Abstract
Uniparental inheritance (UPI) of mitochondria is common among eukaryotes. The underlying molecular basis by which the sexes of the parents control this non-Mendelian pattern of inheritance is yet to be fully understood. Two major factors have complicated the understanding of the role of sex-specific genes in the UPI phenomenon: in many cases (i) fusion occurs between cells of unequal size or (ii) mating requires a large region of the genome or chromosome that includes genes unrelated to sex determination. The fungus Phycomyces blakesleeanus is a member of the Mucoromycotina and has a simple mating type locus encoding only one high-mobility group (HMG) domain protein, and mating occurs by fusion of isogamous cells, thus providing a model system without the limitations mentioned above. Analysis of more than 250 progeny from a series of genetic crosses between wild-type strains of Phycomyces revealed a correlation between the individual genes in the mating type locus and UPI of mitochondria. Inheritance is from the plus (+) sex type and is associated with degradation of the mtDNA from the minus (−) parent. These findings suggest that UPI can be directly controlled by genes that determine sex identity, independent of cell size or the complexity of the genetic composition of a sex chromosome.
INTRODUCTION
The uniparental inheritance (UPI) of organelle genomes during sexual reproduction is prevalent in eukaryotic lineages. The physical processes accounting for this pattern of inheritance include the selective degradation of organelles of one genotype and the disproportionate contributions of the cytoplasmic contents, together with an underlying selective benefit of homoplasmy, whereby cells house a single mitochondrial genotype (1, 2). In most cases, the maternal parent contributes the mitochondria into the zygote. However, whether or not the genes on sex chromosomes or in mating-type loci that specifically determine the sex of the parent are directly required for UPI is untested. This is because it has not been possible to uncouple the effects of differing cell sizes (anisogamy) or the numerous genes present on sex chromosomes from their potential contribution to UPI. To answer this fundamental question requires analysis in an organism with equally sized gametes (isogamy) and a simple single-gene sex-determining system.
Members of the fungal kingdom may undergo a sexual cycle that involves the fusion of equally sized cells and whose sex types are determined by a small region of the genome known as the mating type (MAT) locus. However, many of these species, like the model ascomycete yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe (3–6), inherit the mitochondrial genotypes from either parent. Similarly, some basidiomycete mushroom species initiate their sexual cycle through the fusion of equally sized cells, and depending on the species they can exhibit the inheritance of the mitochondrial genome from just one or either parent. In species with uniparental inheritance, the complex genetic makeup of their mating type systems have prevented implicating sex-determining genes directly, with some exceptions that are discussed below (7, 8). Phycomyces blakesleeanus is a fungus in the subphylum Mucoromycotina that is a distant relative of the ascomycete and basidiomycete species. Mating involves the fusion of two identically sized gametangium cells. Without any features to distinguish between the two parents, the terms “(+)” (plus) and “(−)” (minus) were thus coined arbitrarily for Mucoromycotina species to designate the sex or mating type of strains. Sex determination is governed by a small genetic locus, which in Phycomyces contains a single gene for each allele, either sexM or sexP (9). The alleles share no DNA sequence similarity, although both SexM and SexP proteins contain a high-mobility group (HMG) domain found in transcription factors. This scenario resembles the SOX3/SRY HMG-domain proteins encoded by the X and Y chromosomes of humans and other mammals but entails a small (<6-kb) locus encoding one protein rather than a sex chromosome.
The expectation was that Phycomyces would exhibit biparental inheritance of its mitochondrial genome, as seen in other fungi that undergo fusion of isogamete-like cells. Instead, in this study uniparental inheritance was observed. Our findings provide a strong correlation between the mating type locus genes and UPI, with the implication that similar regulation may exist in other eukaryotes or was important at one stage of their evolution in establishing this system of inheritance.
MATERIALS AND METHODS
Strains and growth conditions.
Phycomyces blakesleeanus strains were grown on potato dextrose agar (PDA) plates at room temperature. Strains were NRRL1555 sex type (−), NRRL1554 (+), A56 (+), SC10 -1a (+), UBC21 (+), and UBC33 (−). These strains are wild isolates, except for A56. NRRL1555 is the standard laboratory strain, which has been sequenced, and A56 is isogenic to NRRL1555, after 11 backcrosses to introgress the (+) sex allele into the NRRL1555 background (10). KC10B1 (−) and KC10B6 (+) are progeny isolated from a SC10-1a × NRRL1555 cross; they contain the SC10-1a mitochondrial DNA (mtDNA) genotype. KC10F1 (+) and KC10F6 (−) are from an A56 × NRRL1555 cross; they contain the A56 mtDNA genotype. For preparing mycelia for DNA extraction, sporangiospores were inoculated in yeast extract-peptone-dextrose (YPD) medium and incubated for 3 to 5 days.
Crosses.
Crosses were performed on V8 medium plates (5% V8 juice, 0.5 g/liter KH2PO4, 1 mg/liter thiamine, 4% agar; pH 6). Heat-activated spores from strains of opposite mating types were inoculated 6 to 7 cm apart on 10-cm-diameter petri dishes and incubated in darkness at room temperature until the zygospores formed (5 to 7 days). The zygospores were isolated with forceps, placed on wet filter paper, and incubated 2 to 4 months until germination. After the germination of zygospores, individual germspores were streaked onto PDA to isolate single colonies. Each progeny was isolated from a different zygospore, to ensure that each was the result of an independent meiotic event. The sexes of the progeny were tested by inoculating them adjacent to the standard wild-type (+) and (−) strains.
DNA extraction.
Mycelium was dried in a freeze-dryer, and genomic DNA was extracted from mycelium broken apart with 2-mm glass beads with a CTAB (cetyltrimethylammonium bromide) extraction buffer (11).
Mitochondrial genotyping.
Primers ALID0933 (5′-CGAATGACCCGAGAAGCC-3′) and ALID0934 (5′-GGAGTGACTCATCTTTCG-3′) were used to amplify a polymorphic region of the mitochondrial genome, identified by comparing genome sequences of strains NRRL1555 and UBC21 (http://genome.jgi-psf.org/Phybl2/Phybl2.home.html) and suitable for comparing the different mitochondrial genotypes in the other wild-type strains. ALID0933 is within the orf511 gene, and ALID0934 is in the intergenic region between orf511 and nad4L. PCR products were digested with TaqαI restriction enzyme at 65°C and separated by electrophoresis on 2% agarose/1× Tris-acetate-EDTA gels. For PCR from zygospores, a single zygospore was crushed in 10 μl of dilution buffer (Thermo Scientific Direct PCR kit) and incubated for 10 min at room temperature; 2 μl of this was used as the template for the PCRs. Products were either digested with TaqαI and resolved on agarose gels or sequenced.
Another mitochondrial polymorphism between the atp9 and trnG genes was examined in 12 randomly selected progenies from cross 1 and the two parents (NRRL1555 and UBC21). The region was amplified by primers ALID2131 (5′-AATTGTTGCTAATCCAGCTC-3′) and ALID2132 (5′-AAAAGGTCGTCACCTTCGTC-3′). The PCR products were gel purified and sequenced, and the sequences were compared for the 14 strains.
RESULTS AND DISCUSSION
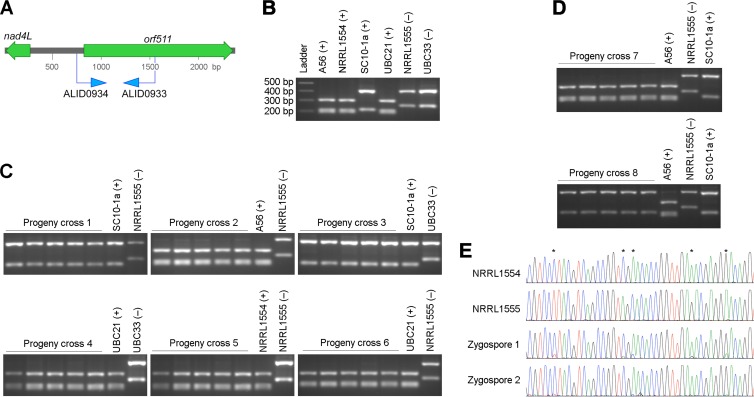
A genetic map of Phycomyces was recently created using nuclear molecular markers in progeny after crossing two wild-type parents (12). Here, oligonucleotide primers were designed to amplify a variable part of the mitochondrial genome (mtDNA) (Fig. 1A) and used on the 121 progeny from this cross. The progeny were derived from independent meiotic reduction events, because each zygospore forms independently of the others, and a single strain was isolated from the zygospores germinated. The difference in PCR-restriction fragment length polymorphism (RFLP) patterns between the strains was used to track the source of the mitochondrial genome in the progeny (Fig. 1B). Rather than exhibiting biparental inheritance, all 121 progeny had inherited their mitochondrial genome from one parent (Fig. 1C and Table 1). To ensure that this result was not a peculiarity of the molecular marker employed, an additional polymorphic site in the mitochondrial genome was identified. This region was amplified and sequenced from 12 randomly selected progeny: all had inherited the allele from the (+) parent. This result suggests that there is transmission of the full mitochondrial genome from the (+) parent into the progeny.
FIG 1.
UPI of mitochondria occurs in Phycomyces. (A) Positions of the oligonucleotide primers used to amplify a variable part of the mitochondrial genome. (B) Agarose gel showing the sizes of the PCR products of an mtDNA fragment cut with TaqαI restriction enzyme from the parental strains. (C) Gels of the PCR products of the mtDNA fragment cut with TaqαI, from parents and five representative progeny from eight crosses (Table 1). (D) The mitochondria are inherited from the (+) parent regardless of the origin of the mitochondria. Agarose gels show the mtDNA fragment cut with TaqαI from the strains in which the mitochondrial genome was switched into the other sex background. (E) The mtDNA from the (−) parent is degraded in the zygospore. Chromatograms of a fragment of the mitochondrial genome amplified from the parents NRRL1554 (+) and NRRL1555 (−) and two single zygospores (one week old). The asterisks show the nucleotide sequences that differ between the parents. A trace of the mtDNA from the (−) parent is still amplified from zygospore 1.
TABLE 1.
Crosses and number of progeny analyzed
| Cross no. | Parents [(+) × (−)] | No. of progeny analyzed |
|---|---|---|
| 1 | UBC21 × NRRL1555 | 121 |
| 2 | SC10-1a × NRRL1555 | 17 |
| 3 | SC10-1a × UBC33 | 26 |
| 4 | NRRL1554 × NRRL1555 | 16 |
| 5 | UBC21 × UBC33 | 12 |
| 6 | A56 × NRRL1555 | 28 |
| 7 | KC10F1 × KC10B1 | 22 |
| 8 | KC10B6 × KC10F6 | 10 |
Segregation of the mitochondrial genome in progeny from a series of crosses shows that UPI is consistently observed in Phycomyces (Fig. 1C and Table 1). First, in all crosses UPI is from the (+) parent, including in crosses between a selection of different wild-type isolates. These wild-type isolates are all unique and North American in origin (from Canada for UBC21 and UBC33 and from the United States for the others), and their isolation dates span over a century, from Albert Blakeslee's collection from the turn of the 20th century to an isolate from South Carolina collected in 2010. Second, reciprocal crosses between strains, in which the mtDNA was switched into the opposite mating type background, establish that the mitochondrial genome itself does not contribute to UPI (Fig. 1D, crosses 7 and 8). Third, the sex locus controls inheritance (cross 6) rather than other elements in the genome, because strain A56 is isogenic with strain NRRL1555 except for its sex allele (10).
The meiotic reduction process in Phycomyces is not always perfect, with the emergence of heterokaryons that can be easily identified by their morphology if they carry the two alleles of the sex locus. Three such strains were examined, and all had inherited the (+) parent mitochondria. This indicates that the selection process occurs with fidelity even if nuclear fusion or chromosome disjunction does not and that UPI is initiated early in the mating process. Since both mating types were present in these heterokaryons, this also indicates that selection is most likely ensured by the degradation of (−) mitochondria rather than the protection and preferential replication of (+) mitochondria. Degradation of organelles is a common feature for UPI in eukaryotes (1). For example, recent findings in the nematode Caenorhabditis elegans establish the degradation of sperm mitochondria by induction of autophagy (13–15).
Electron microscopy of the developing zygospores of Phycomyces has illustrated that mitochondria lie adjacent on either side of the cell walls as the walls dissolve between the two gametangia, suggesting that the events leading to UPI must occur postfusion (16). The timing of mitochondrial degradation was assessed by PCR on zygospores. Zygospores are the single cells that result from the sexual fusion of (+) and (−) gametangia and also act as resting structures that are dormant for 2 months to over a year. Analysis of the mitochondrial genotypes demonstrates that degradation of mtDNA from the (−) parent occurs early during the process of zygospore maturation, since this is observed within a week after the formation of the zygospore (Fig. 1E).
Two other species of fungi with UPI linked to their mating type alleles are worth comparing with Phycomyces (8). In the first, Ustilago maydis, the pheromone MAT locus controls uniparental inheritance; however, this is mediated by two genes within the locus, neither of which is required for pheromone biosynthesis or in determining the cell type (17, 18). The second species is Cryptococcus neoformans, in which mitochondrial inheritance is governed at the pre- and postzygotic stages of the sexual cycle (19, 20). The postzygotic control is in part conferred by the Sxi1α and Sxi2a homeodomain proteins, which also specify the mating types of the parents. A similar situation likely occurs in the related species Cryptococcus gattii. Crosses between congenic strains differing only in mating type exhibit UPI, while in crosses using strains with different genetic background both parents can contribute their mitochondrial DNA to the progeny (21, 22). In addition, in C. neoformans the environmental and genetic conditions alter the proportion of uniparental versus biparental inheritance (20, 23–25), and cytological analysis indicates that the mating type α cell delivers its nucleus to the a cell (26). Thus, neither U. maydis nor the Cryptococcus species rely on sex-determining genes for the sole determinant of UPI as is the case in Phycomyces.
The mechanism whereby SexM or SexP, encoded by the sex locus of Phycomyces, contributes to UPI remains to be elucidated. The absence of a stable transformation system for this species prevents gene manipulation approaches to explore this trait (27). Nevertheless, finding this correlation between mating type and UPI in Phycomyces provides research directions in more tractable eukaryotes, to test if the genes required for sex determination also control UPI more widely.
We hypothesize that the equivalent genes that control the sex of an organism also control UPI or may have done so in the past. In some organisms, the original roles of the sex-determining factor in UPI may have been superseded by other genes. In U. maydis, the lga2 and rga2 genes in the a mating type locus regulate UPI of mitochondria (17). These two genes have been incorporated into a locus that controls the sex of the cell but are not involved in determining sex of the mating partner. A similar scenario may have occurred in the green alga Chlamydomonas reinhardtii, which has a large mating type (MT) locus. In this organism, isogametes fuse, and in the progeny the mitochondrial genotype is from the minus parent and, conversely, the chloroplast genotype from the plus parent (28). The ezy-1 and ezy-2 genes that are located within MT have been implicated in chloroplast inheritance (29, 30). If so, this represents another example in which the control of UPI is through genetic factors that have been recruited into the region of the genome that includes the sex-determining genes. Even more compelling is evidence that the homeodomain protein Gsp1 is also involved in C. reinhardtii, because codeletion of this gene and the adjacent homolog of an inositol monophosphatase impairs UPI (31).
In summary, the present study in Phycomyces provides an example in which genes located within the sex-determining region of the genome likely have two functions. One is to determine the mating type or sex of the cells, and the other is to direct uniparental inheritance during mating. Given that those two processes are intimately linked in eukaryotes, we propose that this represents an ancestral state in eukaryotes that coordinated the distribution of both nuclear and mitochondrial genetic material during sexual reproduction.
ACKNOWLEDGMENTS
We thank Joseph Heitman and Xiaorong Lin for comments on the manuscript.
This research was supported by NSF grant MCB-0920581.
Footnotes
Published ahead of print 15 November 2013
REFERENCES
- 1.Sato M, Sato K. 2013. Maternal inheritance of mitochondrial DNA by diverse mechanisms to eliminate paternal mitochondrial DNA. Biochim. Biophys. Acta 1833:1979–1984. 10.1016/j.bbamcr.2013.03.010 [DOI] [PubMed] [Google Scholar]
- 2.Birky CW., Jr 2001. The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu. Rev. Genet. 35:125–148. 10.1146/annurev.genet.35.102401.090231 [DOI] [PubMed] [Google Scholar]
- 3.Strausberg RL, Perlman PS. 1978. The effect of zygotic bud position on the transmission of mitochondrial genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 163:131–144. 10.1007/BF00267404 [DOI] [PubMed] [Google Scholar]
- 4.Seitz-Mayr G, Wolf K, Kaudewitz F. 1978. Extrachromosomal inheritance in Schizosaccharomyces pombe. VII. Studies by zygote clone analysis on transmission, segregation, recombination, and uniparental inheritance of mitochondrial markers conferring resistance to antimycin, chloramphenicol, and erythromycin. Mol. Gen. Genet. 164:309–320 [DOI] [PubMed] [Google Scholar]
- 5.Dujon B, Slonimski PP, Weill L. 1974. Mitochondrial genetics IX: a model for recombination and segregation of mitochondrial genomes in Saccharomyces cerevisiae. Genetics 78:415–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolf K, Seitz-Mayr G, Kaudewitz F. 1978. Extrachromosomal inheritance in Schizosaccharomyces pombe. VIII. Extent of cytoplasmic mixing in zygotes estimated by tetrad analysis of crosses involving mitochondrial markers conferring resistance to antimycin, chloramphenicol, and erythromycin. Mol. Gen. Genet. 164:321–329 [DOI] [PubMed] [Google Scholar]
- 7.Griffiths AJF. 1996. Mitochondrial inheritance in filamentous fungi. J. Genet. 75:403–414. 10.1007/BF02966318 [DOI] [Google Scholar]
- 8.Basse CW. 2010. Mitochondrial inheritance in fungi. Curr. Opin. Microbiol. 13:712–719. 10.1016/j.mib.2010.09.003 [DOI] [PubMed] [Google Scholar]
- 9.Idnurm A, Walton FJ, Floyd A, Heitman J. 2008. Identification of the sex genes in an early diverged fungus. Nature 451:193–196. 10.1038/nature06453 [DOI] [PubMed] [Google Scholar]
- 10.Alvarez MI, Eslava AP. 1983. Isogenic strains of Phycomyces blakesleeanus suitable for genetic analysis. Genetics 105:873–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pitkin JW, Panaccione DG, Walton JD. 1996. A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 142:1557–1565. 10.1099/13500872-142-6-1557 [DOI] [PubMed] [Google Scholar]
- 12.Chaudhary S, Polaino S, Shakya VPS, Idnurm A. 2013. A new genetic map for the zygomycete fungus Phycomyces blakesleeanus. PLoS One 8:e58931. 10.1371/journal.pone.0058931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al Rawi S, Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. 2011. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 334:1144–1147. 10.1126/science.1211878 [DOI] [PubMed] [Google Scholar]
- 14.Sato M, Sato K. 2011. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 334:1141–1144. 10.1126/science.1210333 [DOI] [PubMed] [Google Scholar]
- 15.Al Rawi S, Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. 2012. Allophagy: a macroautophagic process degrading spermatozoid-inherited organelles. Autophagy 8:421–423. 10.4161/auto.19242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Donnell KL, Hooper GR, Fields WG. 1976. Zygosporogenesis in Phycomyces blakesleeanus. Can. J. Bot. 54:2573–2586. 10.1139/b76-277 [DOI] [Google Scholar]
- 17.Fedler M, Luh K-S, Stelter K, Nieto-Jacobo F, Basse CW. 2009. The a2 mating-type locus genes lga2 and rga2 direct uniparental mitochondrial DNA (mtDNA) inheritance and constrain mtDNA recombination during sexual development of Ustilago maydis. Genetics 181:847–860. 10.1534/genetics.108.096859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahlert M, Vogler C, Stelter K, Hause G, Basse CW. 2009. The a2 mating-type-locus gene lga2 of Ustilago maydis interferes with mitochondrial dynamics and fusion, partially in dependence on a Dnm1-like fission component. J. Cell Sci. 122:2402–2412. 10.1242/jcs.039354 [DOI] [PubMed] [Google Scholar]
- 19.Gyawali R, Lin X. 2011. Mechanisms of uniparental mitochondrial DNA inheritance in Cryptococcus neoformans. Mycobiology 39:235–242. 10.5941/MYCO.2011.39.4.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gyawali R, Lin X. 2013. Prezygotic and postzygotic control of uniparental mitochondrial DNA inheritance in Cryptococcus neoformans. mBio 4:e00112–00113. 10.1128/mBio.00112-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhai B, Zhu P, Lin X, Idnurm A. 2013. Cryptococcus gattii congenic strains for understanding the evolution of virulence in human pathogenic fungi. Infect. Immun. 81:2626–2637. 10.1128/IAI.00259-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voelz K, Ma H, Byrnes EJ, Phadke S, Zhu P, Mueller O, Farrer RA, Henk DA, Lewit Y, Hsueh Y-P, Fisher MC, Idnurm A, Heitman J, May RC. 2013. Transmission of hypervirulence traits via sexual reproduction within and between lineages of the human fungal pathogen Cryptococcus gattii. PLoS Genet. 9:e1003771. 10.1371/journal.pgen.1003771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Z, Hull CM, Heitman J, Sun S, Xu J. 2004. SXI1α controls uniparental mitochondrial inheritance in Cryptococcus neoformans. Curr. Biol. 14:R743–R744. 10.1016/j.cub.2004.09.008 [DOI] [PubMed] [Google Scholar]
- 24.Yan Z, Sun S, Shahid M, Xu J. 2007. Environment factors can influence mitochondrial inheritance in the fungus Cryptococcus neoformans. Fungal Genet. Biol. 44:315–322. 10.1016/j.fgb.2006.10.002 [DOI] [PubMed] [Google Scholar]
- 25.Toffaletti DL, Nielsen K, Dietrich F, Heitman J, Perfect JR. 2004. Cryptococcus neoformans mitochondrial genomes from serotype A and D strains do not influence virulence. Curr. Genet. 46:193–204. 10.1007/s00294-004-0521-9 [DOI] [PubMed] [Google Scholar]
- 26.McClelland CM, Chang YC, Varma A, Kwon-Chung KJ. 2004. Uniqueness of the mating system in Cryptococcus neoformans. Trends Microbiol. 12:208–212. 10.1016/j.tim.2004.03.003 [DOI] [PubMed] [Google Scholar]
- 27.Obraztsova IN, Prados N, Holzmann K, Avalos J, Cerdá-Olmedo E. 2004. Genetic damage following introduction of DNA in Phycomyces. Fungal Genet. Biol. 41:168–180. 10.1016/j.fgb.2003.09.007 [DOI] [PubMed] [Google Scholar]
- 28.Goodenough U, Lin H, Lee JH. 2007. Sex determination in Chlamydomonas. Semin. Cell Dev. Biol. 18:350–361. 10.1016/j.semcdb.2007.02.006 [DOI] [PubMed] [Google Scholar]
- 29.Armbrust EV, Ferris PJ, Goodenough UW. 1993. A mating type-linked gene cluster expressed in Chlamydomonas zygotes participates in the uniparental inheritance of the chloroplast genome. Cell 74:801–811. 10.1016/0092-8674(93)90460-8 [DOI] [PubMed] [Google Scholar]
- 30.Ferris PJ, Armbrust EV, Goodenough UW. 2002. Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics 160:181–200 http://www.genetics.org/content/160/1/181.full.pdf+html [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishimura Y, Shikanai T, Nakamura S, Kawai-Yamada M, Uchimiya H. 2012. Gsp1 triggers the sexual developmental program including inheritance of chloroplast DNA and mitochondrial DNA in Chlamydomonas reinhardtii. Plant Cell 24:2401–2414. 10.1105/tpc.112.097865 [DOI] [PMC free article] [PubMed] [Google Scholar]