloodstream form 221 cell line according to our published procedures (31, 32). The pC-TbMAPK6-3HA-PAC construct was also transfected into the pStemLoop-TbMAPK6 bloodstream cell line to monitor the TbMAPK6 protein level after RNAi. Stable transfectants were selected under 1 μg/ml puromycin, and clonal cell lines were obtained by limiting dilution. Correct in situ tagging of one of the two TbMAPK6 alleles was confirmed by PCR and subsequent sequencing of the PCR fragment, as well as by Western blotting with anti-hemagglutinin (HA) antibody to detect triple-HA-epitope (3HA)-tagged TbMAPK6.
Immunofluorescence microscopy.
Trypanosome cells were washed with PBS and fixed in 4% paraformaldehyde. The fixed cells were allowed to adhere to poly-l-lysine-treated coverslips. The cells were then incubated with YL 1/2 monoclonal antibody (MAb) for tyrosinated α-tubulin (1:2,000 dilution) (33). After three washes, the cells were incubated with fluorescein isothiocyanate (FITC)-conjugated anti-rat IgG at room temperature for 1 h. For anti-HA immunofluorescence, the fixed cells were incubated with FITC-conjugated anti-HA MAb for 1 h at room temperature. The slides were mounted in VectaShield mounting medium (Vector Laboratories) containing DAPI (4′,6-diamidino-2-phenylindole) and examined using an inverted microscope (model IX71; Olympus) equipped with a cooled charge-coupled-device (CCD) camera (model Orca-ER; Hamamatsu) and a PlanApo N 60× 1.42-numerical-aperture (NA) differential interference contrast (DIC) objective. Images were acquired and processed using Slidebook5 software (Intelligent Imaging Innovations, Inc.).
RESULTS
Subcellular localization of TbMAPK6 in the procyclic and bloodstream forms.
To determine the subcellular localization of TbMAPK6 in the procyclic and bloodstream forms, endogenous TbMAPK6 was tagged at the C terminus with a triple HA epitope. Western blotting with anti-HA antibody detected a single protein band of ∼50 kDa, similar to the calculated molecular mass (49.7 kDa) of TbMAPK6, in both life cycle forms (Fig. 1A). The levels of TbMAPK6 protein appear to be similar in the two life cycle forms (Fig. 1A). To determine the subcellular localization of TbMAPK6, we carried out immunofluorescence microscopy with FITC-conjugated anti-HA antibody. In the procyclic form, TbMAPK6::3HA was detected in a punctate pattern throughout the cytosol at all cell cycle stages (Fig. 1B). TbMAPK6::3HA likely is also present in the nucleus, but the level of TbMAPK6::3HA protein is much reduced in the nucleus (Fig. 1B). In the bloodstream form, TbMAPK6::3HA was also detected in a punctate pattern that was distributed throughout the cytosol and the nucleus at all cell cycle stages examined (Fig. 1C). However, there appeared to be no significant reduction in the level of TbMAPK6::3HA protein in the nuclei of bloodstream cells compared to the procyclic cells (Fig. 1B and C). These observations suggest that TbMAPK6 displays distinct subcellular localization patterns in the two life cycle forms, although the mechanism underlying this distinction remains elusive.
FIG 1.
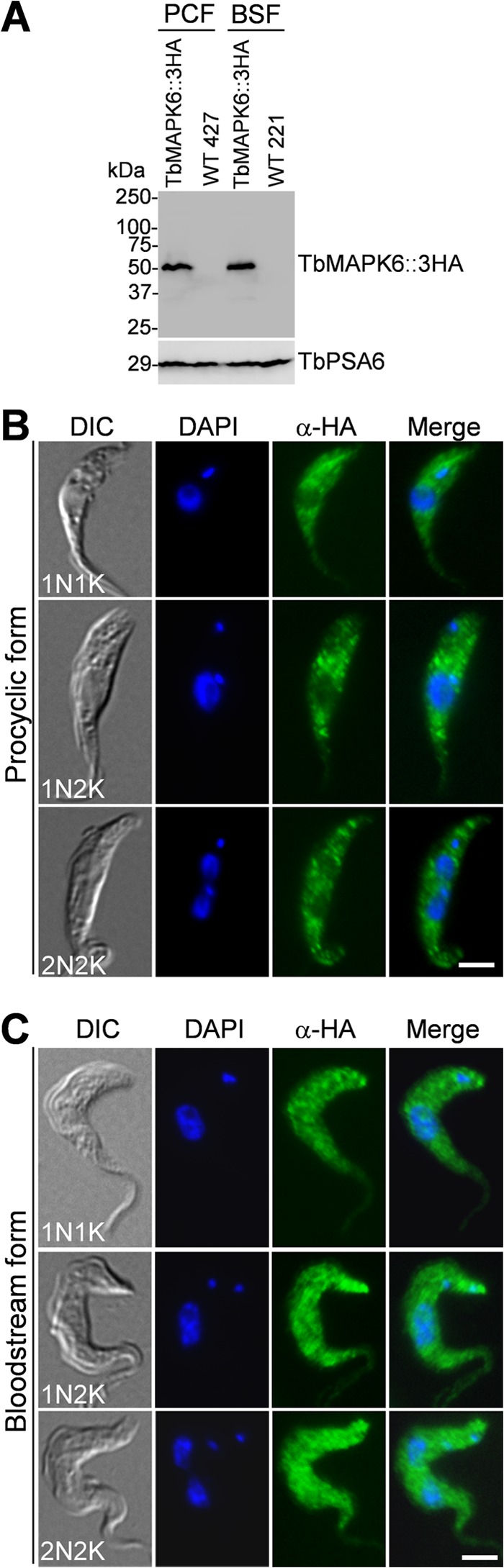
Subcellular localization of TbMAPK6 in the procyclic and bloodstream forms of T. brucei. (A) Western blot to detect 3HA-tagged TbMAPK6 in the procyclic form (PCF) and the bloodstream form (BSF). The same membrane was reprobed with anti-TbPSA6, the α6 subunit of the 26S proteasome, as the loading control. (B and C) Subcellular localization of 3HA-tagged TbMAPK6 in the procyclic form (B) and the bloodstream form (C) at different cell cycle stages. TbMAPK6::3HA was expressed from the endogenous locus and was visualized by indirect immunofluorescence microscopy. Bars, 2 μm. WT, wild type.
RNAi of TbMAPK6 in the procyclic form slightly slows cell proliferation.
To investigate the function of TbMAPK6 in trypanosomes, RNAi was first carried out in the procyclic form. To examine the efficiency of RNAi, Northern blotting was performed to monitor TbMAPK6 mRNA levels before and after RNAi induction, and the result showed that upon RNAi induction for 1 day, TbMAPK6 mRNA was significantly decreased in the cells (Fig. 2A). This depletion of TbMAPK6 mRNA resulted in moderate depression of cell proliferation (Fig. 2B), which might be attributed to cell cycle defects. To ascertain whether cell cycle progression was affected by TbMAPK6 RNAi, we tabulated the numbers of nuclei and kinetoplasts in the cells. This was based on the fact that trypanosome cells at different cell cycle stages generally contain distinct numbers of nuclei and kinetoplasts. The G1- and S-phase cells always possess one nucleus and one kinetoplast (1N1K), and the G2 cells and early mitotic cells, such as prometaphase and metaphase cells, usually contain one nucleus and two kinetoplasts (1N2K). Cells at late stages of mitosis, such as anaphase and telophase, and during cytokinesis always contain two nuclei and two kinetoplasts (2N2K). After TbMAPK6 RNAi induction, the number of 1N1K cells gradually decreased, which was accompanied by a slight increase in the number of 2N2K cells and the emergence of cells with abnormal numbers of nuclei and kinetoplasts, such as 2N1K cells, XN1K (>2 nuclei and one kinetoplast) cells, XN2K (>2 nuclei and two kinetoplasts) cells, and XNXK (>2 nuclei and >2 kinetoplasts) cells (Fig. 2C and D). The number of anucleate (0N1K) cells, also called zoid cells, only slightly increased up to 2% of the total cell population (Fig. 2C). After RNAi induction for 6 days, all these abnormal cell types constituted about 40% of the total cell population (Fig. 2C). The increase of 2N2K cells and the emergence of multinucleated (XN1K, XN2K, and XNXK) cells suggest that cytokinesis was somewhat defective upon TbMAPK6 RNAi.
FIG 2.
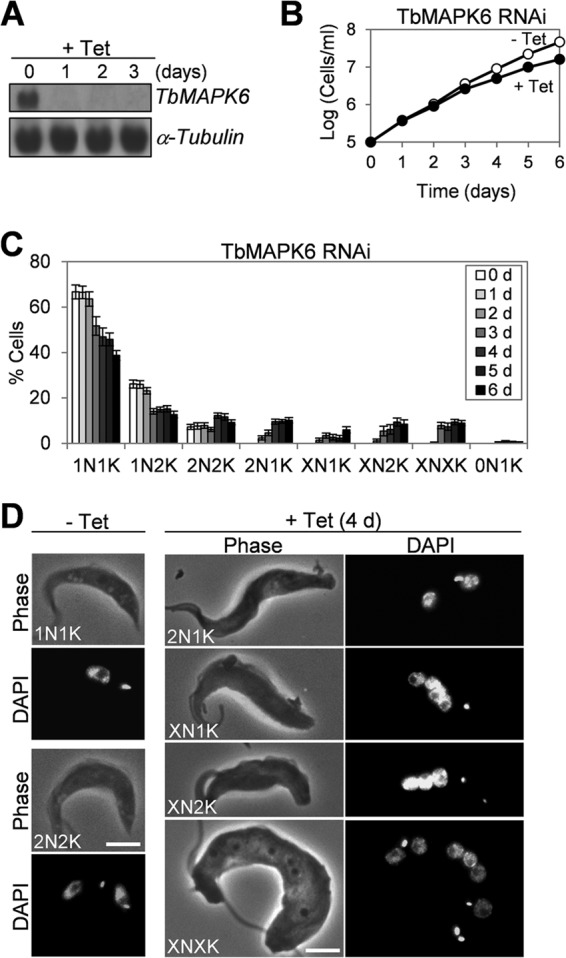
RNAi of TbMAPK6 in the procyclic form of T. brucei. (A) Northern blot to monitor TbMAPK6 mRNA levels before and after tetracycline (Tet) induction for 3 days. The α-tubulin mRNA level served as the loading control. (B) RNAi of TbMAPK6 slows cell proliferation. (C) Tabulation of cells with different numbers of nuclei (N) and kinetoplasts (K) upon TbMAPK6 knockdown. The data are presented as the mean percentages ± standard deviations (SD) of 300 cells counted for each time point (d, day[s]) from three independent experiments. X > 2. (D) Morphology of TbMAPK6 RNAi cells. Bars, 2 μm.
Effect of TbMAPK6 RNAi on cytokinesis progression and completion in the procyclic form.
Cytokinesis in trypanosomes is known to initiate from the anterior tip of the new FAZ and proceeds toward the posterior end of the cell, with the cytokinesis cleavage furrow placed between the new flagellum/FAZ and the old flagellum/FAZ (32). At the final stage of cytokinesis, the two daughter cells are connected through the posterior tips of the cells (32).
Among the 2N2K, XN2K, and XNXK cells produced after TbMAPK6 RNAi induction, a certain percentage of the cells appeared to be arrested midway to the completion of cytokinesis, with the cleavage furrow clearly visible (Fig. 3A, arrows), or at the late stage of cytokinesis, with the two daughter cells connected through the posterior ends and failing to be separated (Fig. 3A, arrowheads). These cells constituted ∼13% of the 2N2K cell type, ∼18% of the XN2K cell type, and ∼25% of the XNXK cell type (Fig. 3B). In the uninduced control cells, however, the 2N2K cells exhibiting a visible cleavage furrow were very rare, and no 2N2K cells at the late stage of cytokinesis could be found among the more than 600 cells examined (Fig. 3B). These observations suggest that TbMAPK6 RNAi likely did not compromise the initiation of cytokinesis but instead impaired furrow ingression and cell abscission in the procyclic form.
FIG 3.
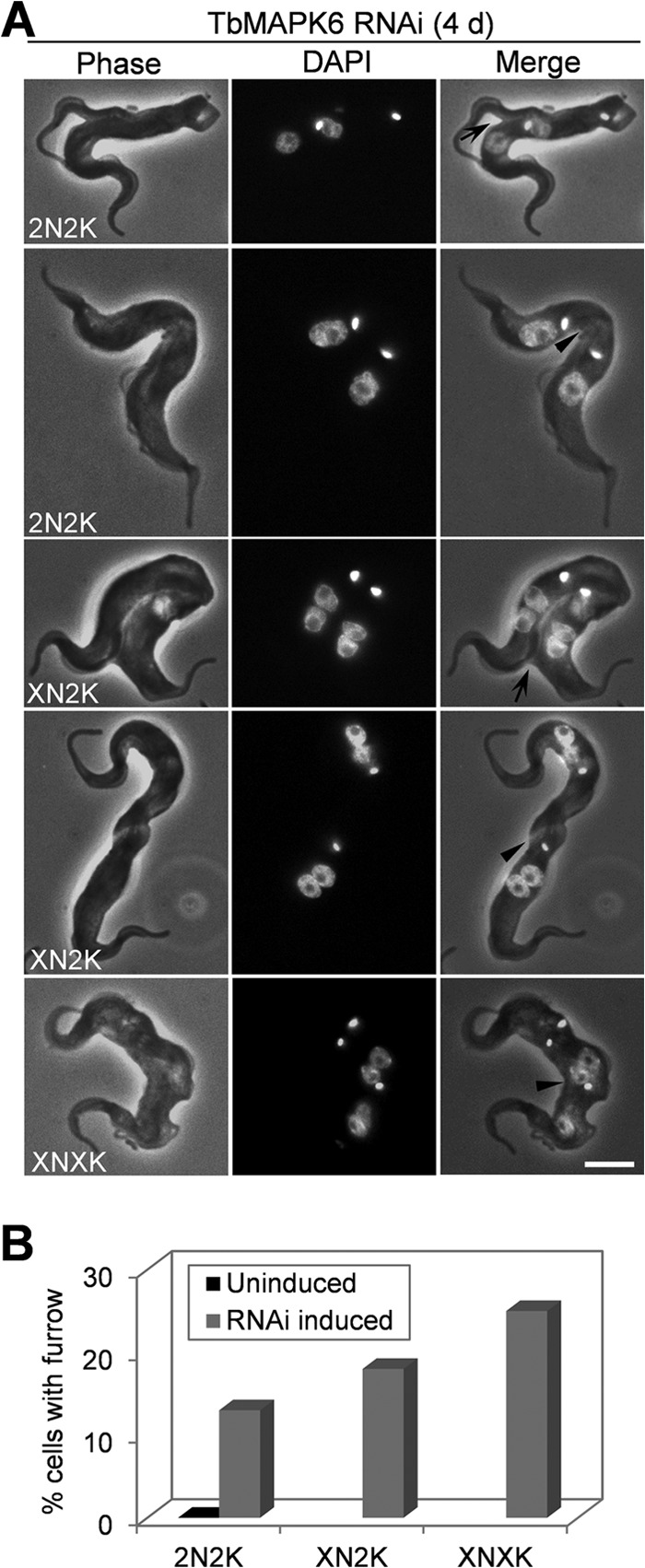
TbMAPK6 is required for furrow ingression and the completion of cytokinesis in the procyclic form. (A) TbMAPK6 RNAi cells undergoing cytokinesis. The arrows indicate the cleavage furrow. The arrowheads indicate potential cleavage furrows. Bar, 2 μm. (B) Percentages of the uninduced control and TbMAPK6 RNAi cells with the indicated complement of nuclei and kinetoplasts showing a visible cleavage furrow. The data are presented as the mean percentages of 200 cells counted for each cell type from three independent experiments. X > 2.
The accumulation of a small percentage of 2N1K and XN1K cells upon TbMAPK6 RNAi (Fig. 2C and D) suggests that kinetoplast replication and/or segregation was likely inhibited in these cells. It is known that kinetoplast segregation in trypanosomes is mediated by basal body segregation (34). Therefore, the production of 2N1K and XN1K cells upon TbMAPK6 RNAi induction might be attributed to defective basal body duplication and/or segregation in these cells. To test this possibility, we immunostained the cells with the YL 1/2 antibody, which labels the tyrosinated α-tubulin in the mature basal bodies and in the posterior portion of the trypanosome cell (35). The antibody stains only the mature basal body and not the pro-basal body in trypanosomes. While the uninduced control 2N2K cells all possessed two well-segregated mature basal bodies, the TbMAPK6-deficient 2N1K cells contained either a single mature basal body, which constituted ∼65% of the 2N1K population, or two closely associated mature basal bodies next to the single kinetoplast, which represented about 35% of the 2N1K cell population (Fig. 4). Of the XN1K cells, about 75% contained one mature basal body, and the rest contained two closely associated mature basal bodies (Fig. 4). Among the XN2K cells, however, about 60% possessed two well-segregated mature basal bodies (Fig. 4), indicating that these cells were likely derived from the 2N2K cells that had undergone multiple rounds of mitosis in the absence of cytokinesis. Intriguingly, the remaining XN2K cells (∼40% of the total XN2K cell population) all contained two pairs of basal bodies, with the two basal bodies in each pair closely associated with each other (Fig. 4). The emergence of four mature basal bodies in these XN2K cells indicates that each of the two original mature basal bodies was further duplicated or the two pro-basal bodies that were associated with the two original mature basal bodies became mature and thus were detected by the YL 1/2 antibody. Nevertheless, these results suggest that segregation, but not duplication and maturation, of basal bodies was impaired in these RNAi cells. Indeed, among all the XNXK cells, multiple (>4) mature basal bodies were always detected by YL 1/2 (Fig. 4). However, the presence of a single mature basal body in the majority of the 2N1K and XN1K cells argues that basal body duplication was likely also compromised in these RNAi cells. Together, these results suggest that RNAi of TbMAPK6 resulted in defects in basal body duplication and segregation. However, given that only a small portion of the RNAi cells exhibit basal body duplication/segregation defects, it is not clear whether these defects are due to direct or indirect effects of TbMAPK6 RNAi.
FIG 4.
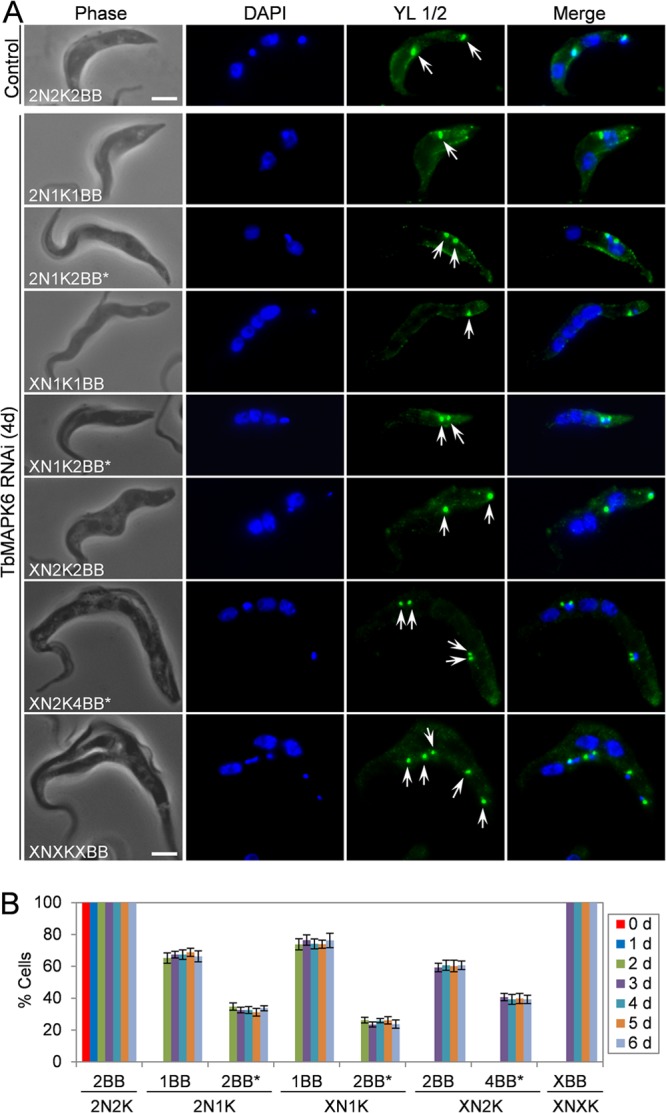
Effects of TbMAPK6 RNAi on basal body duplication and segregation in the procyclic form. (A) Immunofluorescence microscopic analysis of the basal body in uninduced control and TbMAPK6 RNAi cells. The cells were immunostained with the YL 1/2 antibody and counterstained with DAPI for nuclear and kinetoplast DNA. The arrows indicate the mature basal bodies labeled by YL 1/2. The asterisks indicate closely associated basal bodies. Bars, 2 μm. (B) Percentages of TbMAPK6 RNAi cells with different numbers of basal bodies. The data are presented as the mean percentages ± SD of 200 cells counted for each time point from three independent experiments. Note that no 2N1K, XN1K, XN2K, and XNXK cells were detected at day 0 of TbMAPK6 RNAi. X > 2.
RNAi of TbMAPK6 in the bloodstream form arrests cell proliferation and leads to rapid cell death.
One of the most unusual features of cell cycle regulation in trypanosomes is the existence of distinct cell cycle control mechanisms between the procyclic and bloodstream forms (10, 11, 36, 37). To investigate the function of TbMAPK6 in the bloodstream form, RNAi was also carried out, and quantitative RT-PCR showed that upon RNAi induction for 2 days, TbMAPK6 mRNA was reduced only ∼55% (Fig. 5A), which appeared to be less effective than in the procyclic form (Fig. 2A). To monitor TbMAPK6 protein levels before and after RNAi, we tagged the endogenous TbMAPK6 with a C-terminal triple HA epitope in the TbMAPK6 RNAi cell line. Western blotting with anti-HA antibody showed that the level of TbMAPK6 protein was gradually decreased after RNAi, and at day 2 of RNAi, the protein level was reduced to about 30% of the level in the uninduced control cells (Fig. 5B). Despite the moderate RNAi efficiency, however, TbMAPK6-deficient cells exhibited severe growth defects and eventually died after RNAi induction for 4 days (Fig. 5C), suggesting that TbMAPK6 is essential for cell viability in the bloodstream form.
FIG 5.
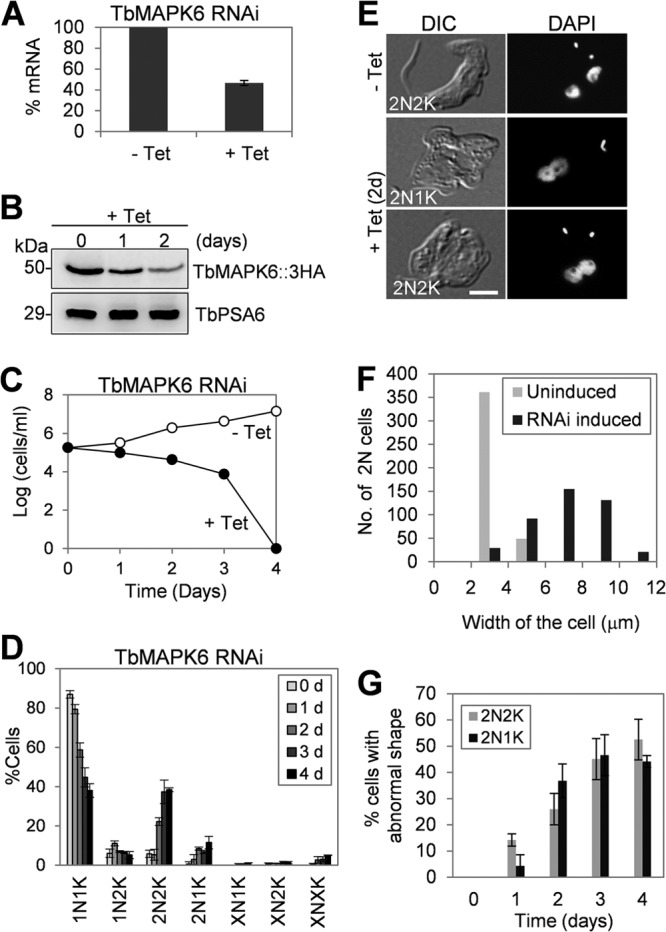
RNAi of TbMAPK6 in the bloodstream form abrogates cytokinesis initiation. (A) TbMAPK6 mRNA levels before and after RNAi induction for 2 days monitored by quantitative RT-PCR. (B) TbMAPK6 protein levels before and after RNAi induction for 1 and 2 days measured by Western blotting. TbMAPK6 was tagged at the C terminus with a triple HA epitope in one of its endogenous loci in the TbMAPK6 RNAi cell line. The protein level of TbPSA6, the α6 subunit of the 26S proteasome, is presented as the loading control. (C) Effect of TbMAPK6 RNAi on bloodstream cell proliferation. (D) Tabulation of cells with different numbers of nuclear (N) and kinetoplast (K) DNAs upon TbMAPK6 RNAi. The data are presented as the mean percentages ± SD of 200 cells counted for each time point from three independent experiments. X > 2. (E) Morphology of TbMAPK6 RNAi cells. Bar, 2 μm. (F) Cell widths of uninduced control and TbMAPK6 RNAi cells (day 2). About 400 dinucleate cells (2N2K and 2N1K) were measured for cell width and plotted. Cell width was measured from the middle of the cell between the two nuclei. (G) Percentages of TbMAPK6 RNAi cells exhibiting abnormal cell morphology. The data are presented as the mean percentages ± SD of 200 cells counted for each time point from three independent experiments.
To examine whether TbMAPK6 RNAi affected cell cycle progression in the bloodstream form, we tabulated the numbers of nuclei and kinetoplasts in the cells. After RNAi induction for 4 days, the number of 1N1K cells gradually decreased from ∼86% to ∼37%, which was accompanied by an increase of 2N2K cells from ∼6% to ∼38% (Fig. 5D). Additionally, a small proportion of 2N1K cells (∼7%) and multinucleated (XN1K, XN2K, and XNXK) cells (∼9%) also emerged after RNAi for 4 days (Fig. 5D). These observations suggest that cytokinesis in many of the TbMAPK6 RNAis was arrested. Moreover, the two nuclei in the 2N2K and 2N1K cells accumulated upon TbMAPK6 RNAi appeared to associate with each other (Fig. 5E). Since TbMAPK6 is also localized to the nucleus in the bloodstream form (Fig. 1C), this observation suggests that TbMAPK6 RNAi may also cause defects in nuclear segregation.
Effect of TbMAPK6 on cytokinesis initiation in the bloodstream form.
A closer examination of the 2N1K and 2N2K cells accumulated upon TbMAPK6 RNAi showed that these cells apparently lost their normal morphology (Fig. 5E). The cell bodies of these RNAi cells became much wider than those of the uninduced control cells (Fig. 5E). The average width of the cell body in the RNAi cells was calculated to be ∼7.2 μm, which was about 2 times the average width (∼3.5 μm) of the uninduced control 2N2K cells (Fig. 5F). These abnormally shaped cells emerged after only 1 day of RNAi induction and further accumulated to about 50% of the total population after 4 days of RNAi (Fig. 5G). The increase in the width of the cell body in the 2N1K and 2N2K cells in the RNAi population appeared to be attributable to the inhibited cytokinesis initiation, because no ingressing cleavage furrow was clearly visible in the cells (Fig. 5E). Altogether, these results may argue that TbMAPK6 RNAi arrests the bloodstream form before the initiation of cytokinesis. This is in contrast to the TbMAPK6-deficient procyclic cells, which appeared to be arrested midway to the completion of cytokinesis (Fig. 3). However, the underlying mechanism behind this distinction remains to be explored.
The emergence of a small percentage of 2N1K and XN1K cells after TbMAPK6 RNAi suggests that kinetoplast replication/segregation was likely disrupted in those cells (Fig. 5E), which could be attributed to defective basal body duplication or inhibited segregation of duplicated basal bodies. To examine whether basal body duplication and/or segregation was defective in the cells, the RNAi cells were immunostained with YL 1/2 antibody (38). We found that an average of 86% of the 2N1K cells contained only one mature basal body and the remaining 2N1K cells (∼14%) contained two closely associated mature basal bodies (Fig. 6), suggesting that in the majority of the 2N1K cells, basal body duplication was compromised. The presence of two closely associated basal bodies in some 2N1K cells suggests that segregation of basal bodies was impaired after the basal bodies were duplicated. In the XN1K cells, two closely associated basal bodies were always detected, and in the XNXK cells, multiple (>2) basal bodies were found, with some of the basal bodies positioned close to each other (Fig. 6), suggesting defective basal body segregation. Altogether, these results indicated that RNAi of TbMAPK6 also caused defects in basal body duplication/segregation in the bloodstream form. Nevertheless, given that the 2N1K and XN1K cells constitute only a small percentage of the total cell population after TbMAPK6 RNAi for 4 days (Fig. 5D), it suggests that the primary role of TbMAPK6 is not to control basal body duplication/segregation, but rather, to promote cytokinesis initiation in the bloodstream form.
FIG 6.
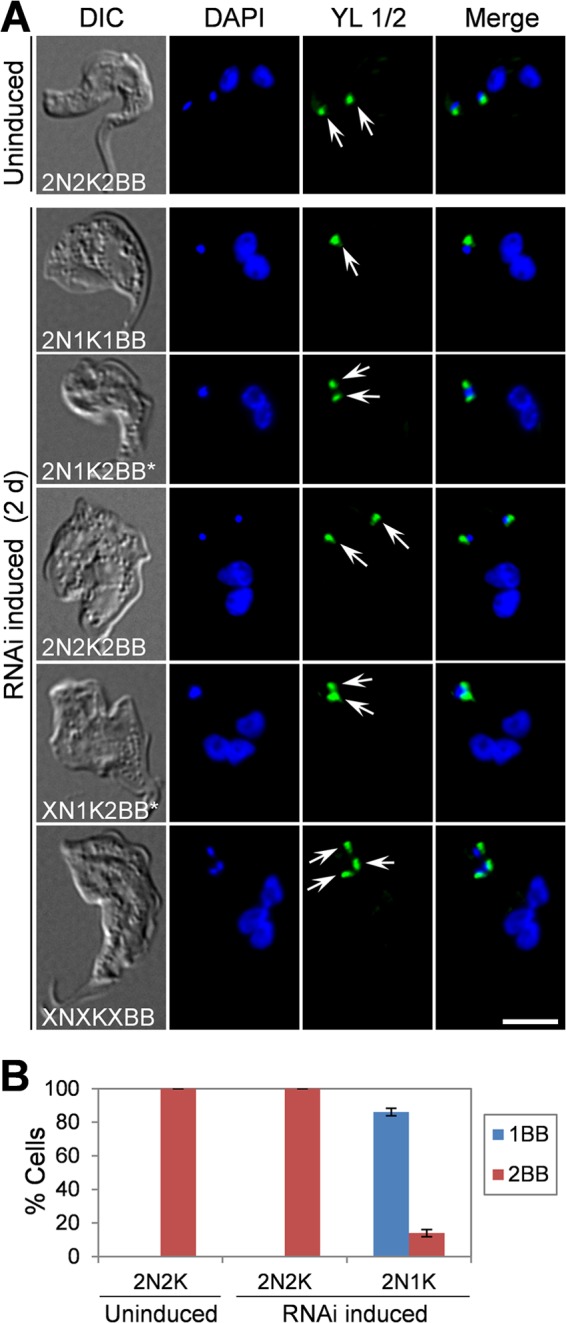
Effects of TbMAPK6 RNAi on basal body duplication and segregation in the bloodstream form. (A) Control cells and TbMAPK6 RNAi cells were immunostained with YL 1/2 antibody, which labels the tyrosinated α-tubulin in the mature basal bodies (arrows), and counterstained with DAPI for nuclear and kinetoplast DNA. X > 2. The asterisks indicate closely associated basal bodies. Bar, 2 μm. (B). Tabulation of the numbers of basal bodies (BB) in control cells and TbMAPK6 RNAi cells. The data are presented as the mean percentages ± SD of 200 cells counted for each time point from three independent experiments.
DISCUSSION
We report in this paper the functional characterization of a MAP kinase homolog and demonstrate its distinct roles in cytokinesis in different life cycle stages of T. brucei. In the procyclic form of T. brucei, RNAi of TbMAPK6 resulted in moderate growth defects and defective cleavage furrow ingression and cell abscission (Fig. 2 and 3). In the bloodstream form, however, RNAi of TbMAPK6 led to severe growth inhibition and rapid cell death and arrested the cells before cytokinesis initiation, thus accumulating cells with two mature basal bodies, two kinetoplasts, and two nuclei (Fig. 5). The observation that RNAi of TbMAPK6 in both forms also produced a small population of 2N1K cells containing a single mature basal body (Fig. 4 and 6) is interesting and seems to argue that basal body duplication and/or the segregation of duplicated basal bodies is defective in these cells. It has been generally acknowledged that basal body duplication and segregation constitute the first cytoskeletal event during the trypanosome cell division cycle (6), and a number of studies have shown that defects in basal body duplication and segregation lead to cytokinesis arrest (39–43). While the defective basal body duplication and/or segregation caused by TbMAPK6 RNAi likely contributed, at least partly, to cytokinesis defects in a small cell population, additional evidence showed that TbMAPK6 is primarily implicated in cytokinesis in both the procyclic and bloodstream forms (Fig. 3 and 5). Moreover, given that the two nuclei in TbMAPK6-deficient bloodstream cells were not well separated (Fig. 5E and 6A) and given the localization of TbMAPK6 in the nuclei of the bloodstream cells (Fig. 1C), a potential role for TbMAPK6 in regulating mitosis in the bloodstream form cannot be ruled out.
Cytokinesis in trypanosomes is unusual in that it is initiated from the anterior tip of the new FAZ and proceeds longitudinally toward the posterior end of the cell without the involvement of the actomyosin contractile ring, the cytokinesis apparatus that is essential and well conserved from fungi to humans (2). Although the regulatory pathways driving the initiation and completion of cytokinesis in trypanosomes remain to be delineated, two protein kinase homologs, the Aurora B kinase (TbAUK1) and the Polo-like kinase (TbPLK), likely function as upstream regulators of the cytokinesis initiation pathway(s) in trypanosomes, because both proteins are enriched at the anterior tip of the new FAZ prior to cytokinesis initiation and both are required for cytokinesis initiation (44–46). However, the downstream factors of TbAUK1 and TbPLK in the cytokinesis regulatory pathway(s) have not been identified. In addition to TbAUK1 and TbPLK, a number of other proteins have also been identified as potential regulators of cytokinesis due to the fact that deficiency in these proteins leads to cytokinesis defects (13, 39, 41, 43, 47–55). Nevertheless, precisely how these proteins are involved in controlling cytokinesis has not been well established. It should be noted that none of the proteins are enriched at the anterior tip of the new FAZ or on the cleavage furrow, making them less likely to be the direct regulators of cytokinesis. These proteins are associated with basal bodies and/or the flagellum (39, 41, 47) or the microtubule cytoskeleton (53, 54) or are localized predominantly in the cytosol (13, 43, 48–52). Like many of the above-mentioned proteins, TbMAPK6 is also distributed throughout the cytosol in both life cycle forms (Fig. 1). Therefore, its involvement in cytokinesis might be through regulating the downstream factor(s) that is localized at the initiation site of cytokinesis and/or the cleavage furrow and plays a direct role in cytokinesis. Future efforts will be directed to the identification of the TbMAPK6 downstream factor(s) and to the delineation of the signaling pathway mediated by TbMAPK6.
A number of studies have demonstrated the existence of stage-specific differences in cell cycle regulation in trypanosomes (10, 11, 13, 37, 56). Similarly, TbMAPK6 RNAi also revealed drastic differences in cytokinesis between the two life cycle forms: the procyclic form was arrested midway to the completion of cytokinesis, and the bloodstream form was halted before the initiation of cytokinesis (Fig. 3 and 5). However, the molecular basis underlying this distinction is not clear. Presumably, TbMAPK6 could regulate distinct cytokinesis-related genes in the two life cycle forms. Unfortunately, none of these downstream cytokinesis genes are known. The NRK1 substrate MAP65 and Pmk1-regulated myosin II apparently do not have homologs in the trypanosome genome, suggesting that the TbMAPK6-mediated cytokinesis pathway in trypanosomes likely differs from that in plants and yeast. Moreover, although TbMAPK6, NRK1, Pmk1, and ERK1/2 are all required for cytokinesis in the respective organisms, TbMAPK6 appears to belong to another family of MAP kinases, because TbMAPK6 possesses a TDY motif at the activation lip rather than the TEY motif found in NRK1, Pmk1, and ERK1/2 (see Fig. S1 in the supplemental material). Different MAP kinases can be classified based on the central residue of the activation lip TXY motif (57), and it is generally accepted that MAP kinases with a TEY motif all belong to the family of extracellular growth factor-regulated kinases (ERKs) in animals, yeast, and plants (19). MAP kinases containing the activation lip TDY motif are poorly characterized in eukaryotes, and their functions remain elusive. Further investigation of the regulation and function of TbMAPK6 will contribute to a better understanding of the regulatory circuit that controls cytokinesis in trypanosomes.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to George A. M. Cross of Rockefeller University for providing the procyclic 29-13 cell line and the bloodstream form SM cell line and to Paul Englund of Johns Hopkins School of Medicine for providing the pZJM vector. We also thank Arthur Günzl of the University of Connecticut Health Center for providing the pC-PTP-NEO and pC-HA-BSD vectors.
This work was supported by NIH grant R01AI101437 to Z.L. Y.W. is supported by a fellowship from the China Scholarship Council.
Footnotes
Published ahead of print 8 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00258-13.
REFERENCES
- 1.Straight AF, Field CM. 2000. Microtubules, membranes and cytokinesis. Curr. Biol. 10:R760–R770. 10.1016/S0960-9822(00)00746-6 [DOI] [PubMed] [Google Scholar]
- 2.Balasubramanian MK, Bi E, Glotzer M. 2004. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr. Biol. 14:R806–R818. 10.1016/j.cub.2004.09.022 [DOI] [PubMed] [Google Scholar]
- 3.Glotzer M. 2005. The molecular requirements for cytokinesis. Science 307:1735–1739. 10.1126/science.1096896 [DOI] [PubMed] [Google Scholar]
- 4.Oliferenko S, Chew TG, Balasubramanian MK. 2009. Positioning cytokinesis. Genes Dev. 23:660–674. 10.1101/gad.1772009 [DOI] [PubMed] [Google Scholar]
- 5.Pollard TD, Wu JQ. 2010. Understanding cytokinesis: lessons from fission yeast. Nat. Rev. Mol. Cell Biol. 11:149–155. 10.1038/nrm2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaughan S, Gull K. 2008. The structural mechanics of cell division in Trypanosoma brucei. Biochem. Soc. Trans. 36:421–424. 10.1042/BST0360421 [DOI] [PubMed] [Google Scholar]
- 7.Vaughan S, Kohl L, Ngai I, Wheeler RJ, Gull K. 2008. A repetitive protein essential for the flagellum attachment zone filament structure and function in Trypanosoma brucei. Protist 159:127–136. 10.1016/j.protis.2007.08.005 [DOI] [PubMed] [Google Scholar]
- 8.Kohl L, Robinson D, Bastin P. 2003. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 22:5336–5346. 10.1093/emboj/cdg518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Salcedo JA, Perez-Morga D, Gijon P, Dilbeck V, Pays E, Nolan DP. 2004. A differential role for actin during the life cycle of Trypanosoma brucei. EMBO J. 23:780–789. 10.1038/sj.emboj.7600094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammarton TC, Clark J, Douglas F, Boshart M, Mottram JC. 2003. Stage-specific differences in cell cycle control in Trypanosoma brucei revealed by RNA interference of a mitotic cyclin. J. Biol. Chem. 278:22877–22886. 10.1074/jbc.M300813200 [DOI] [PubMed] [Google Scholar]
- 11.Tu X, Wang CC. 2004. The involvement of two cdc2-related kinases (CRKs) in Trypanosoma brucei cell cycle regulation and the distinctive stage-specific phenotypes caused by CRK3 depletion. J. Biol. Chem. 279:20519–20528. 10.1074/jbc.M312862200 [DOI] [PubMed] [Google Scholar]
- 12.Gruneberg U, Nigg EA. 2003. Regulation of cell division: stop the SIN! Trends Cell Biol. 13:159–162. 10.1016/S0962-8924(03)00034-5 [DOI] [PubMed] [Google Scholar]
- 13.Hammarton TC, Lillico SG, Welburn SC, Mottram JC. 2005. Trypanosoma brucei MOB1 is required for accurate and efficient cytokinesis but not for exit from mitosis. Mol. Microbiol. 56:104–116. 10.1111/j.1365-2958.2005.04542.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Widmann C, Gibson S, Jarpe MB, Johnson GL. 1999. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79:143–180 [DOI] [PubMed] [Google Scholar]
- 15.Sasabe M, Machida Y. 2012. Regulation of organization and function of microtubules by the mitogen-activated protein kinase cascade during plant cytokinesis. Cytoskeleton 69:913–918. 10.1002/cm.21072 [DOI] [PubMed] [Google Scholar]
- 16.Toda T, Dhut S, Superti-Furga G, Gotoh Y, Nishida E, Sugiura R, Kuno T. 1996. The fission yeast pmk1+ gene encodes a novel mitogen-activated protein kinase homolog which regulates cell integrity and functions coordinately with the protein kinase C pathway. Mol. Cell. Biol. 16:6752–6764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satoh R, Morita T, Takada H, Kita A, Ishiwata S, Doi A, Hagihara K, Taga A, Matsumura Y, Tohda H, Sugiura R. 2009. Role of the RNA-binding protein Nrd1 and Pmk1 mitogen-activated protein kinase in the regulation of myosin mRNA stability in fission yeast. Mol. Biol. Cell 20:2473–2485. 10.1091/mbc.E08-09-0893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasahara K, Nakayama Y, Nakazato Y, Ikeda K, Kuga T, Yamaguchi N. 2007. Src signaling regulates completion of abscission in cytokinesis through ERK/MAPK activation at the midbody. J. Biol. Chem. 282:5327–5339. 10.1074/jbc.M608396200 [DOI] [PubMed] [Google Scholar]
- 19.Wiese M. 2007. Leishmania MAP kinases—familiar proteins in an unusual context. Int. J. Parasitol. 37:1053–1062. 10.1016/j.ijpara.2007.04.008 [DOI] [PubMed] [Google Scholar]
- 20.Hua SB, Wang CC. 1997. Interferon-gamma activation of a mitogen-activated protein kinase, KFR1, in the bloodstream form of Trypanosoma brucei. J. Biol. Chem. 272:10797–10803. 10.1074/jbc.272.16.10797 [DOI] [PubMed] [Google Scholar]
- 21.Muller IB, Domenicali-Pfister D, Roditi I, Vassella E. 2002. Stage-specific requirement of a mitogen-activated protein kinase by Trypanosoma brucei. Mol. Biol. Cell 13:3787–3799. 10.1091/mbc.E02-02-0093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellis J, Sarkar M, Hendriks E, Matthews K. 2004. A novel ERK-like, CRK-like protein kinase that modulates growth in Trypanosoma brucei via an autoregulatory C-terminal extension. Mol. Microbiol. 53:1487–1499. 10.1111/j.1365-2958.2004.04218.x [DOI] [PubMed] [Google Scholar]
- 23.Guttinger A, Schwab C, Morand S, Roditi I, Vassella E. 2007. A mitogen-activated protein kinase of Trypanosoma brucei confers resistance to temperature stress. Mol. Biochem. Parasitol. 153:203–206. 10.1016/j.molbiopara.2007.02.001 [DOI] [PubMed] [Google Scholar]
- 24.Domenicali Pfister D, Burkard G, Morand S, Renggli CK, Roditi I, Vassella E. 2006. A mitogen-activated protein kinase controls differentiation of bloodstream forms of Trypanosoma brucei. Eukaryot. Cell 5:1126–1135. 10.1128/EC.00094-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandal G, Sharma M, Kruse M, Sander-Juelch C, Munro LA, Wang Y, Vilg JV, Tamas MJ, Bhattacharjee H, Wiese M, Mukhopadhyay R. 2012. Modulation of Leishmania major aquaglyceroporin activity by a mitogen-activated protein kinase. Mol. Microbiol. 85:1204–1218. 10.1111/j.1365-2958.2012.08169.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wirtz E, Leal S, Ochatt C, Cross GA. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101. 10.1016/S0166-6851(99)00002-X [DOI] [PubMed] [Google Scholar]
- 27.Dang HQ, Li Z. 2011. The Cdc45.Mcm2-7.GINS protein complex in trypanosomes regulates DNA replication and interacts with two Orc1-like proteins in the origin recognition complex. J. Biol. Chem. 286:32424–32435. 10.1074/jbc.M111.240143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Morris JC, Drew ME, Englund PT. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174–40179. 10.1074/jbc.M008405200 [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Li Z, Wang CC. 2003. Differentiation of Trypanosoma brucei may be stage non-specific and does not require progression of cell cycle. Mol. Microbiol. 49:251–265. 10.1046/j.1365-2958.2003.03575.x [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Hu H, Li Z. 2013. The cooperative roles of PHO80-like cyclins in regulating the G1/S transition and posterior cytoskeletal morphogenesis in Trypanosoma brucei. Mol. Microbiol. 90:130–146. 10.1111/mmi.12352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu H, Hu L, Yu Z, Chasse AE, Chu F, Li Z. 2012. An orphan kinesin in trypanosomes cooperates with a kinetoplastid-specific kinesin to maintain cell morphology by regulating subpellicular microtubules. J. Cell Sci. 125:4126–4136. 10.1242/jcs.106534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, Umeyama T, Wang CC. 2008. The chromosomal passenger complex and a mitotic kinesin interact with the Tousled-like kinase in trypanosomes to regulate mitosis and cytokinesis. PLoS One 3:e3814. 10.1371/journal.pone.0003814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kilmartin JV, Wright B, Milstein C. 1982. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J. Cell Biol. 93:576–582. 10.1083/jcb.93.3.576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson DR, Gull K. 1991. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature 352:731–733. 10.1038/352731a0 [DOI] [PubMed] [Google Scholar]
- 35.Sherwin T, Schneider A, Sasse R, Seebeck T, Gull K. 1987. Distinct localization and cell cycle dependence of COOH terminally tyrosinolated alpha-tubulin in the microtubules of Trypanosoma brucei brucei. J. Cell Biol. 104:439–446. 10.1083/jcb.104.3.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tu X, Kumar P, Li Z, Wang CC. 2006. An aurora kinase homologue is involved in regulating both mitosis and cytokinesis in Trypanosoma brucei. J. Biol. Chem. 281:9677–9687. 10.1074/jbc.M511504200 [DOI] [PubMed] [Google Scholar]
- 37.Li Z, Wang CC. 2006. Changing roles of aurora-B kinase in two life cycle stages of Trypanosoma brucei. Eukaryot. Cell 5:1026–1035. 10.1128/EC.00129-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherwin T, Gull K. 1989. Visualization of detyrosination along single microtubules reveals novel mechanisms of assembly during cytoskeletal duplication in trypanosomes. Cell 57:211–221. 10.1016/0092-8674(89)90959-8 [DOI] [PubMed] [Google Scholar]
- 39.Li Z, Wang CC. 2008. KMP-11, a basal body and flagellar protein, is required for cell division in Trypanosoma brucei. Eukaryot. Cell 7:1941–1950. 10.1128/EC.00249-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pradel LC, Bonhivers M, Landrein N, Robinson DR. 2006. NIMA-related kinase TbNRKC is involved in basal body separation in Trypanosoma brucei. J. Cell Sci. 119:1852–1863. 10.1242/jcs.02900 [DOI] [PubMed] [Google Scholar]
- 41.Selvapandiyan A, Kumar P, Morris JC, Salisbury JL, Wang CC, Nakhasi HL. 2007. Centrin1 is required for organelle segregation and cytokinesis in Trypanosoma brucei. Mol. Biol. Cell 18:3290–3301. 10.1091/mbc.E07-01-0022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgan GW, Denny PW, Vaughan S, Goulding D, Jeffries TR, Smith DF, Gull K, Field MC. 2005. An evolutionarily conserved coiled-coil protein implicated in polycystic kidney disease is involved in basal body duplication and flagellar biogenesis in Trypanosoma brucei. Mol. Cell. Biol. 25:3774–3783. 10.1128/MCB.25.9.3774-3783.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gourguechon S, Wang CC. 2009. CRK9 contributes to regulation of mitosis and cytokinesis in the procyclic form of Trypanosoma brucei. BMC Cell Biol. 10:68. 10.1186/1471-2121-10-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z, Lee JH, Chu F, Burlingame AL, Gunzl A, Wang CC. 2008. Identification of a novel chromosomal passenger complex and its unique localization during cytokinesis in Trypanosoma brucei. PLoS One 3:e2354. 10.1371/journal.pone.0002354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar P, Wang CC. 2006. Dissociation of cytokinesis initiation from mitotic control in a eukaryote. Eukaryot. Cell 5:92–102. 10.1128/EC.5.1.92-102.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Graffenried CL, Ho HH, Warren G. 2008. Polo-like kinase is required for Golgi and bilobe biogenesis in Trypanosoma brucei. J. Cell Biol. 181:431–438. 10.1083/jcb.200708082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price HP, Peltan A, Stark M, Smith DF. 2010. The small GTPase ARL2 is required for cytokinesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 173:123–131. 10.1016/j.molbiopara.2010.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fisk JC, Zurita-Lopez C, Sayegh J, Tomasello DL, Clarke SG, Read LK. 2010. TbPRMT6 is a type I protein arginine methyltransferase that contributes to cytokinesis in Trypanosoma brucei. Eukaryot. Cell 9:866–877. 10.1128/EC.00018-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma J, Benz C, Grimaldi R, Stockdale C, Wyatt P, Frearson J, Hammarton TC. 2010. Nuclear DBF-2-related kinases are essential regulators of cytokinesis in bloodstream stage Trypanosoma brucei. J. Biol. Chem. 285:15356–15368. 10.1074/jbc.M109.074591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodgers MJ, Albanesi JP, Phillips MA. 2007. Phosphatidylinositol 4-kinase III-beta is required for Golgi maintenance and cytokinesis in Trypanosoma brucei. Eukaryot. Cell 6:1108–1118. 10.1128/EC.00107-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rothberg KG, Burdette DL, Pfannstiel J, Jetton N, Singh R, Ruben L. 2006. The RACK1 homologue from Trypanosoma brucei is required for the onset and progression of cytokinesis. J. Biol. Chem. 281:9781–9790. 10.1074/jbc.M600133200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benz C, Clucas C, Mottram JC, Hammarton TC. 2012. Cytokinesis in bloodstream stage Trypanosoma brucei requires a family of katanins and spastin. PLoS One 7:e30367. 10.1371/journal.pone.0030367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.May SF, Peacock L, Almeida Costa CI, Gibson WC, Tetley L, Robinson DR, Hammarton TC. 2012. The Trypanosoma brucei AIR9-like protein is cytoskeleton-associated and is required for nucleus positioning and accurate cleavage furrow placement. Mol. Microbiol. 84:77–92. 10.1111/j.1365-2958.2012.08008.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu L, Hu H, Li Z. 2012. A kinetoplastid-specific kinesin is required for cytokinesis and for maintenance of cell morphology in Trypanosoma brucei. Mol. Microbiol. 83:565–578. 10.1111/j.1365-2958.2011.07951.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benz C, Clayton CE. 2007. The F-box protein CFB2 is required for cytokinesis of bloodstream-form Trypanosoma brucei. Mol. Biochem. Parasitol. 156:217–224. 10.1016/j.molbiopara.2007.08.005 [DOI] [PubMed] [Google Scholar]
- 56.Tu X, Mancuso J, Cande WZ, Wang CC. 2005. Distinct cytoskeletal modulation and regulation of G1-S transition in the two life stages of Trypanosoma brucei. J. Cell Sci. 118:4353–4364. 10.1242/jcs.02567 [DOI] [PubMed] [Google Scholar]
- 57.Kultz D. 1998. Phylogenetic and functional classification of mitogen- and stress-activated protein kinases. J. Mol. Evol. 46:571–588. 10.1007/PL00006338 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


