Abstract
Chlamydomonas reinhardtii is a model alga for studying triacylglycerol (TAG) accumulation in the photosynthetic production of biofuel. Previous studies were conducted under photoheterotrophic growth conditions in medium supplemented with acetate and/or ammonium. We wanted to demonstrate TAG accumulation under truly photoautotrophic conditions without reduced elements. We first reidentified all lipid components and fatty acids by mass spectrometry, because the currently used identification knowledge relies on data obtained in the 1980s. Accordingly, various isomers of fatty acids, which are potentially useful in tracing the flow of fatty acids leading to the accumulation of TAG, were detected. In strain CC1010 grown under photoautotrophic conditions, TAG accumulated to about 57.5 mol% of total lipids on a mole fatty acid basis after the transfer to nitrogen-deficient conditions. The content of monogalactosyl diacylglycerol, sulfoquinovosyl diacylglycerol, and phosphatidylglycerol decreased drastically. The accumulated TAG contained 16:0 as the major acid and 16:4(4,7,10,13), 18:2(9,12), and 18:3(9,12,15), which are typically found in chloroplast lipids. Additionally, 18:1(11) and 18:3(5,9,12), which are specific to extrachloroplast lipids, were also abundant in the accumulated TAG. Photosynthesis and respiration slowed markedly after the shift to nitrogen-deficient conditions. These results suggest that fatty acids for the production of TAG were supplied not only from chloroplast lipids but also from other membranes within the cells, although the possibility of de novo synthesis cannot be excluded. Under nitrogen-replete conditions, supplementation with a high concentration of CO2 promoted TAG production in the cells grown photoautotrophically, opening up the possibility to the continuous production of TAG using CO2 produced by industry.
INTRODUCTION
Photosynthetic production of various forms of reduced carbon is the ultimate source of biological free energy, which enables biological, social, and industrial human activities. Agricultural production has been the major source of human nutrition, but biofuel is now attracting social interest as an alternative to fossil resources. Algal production of bioresources is a new perspective as a means to supply both biofuel and chemical materials, avoiding competition with nutritional resources (1). Among various algal products, triacylglycerol (TAG), hydrocarbons, and carbohydrates are the major targets of bioengineering. In many algae, TAG is known to accumulate under conditions of nutrient deficiency (2). A unicellular green alga, Chlamydomonas reinhardtii, is currently studied as a model organism for elucidating the metabolic pathway and signaling pathway of TAG accumulation. This alga has been used in studies of photosynthesis, reproduction, and motility, mainly because the methodology for genetic analysis is applicable (3, 4). In addition to classical crossing, molecular genetic manipulation (5, 6) is becoming useful, especially on the basis of the genomic information (7). Various genes for the synthesis of lipids have also been identified (8).
Despite the increasing number of tools for molecular biological analysis, one of our concerns was that the chemical analysis of the lipids in C. reinhardtii is outdated. The chemical compositional data on the lipid classes and fatty acids date from the 1980s, when Eichenberger's group (9, 10) and one of us (11, 12) identified the major lipid and fatty acid components. At that time, the presence of various C16 and C18 fatty acids was reported, but the double bond position of the fatty acids was not rigorously established (9, 11, 12), except for commonly occurring components. The chemical structure (13, 14) and biosynthesis (11, 13) of the strange lipid diacylglyceryl-N,N,N-trimethylhomoserine (DGTS) have been determined, and the genes involved in the synthesis of DGTS were later identified (8, 15). An unusual fatty acid, Δ5,9,12-octadecatrienoic acid, was first reported by Eichenberger's group (9) and then confirmed by one of us (11, 12), but the desaturation sequence of this fatty acid remained unclear. In addition, the structures of many fatty acids were determined using the technology available at that time. Therefore, we felt the need to thoroughly reidentify all of the lipid components of C. reinhardtii using the currently available tools, and this analysis must be the first step in analyzing the biosynthetic pathway and metabolic flow of lipids in Chlamydomonas.
Another concern pertains to the strains and growth conditions of the organism. Laboratory strains of C. reinhardtii originate from three lines, namely, the Sager line, the Cambridge line, and the Ebersold/Levine 137c line (16). The strains of the Ebersold/Levine 137c line have a mutation in the nitrate reductase gene and are not able to use nitrate as the sole nitrogen source for growth (17); however, these strains are used for most studies on TAG accumulation in C. reinhardtii, in which TAG production is evaluated under photoheterotrophic conditions (18–22) using Tris-acetate-phosphate (TAP) medium containing ammonium and acetate. In our view, ammonium and acetate are not suitable nutrients for the study of biofuel production because the essential objective for biofuel production is to produce reduced carbon by photosynthesis from carbon dioxide (CO2). Acetate is a reduced form of carbon, and in fact, radioactive acetate has been used as a substrate for the study of fatty acid synthesis (23). Ammonium ion is a highly reduced molecule that should be synthesized by consuming a large amount of reducing equivalent, which could be spared and used for the reduction of CO2 and other compounds within the cell if it is added exogenously. Acetate has been reported to promote TAG production in C. reinhardtii cw15, a strain of the Ebersold/Levine 137c line (18, 24). Theoretically, the net production of biofuel will have to rely on the net reduction of carbon by photosynthesis, because no other way of obtaining reduced carbon from the biosphere exists (25).
In the present study, we focused on the photosynthetic production of TAG from inorganic, nonreduced nutrients. For this purpose, we used C. reinhardtii strain CC1010, a strain of the Cambridge line, which is able to grow photoautotrophically using nitrate as a nitrogen source. First, we reidentified all of the lipid components, including lipid classes and fatty acids. Then, we analyzed the composition of lipids and fatty acids in this strain under normal and nitrogen-deficient conditions. We also monitored the activity of photosynthesis and respiration under these conditions. The effects of the supplementation of CO2 on TAG accumulation were examined under completely photoautotrophic conditions to show that TAG accumulation can be driven by photoautotrophic growth.
MATERIALS AND METHODS
Algal strain.
The green alga Chlamydomonas reinhardtii strain CC1010 was obtained from the Chlamydomonas Resource Center (St. Paul, MN).
Growth conditions.
The algal cells were grown at 25°C in flat culture flasks (capacity, 500 ml with air space) containing 500 ml of modified Bristol's medium (MBM) (26) with continuous aeration by 1% CO2 in air. In some experiments, aeration was provided by either 5% CO2 in air or ordinary air. Light (45 μmol m−2 s−1) was provided by a bank of white fluorescent tubes (color temperature, 6,700 K; model FL20S FR P; Panasonic, Osaka, Japan). MBM contained, per liter, 250 mg KNO3, 7.5 mg MgSO4·7H2O, 25 mg NaCl, 7.5 mg K2HPO4, 175 mg KH2PO4, 13 mg Ca(NO3)·4H2O, 2 ml Fe stock solution, and 1 ml A5 solution (the pH was adjusted to 6.5 with 1 N NaOH). The Fe stock solution contained 0.5 g liter−1 FeSO4·7H2O, whereas the A5 solution contained, per liter, 2.8 g H3BO3, 2.5 g MnSO4·7H2O, 0.2 g ZnSO4·7H2O, 0.07 g CuSO4·5H2O, and 0.02 g Na2MoO4·2H2O. KNO3 was not added in the nitrogen-depleted medium, which we designated MBM-noN; it included 10 mg liter−1 CaCl2·2H2O instead of Ca(NO3)·4H2O.
Counting of cell numbers.
To 1 ml of the culture was added 10 μl of 25% glutaraldehyde solution to fix the cells. Cell numbers were counted in a Fuchs-Rosenthal hemocytometer (Kayagaki, Tokyo, Japan). Experiments were performed in duplicate. Measurements were performed in duplicate for each experiment.
Nile red staining.
To 1 ml of the culture was added 10 μl of 25% glutaraldehyde solution to fix the cells. To 4 μl of the fixed cell suspension was added 4 μl of TAN buffer (0.5 M sucrose, 20 mM Tris-HCl [pH 7.5], 1.2 mM spermidine, 0.5 mM EDTA disodium salt (EDTA), 7 mM 2-mercaptoethanol, 0.4 mM phenylmethylsulfonyl fluoride [PMSF]) containing 1% glutaraldehyde, 1 μl of TAN buffer containing 0.05 mg ml−1 Nile red, and 4 μl of TAN buffer containing 1 μg ml−1 4′,6-diamino-2-phenylindole (DAPI). The stained cells were examined with a BX60 fluorescence microscope (Olympus, Tokyo, Japan). The fluorescence of the Nile red bound to nonpolar lipid was detected with a WIG filter unit (excitation, 520 to 550 nm; dichroic mirror, 565 nm; emission, >580 nm). The fluorescence of DAPI bound to DNA and the autofluorescence of chlorophyll were detected with a WU filter unit (excitation, 330 to 380 nm; dichroic mirror, 400 nm; emission, >420 nm). Under the settings of the present study, the fluorescence of chlorophyll was removed substantially with a WIG filter unit, while the fluorescence of Nile red did not interfere with the observation of the fluorescence of DAPI and chlorophyll.
Measurements of photosynthetic oxygen evolution, respiratory oxygen consumption, and chlorophyll content.
Photosynthetic oxygen evolution and respiratory oxygen consumption were determined using a Clark-type oxygen electrode (OXY1; Hansatech, Norfolk, United Kingdom). A 2-ml aliquot was removed from the culture and transferred to the measuring chamber. Cells were kept in the dark at 25°C for 10 min, and then the oxygen consumption rate (A) was recorded for 2 min. Next, the cells were illuminated by saturating light of 10,000 μmol m−2 s−1 at 25°C for 2 min, and the oxygen evolution rate (B) was recorded. Photosynthetic oxygen evolution was calculated as (A + B). For the determination of the chlorophyll content in the cells of CC1010, a 200-μl aliquot of the culture was mixed with 800 μl of acetone. The mixture was kept on ice for 30 min to extract chlorophyll and then centrifuged at 4°C for 2 min at 15,000 × g. The absorbance values were measured at 710, 663, and 645 nm using a spectrophotometer (model UV-160A; Shimadzu, Kyoto, Japan). Total chlorophyll was determined as described by Porra et al. (27). Experiments were performed in duplicate. Measurements were performed in duplicate for each experiment.
PAM fluorescence measurement.
Pulse amplitude modulation (PAM) fluorescence measurement was performed using a fluorescence monitoring system (model FMS1; Hansatech). A 2-ml aliquot of culture was transferred to the measuring chamber. Cells were kept in the dark at 25°C for 5 min. A standard sequence of measurement for algal cells was used (28). The intensity of the actinic light was 90 μmol m−2 s−1. The intensity of the 0.2-s saturating pulse was 10,000 μmol m−2 s−1. The standard parameters Fv, Fv′, qP, qN, ϕII, and NPQ were calculated as described by Ishikawa et al. (28). Experiments were performed in duplicate, and measurements were performed in duplicate for each experiment.
Extraction of lipids.
The algal cells were harvested by centrifugation (3,000 × g, 10 min, 4°C) at a density of 2 × 106 to 5 × 106 cells ml−1 (about 40 ml). Total lipids were extracted by the method of Bligh and Dyer (29). The chloroform phase was evaporated under vacuum. The lipids were dissolved in 0.4 ml of chloroform-methanol (2:1, by volume) and stored at −20°C until use.
Separation of lipids.
Lipid classes were separated by two-dimensional (2D) thin-layer chromatography (TLC) as described by Sato and Furuya (30). Lipid solution (180 μl) was spotted on a silica gel plate (20 cm by 20 cm; TLC silica gel 60 plate; catalog number 5721; Merck, Darmstadt, Germany) at a position 25 mm by 25 mm from one corner. The first dimension was developed with acetone-benzene-methanol-water (8:3:2:1, by volume) to the top of the plate. After drying for 30 min, the second dimension was developed with chloroform-acetone-methanol-acetic acid-water (10:4:2:3:1, by volume) until the solvent front reached a height of 12.5 cm. After drying again, the plate was further developed in the second dimension with n-hexane–diethyl ether–acetic acid (80:30:1, by volume) to the top of the plate. After drying, the plate was sprayed with 0.01% primuline in 80% aqueous acetone. Lipid spots were detected under ultraviolet light at 365 nm.
Identification of lipid classes.
Each lipid class was scraped off the plate and extracted with chloroform-methanol (2:1, by volume). After filtration with a filter paper, the extract was washed with 1/4 volume of phosphate-buffered saline, and then, the chloroform phase was evaporated to dryness. The lipid was dissolved in chloroform-methanol (2:1, by volume). The lipid solution (5 μl) was mixed with an equal volume of 10 mM 2,5-dihydroxybenzoic acid in acetone. One microliter of this mixture was spotted on a sample plate and allowed to evaporate. Mass spectra were recorded in an AXIMA-CFR Plus matrix-assisted laser desorption ionization (MALDI)–time of flight (TOF) mass spectrometer (Shimadzu) at an irradiation intensity corresponding to the setting at 120 to 125.
Preparation of FAMEs.
Each lipid class was scraped off the plate with a razor blade and transferred to a Pyrex glass tube with a screw cap. Then, 2 ml of 2.5% HCl in anhydrous methanol (Kanto Kagaku, Ltd., Tokyo, Japan) was added. The tube was placed in a heating block at 85°C for 2.5 h. After cooling, fatty acid methyl esters (FAMEs) were extracted four times with 2 ml n-hexane. For the final extraction, 1 ml water was added to achieve complete extraction of the FAMEs into the n-hexane phase. The solvent was removed under vacuum, and the resultant FAME was dissolved in a small volume of n-hexane and stored at −20°C until analysis. Pentadecanoic acid (15:0) was added before the methanolysis as an internal standard for quantification of fatty acid methyl esters by gas chromatography (GC).
Preparation of fatty acid pyrrolidides.
An aliquot of FAME solution was evaporated to dryness and then dissolved in 20 μl of pyrrolidine. After adding 2 μl of acetic acid, the mixture was placed in a heating block at 100°C for 30 min. The reaction mixture was directly analyzed by GC (31).
Preparation of trimethylsilylated hydroxy derivatives.
An aliquot of FAME solution was evaporated to dryness and then dissolved in 500 μl of dioxane-pyridine (8:1, by volume). After adding 50 μl of 5% osmium tetroxide in dioxane, the mixture was kept at ambient temperature for 30 min with continuous stirring. Then, 1.25 ml of methanol and 4.25 ml of 20% aqueous sodium sulfite were added to this mixture. After standing for 1 h, the mixture was centrifuged at 750 × g for 20 min at 15°C. The supernatant was clarified by filtration through glass fiber paper (type GF/F; Whatman International Ltd., Maidstone, Kent, United Kingdom) and then evaporated under vacuum. The residue (completely hydroxylated FAME) was completely dried in a vacuum desiccator for 15 min. The final residue was extracted with diethyl ether and then with methanol. Each of the extracts was dried under vacuum. Trimethylsilylation was performed by adding 20 μl of the reagent TMS-BA (Tokyo Kasei, Ltd., Tokyo, Japan), and the mixture was kept at 80°C for 10 min (32–34).
GC.
FAMEs were analyzed with a gas chromatograph (model GC-2014; Shimadzu) equipped with a flame ionization detector and a capillary column (length, 50 m; internal diameter, 0.25 mm; ULBON HR-SS-10; Shinwa Chemical Co., Kyoto, Japan). The temperature of the column was kept at 180°C for 5 min and then elevated to 230°C at a rate of 3°C min−1. The flow rate of the carrier gas (nitrogen) was 1 ml min−1. Peak areas on the gas chromatogram were used to calculate the relative molar amounts of fatty acids. Absolute amounts were calculated using the internal standard 15:0. The amounts of lipid classes were determined on the basis of the amounts of fatty acids.
GC-mass spectrometry (MS).
FAMEs, fatty acid pyrrolidides, and trimethylated derivatives of hydroxyl fatty acids were analyzed with a gas chromatograph-mass spectrometer (model GCMS-QP2010 Ultra; Shimadzu). High-grade pure helium (He) was used as the carrier gas. The ionization voltage was 70 eV, and the ionization temperature was 200°C. Mass spectra were scanned every 0.2 s.
For the analysis of the FAMEs, a BPX70 column (length,60 m; internal diameter, 0.22 mm; SGE Analytical Science, Victoria, Australia) was used. The column temperature was elevated from 170°C to 250°C at a rate of 3°C min−1 and then kept at 250°C for 5 min. The flow rate of the helium carrier gas was 0.85 ml min−1.
Fatty acid pyrrolidides were also analyzed using the BPX70 column. The column temperature was first kept at 200°C for 1 min, elevated to 250°C at a rate of 10°C min−1, kept at 250°C for 25 min, elevated to 260°C at a rate of 2°C min−1, and finally, kept at 260°C for 5 min. The flow rate of the He carrier was 0.84 ml min−1.
Trimethylsilylated derivatives of hydroxylated fatty acids were analyzed with a nonpolar column (length, 30 m; internal diameter, 0.25 mm; Rtx-5MS; Restek, Bellefonte, PA). The column temperature was initially kept at 150°C for 1 min, elevated to 300°C at a rate of 7°C min−1, kept at 300°C for 10 min, elevated to 330°C at a rate of 7°C min−1, and finally, kept at 330°C for 5 min. The flow rate of the He gas was 1.40 ml min−1.
All of these analytical procedures and typical data are available in Document S1 in the supplemental material.
RESULTS
Critical identification of lipids and fatty acids in C. reinhardtii CC1010.
The total lipid of C. reinhardtii CC1010 grown on MBM was separated by 2D-TLC. Phosphatidylinositol (PI), phosphatidylethanolamine (PE), diacylglyceryl-N,N,N-trimethylhomoserine (DGTS), phosphatidylglycerol (PG), digalactosyl diacylglycerol (DGDG), sulfoquinovosyl diacylglycerol (SQDG), and monogalactosyl diacylglycerol (MGDG) were detected as in the case of C. reinhardtii 137c (9, 10) and UTEX89 (the mating type-minus variant of UTEX90 is CC1010) (12). In addition, triacylglycerol (TAG) was detected under the conditions tested. Each lipid class was recovered from the plate and confirmed by MALDI-TOF MS. Phosphatidic acid (PA) was detected as a very minor spot. Other spots that were not further analyzed included putative wax esters and free fatty acids.
The lipid classes were converted to FAMEs. For critical identification of fatty acids in C. reinhardtii CC1010, fatty acid pyrrolidides and trimethylsilylated hydroxyl derivatives were prepared from the FAMEs and analyzed by GC-MS. In general, the number of carbon atoms and the number of double bonds were identified with the FAMEs, whereas the positions of double bonds were estimated with the pyrrolidides. The exact position of double bonds was determined with trimethylsilylated hydroxyl derivatives (see File S1 in the supplemental material). Previously, strains 137c and UTEX89 have been reported to contain 16:1(7), 16:1(3t), and 16:1(9) (9, 12) as C16 monoenoic acids [note that a fatty acid is represented by X:Y, where X is the number of carbons and Y is the number of double bonds, and the position(s) of the double bond(s) counted from the carboxyl group is shown in parentheses]. In CC1010, 16:1(11) was also identified (see Document S1 in the supplemental material). The fatty acids 16:2(7,10), 16:3(4,7,10), 16:3(7,10,13), and 16:4(4,7,10,13) were identified in CC1010, as reported in C. reinhardtii 137c (9) and UTEX89 (12).
Previously, 137c and UTEX89 has been reported to have 18:1(9) and 18:1(11) as C18 monoenoic acids (9, 12). Now, 18:1(13) was also identified in CC1010 (see Document S1 in the supplemental material). Also, 18:2(9,12), 18:3(5,9,12), 18:3(9,12,15), and 18:4(5,9,12,15) were identified in CC1010, as in the case of 137c and UTEX89 (see Document S1 in the supplemental material) (9, 12). The fatty acid 18:3(5,9,12) was reported to be γ-linolenic acid or 18:3(6,9,12) in a previously published paper (10). In CC1010, 18:3(5,9,12) and 18:3(9,12,15) were unambiguously identified by the three different GC-MS methods. We did not detect 18:3(6,9,12) at all. However, the tetraeoic acid fraction seemed to contain two components, 18:4(5,9,12,15) and 18:4(6,9,12,15) (see Document S1 in the supplemental material). We noticed that 18:4 was described in various previous reports (35, 36), but without detailed identification, probably because they relied on the old identification (9, 12). Because double bonds are hard to identify in such fatty acids with four double bonds, we still need to define the structure carefully. The coexistence of 18:4(5,9,12,15) and 18:4(6,9,12,15) will have to be discussed from the viewpoint of biosynthesis. Longer-chain fatty acids were reported to be present in trace amounts only in C. reinhardtii 137c (9). Now, 20:1(11) and 22:1(13) have been identified in CC1010 (see Document S1 in the supplemental material). In addition, a shorter-chain fatty acid (14:0) and odd-numbered-chain fatty acids 17:0, 17:1(8), 17:1(9), 17:1(10), 17:1(11), 19:0, 19:1(10), and 19:1(13) were detected in the present study in CC1010 (see Document S1 in the supplemental material).
Cell growth and lipid droplet accumulation in photoautotrophically grown C. reinhardtii CC1010.
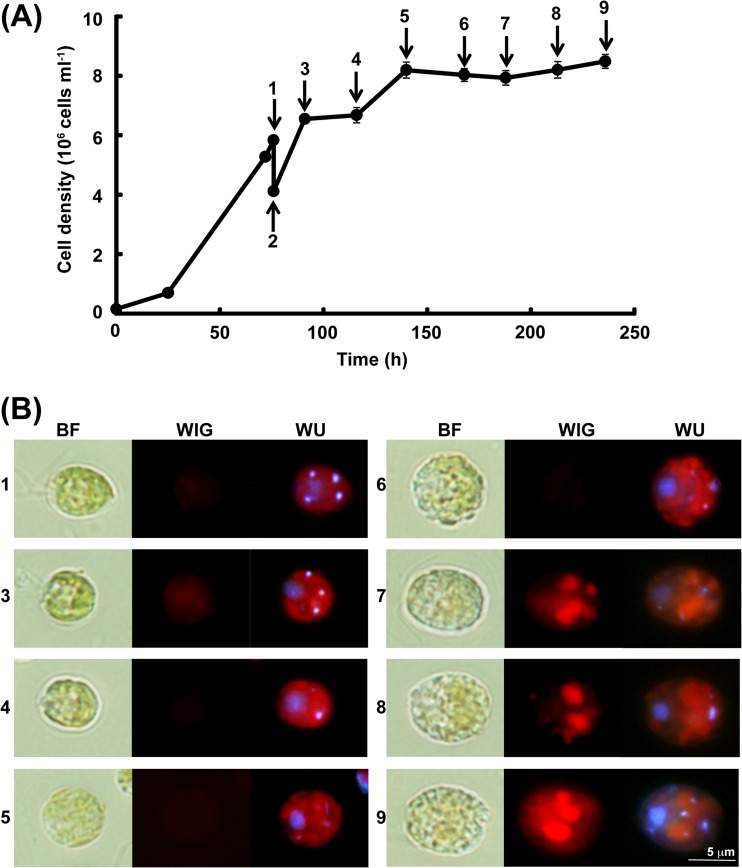
C. reinhardtii CC1010 displayed logarithmic growth when grown photoautotrophically on MBM (Fig. 1A). When the cell density of the culture reached 2 × 106 to 5 × 106 cells ml−1, which corresponded to the early logarithmic phase, the cells were harvested by centrifugation at 4°C for 10 min at 3,000 × g and washed twice with MBM-noN (MBM without a nitrogen source). Cooling was necessary to keep the cells immotile during the manipulation but was kept to a minimum time to avoid adverse effects on the cell physiology. The washed cells were resuspended in MBM-noN. During the continued growth on MBM-noN, the cell density of the culture increased by 2-fold and then reached a plateau (Fig. 1A).
FIG 1.
Growth and nonpolar lipid accumulation of Chlamydomonas reinhardtii strain CC1010 on MBM and MBM-noN. (A) Growth profiles of the strain. Each value represents the mean ± standard error. The arrows labeled 1 to 9 indicate the sampling points for the detection of a nonpolar lipid, as explained in the Results. (B) Detection of the nonpolar lipids that accumulated in the cells by Nile red staining. Panels 1 and 3 to 9 are images for the cells sampled at time points identified by arrows 1 and 3 to 9, respectively, in panel A. Images are representative of two independent cultures. BF, bright-field image; WIG, fluorescence image with WIG filter; WU, fluorescence image with WU filter. With the WIG filter, the fluorescence of Nile red bound to nonpolar lipid was detected as red spots. With the WU filter, the fluorescence of DAPI bound to DNA was detected as blue-white spots. The red color with the WU filter is the autofluorescence of chlorophyll.
Nile red staining was used to monitor the accumulation of lipid droplets consisting of nonpolar lipids such as TAG in the cells grown on MBM-noN (Fig. 1B). The cultures were sampled for Nile red staining at the following time points: prior to the transfer to MBM-noN medium, immediately after the transfer, and at 15 h, 40 h, 64 h, 92 h, 112 h, 137 h, and 160 h after the transfer. Representative images are shown. The fluorescence of Nile red (bound to nonpolar lipid) was detected as a red color with the WIG filter (Fig. 1B). The red color was faintly detected in the cells grown on MBM, indicating a low accumulation of lipid droplets (Fig. 1B, panel 1). In contrast, a marked red color was detected in the cells grown on MBM-noN, indicating that lipid droplets accumulated to a significant level (Fig. 1B, panels 7 to 9). This finding indicated that photoautotrophically grown CC1010 produces nonpolar lipid under conditions of nitrogen deficiency. Apparently, the lipid droplets colocalized with chloroplasts (Fig. 1B, panels 7 to 9), but upon electron microscopic examination, the lipid bodies were found in the cytoplasm between the nucleus and the interior of the cup-shaped chloroplast (results not shown).
Cell viability was checked by observing the cell morphology and measuring the colony-forming efficiency (CFE) (results not shown). Although the value of CFE decreased with time, as described in reference 37, the cells remained healthy, at least until sampling point 8, under the nitrogen-deficient conditions. Some cells were enclosed in a large capsule, and this could be the reason for the lowered CFE. In contrast to the growth in TAP medium without ammonium (results not shown), the cells retained a green color for a longer period of time and looked healthy.
Changes in photosynthetic oxygen evolution, respiratory oxygen consumption, and chlorophyll content after the shift to nitrogen deficiency.
Photosynthetic oxygen evolution, respiratory oxygen consumption, and the chlorophyll content in the cells of strain CC1010 were measured (Fig. 2). The sampling points were identical to those for the Nile red staining. The initial levels of photosynthetic oxygen evolution, respiratory oxygen consumption, and chlorophyll content in the cells grown on MBM-noN were similar to or slightly lower than the respective values for the cells grown on MBM. In the cells grown on MBM-noN, the levels of photosynthetic oxygen evolution and respiratory oxygen consumption decreased markedly within the first 12 h with respect to the initial levels (Fig. 2A to C). The chlorophyll content decreased gradually in a time-dependent manner but did not drop drastically (Fig. 2D). These results suggest that the activities of both photosynthesis and respiration significantly decreased following the removal of combined nitrogen.
FIG 2.
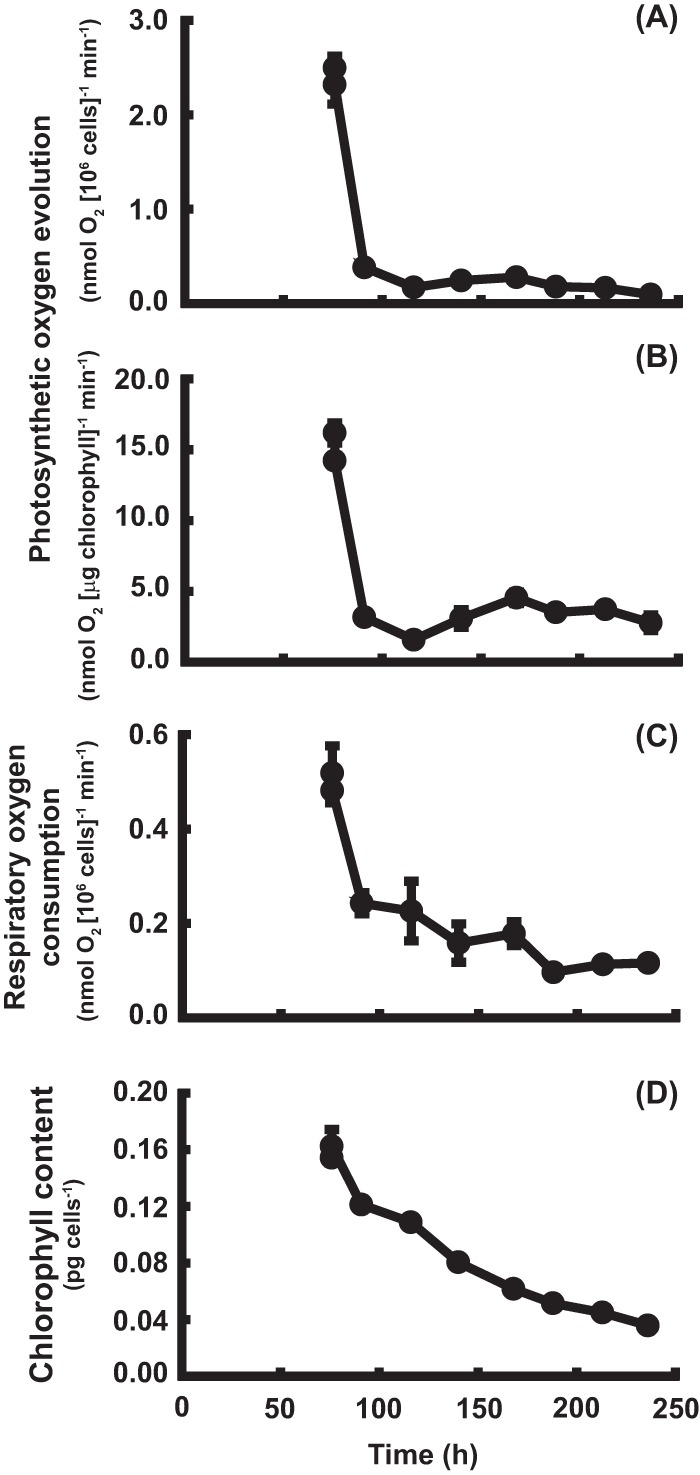
Photosynthetic oxygen evolution, respiratory oxygen consumption, and chlorophyll content of Chlamydomonas reinhardtii strain CC1010 on MBM and MBM-noN. (A, B) Photosynthetic oxygen evolution; (C) respiratory oxygen consumption; (D) chlorophyll content. Each value represents the mean ± standard error.
Changes in photosynthetic performance after the shift to nitrogen deficiency.
PAM fluorescence measurement was performed to analyze the photosynthetic performance of the cells after the transfer to nitrogen-depleted medium. The values of Fv/Fm and Fv′/Fm′ decreased slightly upon transfer to MBN-noN and then remained at high levels during the 24-h period, indicating that photosystem II (PSII) was operative (Fig. 3). The initial rise in qN and NPQ corresponds to the initial decrease in Fv/Fm and Fv′/Fm′ (Fig. 3). The values of qN and NPQ decreased later. In contrast, the values of ϕII and qP decreased significantly 6 h after the transfer to MBM-noN, indicating that electron transport was disturbed by nitrogen deficiency near PSII and between PSII and PSI (Fig. 3).
FIG 3.
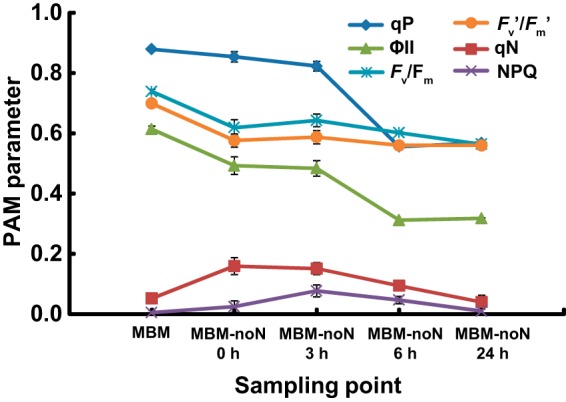
Changes in photosynthetic parameters obtained by pulse amplitude modulation (PAM) fluorescence measurement in the cells of Chlamydomonas reinhardtii strain CC1010 grown on MBM or MBM-noN. Each value represents the mean ± standard error.
Accumulation of TAG in photoautotrophically grown C. reinhardtii CC1010.
Aliquots of the culture were collected prior to the transfer to the MBM-noN condition and at 137 h after the transfer to determine the contents of lipids in CC1010 cells grown on MBM or MBM-noN (Fig. 4 and Table 1). The experiments were performed using four biological replicates for each growth condition. The content of TAG was 57.5 mol% of the total lipids in the cells grown on MBM-noN. It was significantly higher than the content in the cells grown on MBM (P < 0.05). In contrast, the contents of PI, PG, SQDG, DGDG, and MGDG in the cells grown on MBM-noN were lower than the respective contents of the cells grown on MBM at a confidence level of 95% (Table 1). This was consistent with the high accumulation of lipid droplets in the cells grown on MBM-noN (Fig. 1B, panels 7 to 9).
FIG 4.
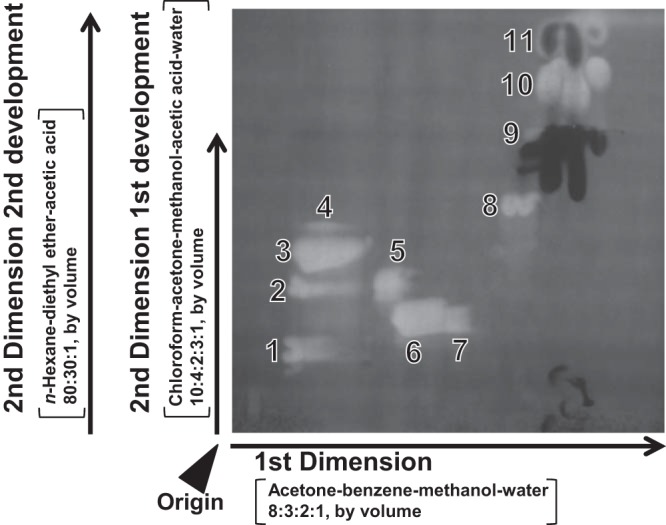
Fractionation of total lipids from cells of Chlamydomonas reinhardtii strain CC1010 grown on MBM-noN by 2D-TLC. Bright fluorescent spots indicate lipids. Dark spots indicate pigments. Data are representative of four independent experiments. 1, phosphatidylinositol (PI); 2, phosphatidylethanolamine (PE); 3, diacylglyceryltrimetylhomoserine (DGTS); 4, phosphatidic acid (PA); 5, phosphatidylglycerol (PG); 6, digalactosyl diacylglycerol (DGDG); 7, sulfoquinovosyl diacylglycerol (SQDG); 8, monogalactosyl diacylglycerol (MGDG); 9, free fatty acids 10, triacylglycerol (TAG); 11, putative waxes.
TABLE 1.
Composition of lipids in cells of Chlamydomonas reinhardtii strain CC1010 grown on MBM or MBM-noN
| Lipid class | Compositionc (mol%) |
|
|---|---|---|
| MBM | MBM-noN | |
| PI | 2.7 ± 0.2 | 1.7 ± 0.3a |
| PE | 4.5 ± 0.4 | 2.9 ± 0.6 |
| DGTS | 14.7 ± 0.6 | 13.7 ± 4.1 |
| PG | 10.7 ± 0.1 | 2.1 ± 0.6a |
| DGDG | 16.0 ± 0.6 | 12.2 ± 1.5a |
| SQDG | 8.6 ± 0.3 | 1.4 ± 0.1a |
| MGDG | 41.3 ± 1.7 | 8.7 ± 1.5a |
| TAG | 1.6 ± 0.4 | 57.5 ± 8.6b |
| Total | 100 | 100 |
Lipid classes that showed decreases in composition on MBM-noN at a confidence level of 95%.
Lipid classes that showed increases in composition on MBM-noN at a confidence level of 95%.
Each value represents the mean of the results obtained from four independent experiments. Composition is indicated as molar percentages of component fatty acids. The contents of total lipids (expressed as amounts of component fatty acids) in CC1010 cells grown on MBM and MBM-noN were 3.55 ± 0.48 pmol cell−1 and 5.12 ± 1.05 pmol cell−1, respectively.
Composition of fatty acids in cells of C. reinhardtii CC1010 grown on MBM and MBM-noN.
The compositions of fatty acids in the cells of C. reinhardtii CC1010 grown on MBM and MBM-noN are shown in Tables 2 and 3, respectively. The specific distribution of fatty acids among lipid classes was found as reported previously (9, 12). The 16:4 fatty acid was concentrated in MGDG, the 18:2 fatty acid was typical for DGDG and PG, the 18:1(11) fatty acid was detected mainly in PI, the 18:3(9,12,15) fatty acid was localized exclusively in MGDG and DGDG, and the 18:3(5,9,12) fatty acid was specific for DGTS and PE. In the current study, we determined that the double bond positions of 16:4 isolated from MGDG are delta 4, 7, 10, and 13 (Table 2). The double bond positions of 18:2, which was isolated from DGDG and PG, were confirmed to be delta 9 and 12 (Table 2). Double bond positions of 18:3(9,12,15) and 18:3(5,9,12), which were isolated from MGDG, DGDG, DGTS, and PE, were confirmed as such (Table 2). In the TAG of C. reinhardtii CC1010 grown on MBM-noN, the contents of 16:4(4,7,10,13), 18:1(11), 18:2(9,12), 18:3(5,9,12), and 18:3(9,12,15) fatty acids increased with respect to the respective values in CC1010 grown on MBM at a confidence level of 95% (Table 3). These results suggest that various different classes of lipids provide fatty acids for the synthesis of TAG.
TABLE 2.
Composition of fatty acids in cells of Chlamydomonas reinhardtii strain CC1010 grown on MBM
| Fatty acid | Contenta (mol%) |
|||||||
|---|---|---|---|---|---|---|---|---|
| PI | PE | DGTS | PG | DGDG | SQDG | MGDG | TAG | |
| 14:0 | 0.6 ± 0.0 | 0.6 ± 0.1 | 0.8 ± 0.0 | 0.2 ± 0.1 | 0.5 ± 0.0 | 1.3 ± 0.2 | 0.1 ± 0.0 | 2.7 ± 0.4 |
| 16: 0 | 59.8 ± 0.8 | 4.7 ± 0.1 | 40.9 ± 0.4 | 36.4 ± 1.2 | 37.5 ± 0.8 | 81.5 ± 0.7 | 2.0 ± 0.2 | 49.6 ± 1.7 |
| 16:1(7) | 0.6 ± 0.3 | 0.8 ± 0.3 | 0.7 ± 0.1 | 0.0 ± 0.0 | 1.4 ± 0.3 | 0.6 ± 0.0 | 1.2 ± 0.1 | 2.4 ± 0.3 |
| 16:1(3t) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 27.4 ± 0.5 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 16:1(9) | 0.1 ± 0.1 | 0.2 ± 0.1 | 0.1 ± 0.0 | 0.9 ± 0.2 | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.2 ± 0.1 | 0.3 ± 0.1 |
| 16:1(11) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.2 ± 0.0 | 0.1 ± 0.1 | 0.1 ± 0.0 | 0.2 ± 0.0 | 0.0 ± 0.0 | 0.7 ± 0.1 |
| 16:2(7,10) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.4 ± 0.2 | 0.0 ± 0.0 | 4.9 ± 0.0 | 0.4 ± 0.1 | 2.6 ± 0.2 | 0.9 ± 0.1 |
| 16:3(4,7,10) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.0 | 0.0 ± 0.0 | 0.2 ± 0.0 | 0.4 ± 0.1 | 2.5 ± 0.1 | 0.6 ± 0.1 |
| 16:3(7,10,13) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.6 ± 0.0 | 0.0 ± 0.0 | 6.7 ± 0.2 | 0.6 ± 0.1 | 4.3 ± 0.1 | 1.0 ± 0.1 |
| 16:4(4,7,10,13) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.8 ± 0.1 | 0.0 ± 0.0 | 0.9 ± 0.1 | 1.9 ± 0.5 | 34.4 ± 0.4 | 2.5 ± 0.2 |
| 17:0 | 0.1 ± 0.1 | 0.6 ± 0.0 | 0.6 ± 0.1 | 0.1 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 1.0 ± 0.3 |
| 18:0 | 1.4 ± 0.1 | 40.1 ± 0.5 | 2.1 ± 0.6 | 0.5 ± 0.0 | 0.4 ± 0.0 | 0.7 ± 0.1 | 0.0 ± 0.0 | 3.0 ± 0.2 |
| 18:1(9) | 0.0 ± 0.0 | 0.9 ± 0.1 | 1.9 ± 0.0 | 4.6 ± 0.6 | 9.4 ± 1.2 | 1.9 ± 0.2 | 2.3 ± 0.6 | 9.0 ± 0.1 |
| 18:1(11) | 35.7 ± 0.5 | 7.6 ± 0.2 | 2.8 ± 0.1 | 5.8 ± 0.1 | 1.9 ± 0.1 | 1.5 ± 0.1 | 0.5 ± 0.2 | 4.0 ± 0.3 |
| 18:2(9,12) | 1.1 ± 0.1 | 3.1 ± 0.2 | 7.7 ± 0.2 | 16.5 ± 0.4 | 18.0 ± 1.0 | 2.8 ± 0.1 | 7.7 ± 0.5 | 8.4 ± 0.4 |
| 18:3(5,9,12) | 0.2 ± 0.1 | 38.2 ± 0.3 | 24.0 ± 0.1 | 0.1 ± 0.1 | 0.7 ± 0.0 | 0.4 ± 0.0 | 0.7 ± 0.0 | 5.5 ± 0.2 |
| 18:3(9,12,15) | 0.4 ± 0.0 | 0.4 ± 0.0 | 5.6 ± 0.3 | 7.4 ± 0.4 | 16.7 ± 0.3 | 4.7 ± 0.4 | 40.5 ± 0.2 | 6.4 ± 0.3 |
| 18:4(5,9,12,15) and (6,9,12,15) | 0.1 ± 0.0 | 2.8 ± 0.2 | 10.7 ± 0.6 | 0.0 ± 0.0 | 0.3 ± 0.0 | 0.5 ± 0.0 | 0.7 ± 0.0 | 2.1 ± 0.2 |
Each value represents the mean ± standard error of the results obtained from four independent experiments. Although 16:1(7) and 16:1(3t) were not completely separated in the GC analysis, only the latter was assigned to PG on the basis of the results of GC-MS.
TABLE 3.
Composition of fatty acids in cells of Chlamydomonas reinhardtii strain CC1010 grown for 6 days in MBM-noN
| Fatty acid | Contentc (mol%) |
|||||||
|---|---|---|---|---|---|---|---|---|
| PI | PE | DGTS | PG | DGDG | SQDG | MGDG | TAG | |
| 14:0 | 0.4 ± 0.1 | 0.4 ± 0.1a | 0.8 ± 0.0a | 0.6 ± 0.1b | 0.6 ± 0.0b | 1.2 ± 0.2 | 0.1 ± 0.0 | 0.7 ± 0.0a |
| 16: 0 | 62.5 ± 1.0b | 8.2 ± 0.5b | 48.4 ± 0.4b | 56.1 ± 0.7b | 61.1 ± 2.5b | 90.9 ± 2.5b | 2.0 ± 0.1 | 36.4 ± 0.4a |
| 16:1(7) | 0.2 ± 0.1 | 0.1 ± 0.1 | 0.6 ± 0.1 | 0.0 ± 0.0 | 0.5 ± 0.1a | 0.6 ± 0.1 | 0.6 ± 0.1a | 1.0 ± 0.0a |
| 16:1(3t) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 15.6 ± 0.5a | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 16:1(9) | 0.1 ± 0.1 | 0.3 ± 0.1b | 0.1 ± 0.0 | 2.5 ± 1.3 | 0.2 ± 0.0 | 0.2 ± 0.1 | 0.3 ± 0.0 | 0.1 ± 0.1 |
| 16:1(11) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.0a | 0.3 ± 0.1 | 0.0 ± 0.0a | 0.1 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.0a |
| 16:2(7,10) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.7 ± 0.4 | 0.0 ± 0.0 | 2.5 ± 0.2a | 0.5 ± 0.3 | 2.9 ± 0.7 | 1.7 ± 0.3b |
| 16:3(4,7,10) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.2 ± 0.1b | 0.0 ± 0.0 | 0.1 ± 0.0 | 0.0 ± 0.0a | 2.3 ± 0.1a | 0.4 ± 0.0 |
| 16:3(7,10,13) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.4 ± 0.2 | 0.0 ± 0.0 | 3.3 ± 0.1a | 0.6 ± 0.3 | 4.4 ± 0.6 | 2.4 ± 0.1b |
| 16:4(4,7,10,13) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.9 ± 0.4 | 0.0 ± 0.0 | 0.6 ± 0.1 | 0.1 ± 0.1a | 35.1 ± 1.1 | 6.5 ± 0.8b |
| 17:0 | 0.1 ± 0.1 | 0.3 ± 0.0a | 0.2 ± 0.1a | 0.2 ± 0.0b | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0a |
| 18:0 | 2.3 ± 0.3b | 48.6 ± 0.3b | 0.9 ± 0.1 | 2.2 ± 0.2b | 0.7 ± 0.1b | 0.6 ± 0.1 | 0.1 ± 0.1 | 2.7 ± 0.2a |
| 18:1(9) | 0.0 ± 0.0 | 0.1 ± 0.1a | 0.3 ± 0.0a | 2.1 ± 0.1a | 2.3 ± 0.2a | 0.5 ± 0.2a | 1.1 ± 0.2a | 7.4 ± 1.3 |
| 18:1(11) | 33.6 ± 0.8a | 4.3 ± 0.4a | 1.7 ± 0.3a | 9.1 ± 0.7b | 1.8 ± 0.2 | 1.4 ± 0.4 | 0.3 ± 0.1 | 5.2 ± 0.3b |
| 18:2(9,12) | 0.4 ± 0.1a | 1.2 ± 0.3a | 7.5 ± 0.6 | 8.4 ± 0.8a | 10.6 ± 1.7a | 1.2 ± 0.5a | 8.3 ± 0.4 | 11.7 ± 1.1b |
| 18:3(5,9,12) | 0.1 ± 0.0 | 35.5 ± 0.3a | 23.5 ± 0.6 | 0.2 ± 0.0 | 1.0 ± 0.2b | 0.2 ± 0.1 | 0.7 ± 0.2 | 9.4 ± 1.0b |
| 18:3(9,12,15) | 0.1 ± 0.1a | 0.3 ± 0.1 | 8.1 ± 1.1 | 2.6 ± 0.2a | 14.5 ± 1.1a | 1.8 ± 0.6a | 41.3 ± 0.4 | 11.8 ± 0.7b |
| 18:4(5,9,12,15) and (6,9,12,15) | 0.0 ± 0.0 | 0.7 ± 0.1a | 5.5 ± 0.5a | 0.0 ± 0.0 | 0.2 ± 0.0a | 0.1 ± 0.1a | 0.3 ± 0.1a | 2.4 ± 0.4 |
Fatty acids that showed a decrease in composition in MBM-noN at a confidence of 95%.
Fatty acids that showed an increase in composition in MBM-noN at a confidence of 95%.
Each value is the mean ± standard error of the results obtained from four independent experiments.
Effects of supplementation of CO2 on accumulation of TAG.
The TAG contents in the cells of C. reinhardtii CC1010 grown with air, 1% CO2, and 5% CO2 were compared to examine the effects of supplementation of CO2 on TAG productivity (Fig. 5). The content of TAG was higher in the cells grown with 5% CO2 than in those grown in air or 1% CO2 (Fig. 5A). This result indicated that supplementation of CO2 can promote TAG accumulation in the cells grown photosynthetically under nitrogen-replete conditions. In the cells grown on MBM-noN, the difference in the TAG content was not significant between the cells grown with air or 1% CO2 (Fig. 5B). The TAG content was, however, significantly lower in the cells grown with 5% CO2 (P < 0.05) (Fig. 5B).
FIG 5.
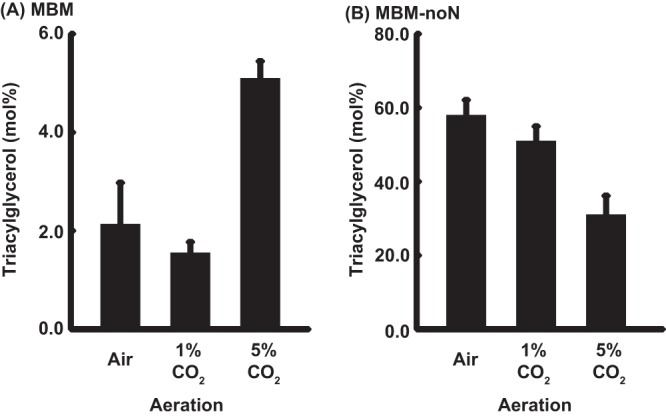
Triacylglycerol content as the percentage of total lipids on a fatty acid basis in the cells of Chlamydomonas reinhardtii strain CC1010 grown with air, 1% CO2, or 5% CO2. (A) MBM: (B) MBM-noN. Experiments were performed in duplicate. Measurements were performed in duplicate for each experiment. Each value represents the mean ± standard error.
We also determined the fatty acid composition of total lipids and TAG in the cells grown with different concentrations of CO2 (see Tables S1 and S2 in the supplemental material). Saturated fatty acids were more abundant in the total lipids under conditions of nitrogen deficiency at different concentrations of CO2. The levels of unsaturated fatty acids were significantly higher with a higher CO2 supply with or without a nitrogen source (P < 0.05). These results are consistent with the general tendency of the fatty acid composition reported previously (12), although TAG was not analyzed before.
DISCUSSION
Reidentification of the lipid components in C. reinhardtii.
We performed the reidentification of lipid classes and fatty acids. For lipid classes, all major spots in 2D-TLC were confirmed by TOF MS. The MS peaks were compatible with the putative molecular species composition. In our analysis, PA was not detected to a significant level. We still do not know whether cardiolipin is present in C. reinhardtii. Phosphatidylcholine was not detected, as described in previous reports (9, 10, 12).
With respect to fatty acids, we obtained several new findings. The double bond positions of the major fatty acids are, in general, consistent with those reported previously, either from our laboratory (12) or from another laboratory (9) (see Document S1 in the supplemental material). In addition, many fatty acids were identified as minor components (see Document S1 in the supplemental material).
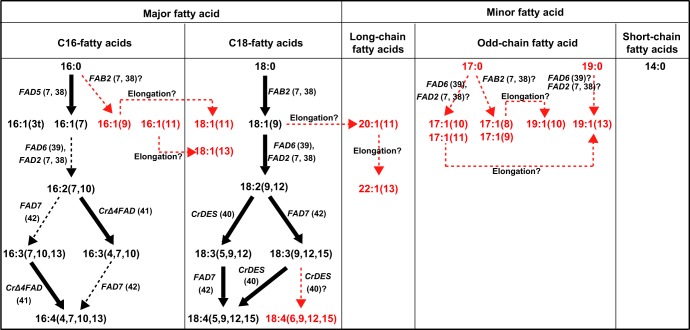
The plausible pathways of fatty acid synthesis in C. reinhardtii CC1010 are depicted in Fig. 6. The genes for the following desaturases are known: stearoyl-ACP Δ9-desaturase (encoded by FAB2) (7, 38), chloroplast Δ12-desaturase (FAD6) (39), Δ5-desaturase acting on C18 fatty acids (an endoplasmic reticulum [ER] enzyme) (40), MGDG palmitate Δ7-desaturase (FAD5) (7, 38), Δ4-desaturase acting on MGDG (chloroplast enzyme) (41), Δ15-desaturase (chloroplast enzyme, FAD7) (42), and oleate Δ12-desaturase (ER enzyme, FAD2) (7, 38). Phosphatidylglycerol palmitate Δ3t-desaturase has not been identified in any plants or algae. The biosynthesis of simple monoenoic acids, such as 16:1(9), 16:1(11), 17:1(8), and 17:1(9), is not easily explained by the known enzymes. We must assume that the enzyme specificity might be flexible; for example, the enzyme acting on C18 acids is capable of acting on C17 or C19 acids. Furthermore, 16:1(9) and 18:1(11) could be produced by the so-called anaerobic pathway of fatty acid synthesis, as in bacteria, but no such evidence has been reported in algae or plants. We believe that the detection of 18:4(6,9,12,15) in addition to 18:4(5,9,12,15) is not an artifact. They were, indeed, separated as TMS derivatives of hydroxylated FAMEs in the GC-MS analysis. The biosynthetic mechanism for 18:4(6,9,12,15), however, is a mystery because no Δ6-desaturase is known to occur in C. reinhardtii (see Document S1 in the supplemental material). A possible explanation is that the Δ5-desaturase, which originated from Δ6-desaturase, according to phylogenetic analysis (N. Sato, unpublished results), can introduce the Δ6 double bond as well, in this special case.
FIG 6.
Plausible pathways of fatty acid synthesis in Chlamydomonas reinhardtii strain CC1010. Fatty acids shown in red were first detected in the current study. Bold arrows indicate the pathways supported by previously reported genes or genes identified on the basis of genome information. References are also shown in parentheses. Hypothetical pathways to explain the results in this study are shown by red arrows. Arrows with dashed tails indicate pathways catalyzed probably by known enzymes.
Genes involved in the elongation of fatty acids have been described (7, 38). Accordingly, 19:1(10), 19:1(13), 18:1(11), 18:1(13), 20:1(11), and 22:1(13) might be produced by the elongation of 17:1(8), 17:1(11), 16:1(9), 16:1(11), 18:1(9), and 20:1(11), respectively. Among these, 20:1(11) and 22:1(13) are likely to be elongation products because no corresponding saturated acids were detected. Various pathways are possible for the synthesis of 18:1(11), but the specific localization of 18:1(11) in PI will have to be explained by a specific mechanism, such as lipid-linked desaturation. This is a new perspective because PI has not been a target of intense study, although it may be an important signaling molecule.
Accumulation of TAG by C. reinhardtii CC1010 under photoautotrophic and nitrogen-deficient conditions.
In the present study, we showed that photoautotrophically grown CC1010 could indeed accumulate TAG, which amounted to about 57.5 mol% of total lipid under nitrogen-deficient conditions (Table 1). In the current study, CC1010 was grown under completely photoautotrophic conditions. Therefore, the carbons of accumulated nonpolar lipid in the cells grown on MBM-noN were all derived from photosynthetically assimilated carbons. This feature of CC1010 is advantageous for the photosynthetic production of TAG as a renewable biofuel. In C. reinhardtii cw15 and BAFJ5 grown on TAP lacking combined nitrogen, which was used in most studies for TAG synthesis in C. reinhardtii, the content of TAG was about 50% (18). The content of TAG in the cells of CC1010 grown under photoautotrophic and nitrogen-deficient conditions was comparable to that in cw15 and BAFJ5 grown under photoheterotrophic and nitrogen-depleted conditions. The current findings indicate that the strains of the Sager and Cambridge lines grown under photoautotrophic growth conditions are suitable in studies of TAG accumulation in C. reinhardtii for the development of systems for biofuel production by algae.
The contents of TAG were about 10% or 75% in C. reinhardtii CC125 grown in high-salt medium or high-salt medium lacking nitrogen, respectively (43). The TAG content was 1.6 mol% of the total lipids in CC1010 grown in MBM (Table 1). Chlamydomonas has been reported to accumulate significant amounts of TAG under stresses of high salinity, low sulfur, low iron, and low oxygen (44–47). In CC125, nitrogen depletion and high salt may induce a high accumulation of TAG in cells grown in high-salt medium lacking nitrogen (43). In cells of C. reinhardtii CC125 grown in TAP lacking combined nitrogen, TAG is thought to be produced by the reconstruction of MGDG (18). A mutant with a mutation of the PGD1 gene encoding galactoglycerolipid lipase showed a reduced TAG content following nitrogen deprivation in C. reinhardtii dw15-1 (37). MGDG, DGDG, and SQDG are known to be chloroplast lipids (38). In cells of CC1010 grown on MBM-noN, remodeling of MGDG to TAG might be preferred over de novo biosynthesis of TAG due to the low activity of photosynthesis.
Sources of fatty acids for TAG production under photoautotrophic and nitrogen-deficient conditions.
In the present study, using C. reinhardtii CC1010, specific esterification of certain fatty acids to certain classes of lipids was evident. Remodeling of MGDG to TAG has been argued in several papers (18, 37). This is supported by the increase in 16:4(4,7,10,13), 18:2(9,12), and 18:3(9,12,15) in TAG (Tables 2 and 3). However, in addition to MGDG, increases of the contents of 18:1(11) and 18:3(5,9,12) in the TAG of CC1010 grown on MBM-noN indicated that fatty acids for the production of TAG were also supplied by DGTS, PE, and PI (Tables 2 and 3). In other words, both the chloroplast and other organelles (notably, the ER) contribute to the production of TAG under conditions of nitrogen deficiency.
However, note that the localization of individual classes of lipids was not seriously studied in Chlamydomonas. An old paper from Eichenberger's laboratory (9) simply assumed that MGDG, DGDG, and SQDG are chloroplast lipids and that DGTS and PE are ER lipids. Although the procedures for preparing intact chloroplasts were described (48), the localization of lipids in isolated membranes was not reported. The fractionation of C. reinhardtii cells is not easy, and isolating large chloroplasts is especially difficult. In addition, avoiding the folding of isolated chloroplasts around themselves or other organelles, which is often the case in isolating large chloroplasts, is also necessary. We will have to develop a suitable procedure for cell fractionation.
Effect of photosynthesis on accumulation of TAG.
Immediately after the shift to nitrogen-deficient conditions, photosynthetic oxygen evolution and respiratory oxygen consumption were markedly repressed. The decrease in chlorophyll content was retarded (Fig. 2). The slowing of photosynthesis seemed to proceed in two steps (Fig. 3). Initially, the light energy was lost as heat by NPQ, perhaps because electron transport was somehow disturbed. Then, from 6 h onward, the values of both ϕII and qP decreased. This second phase of photosynthetic repression could have been caused by the decrease in carbon assimilation, which is normally balanced with nitrogen assimilation (49, 50). The PSII activity, however, was maintained throughout the experiment.
The rapid decrease in respiration rate seemed to be specific to photoautotrophic growth conditions. Philipps et al. (51) reported that respiratory activity in TAP medium is maintained at about 1.4 nmol O2 (106 cells)−1 min−1 after an initial small drop upon the removal of combined nitrogen. The respiratory rate was about 3 times higher than the rate of CC1010 cells on MBM, namely, under normal photosynthetic condition (Fig. 3C). Following nitrogen removal, the respiration of CC1010 became about 10 times lower than the reported value in TAP. Apparently, the high rate of metabolism is maintained in TAP medium containing acetate even after the transfer to nitrogen-deficient conditions. An alternative explanation could be the difference in strains. However, we consider that the results of our experiments could be different from those of previous experiments using TAP medium containing acetate, which is known to support lipid synthesis and respiration (21).
Even though photosynthesis was reduced significantly under conditions of nitrogen deficiency, reported evidence has shown that the contribution of photosynthesis to the production of TAG under nitrogen-deficient conditions was not negligible. The herbicide 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) specifically inhibits photosynthetic electron transport at the acceptor side of PSII (52). DCMU treatment of cells of C. reinhardtii dw15-1 is known to result in a decrease in the TAG level (37), although this experiment used TAP medium.
In C. reinhardtii CC1010, we also have evidence suggesting the de novo synthesis of lipids. The content of 16:0 in PI, PE, DGTS, PG, DGDG, and SQDG was higher in cells grown on MBM-noN than in those grown on MBM. This indicates that the de novo biosynthesis of fatty acids continued under nitrogen-deficient conditions (Table 3), which provides a good reason to assume that reduced photosynthesis might still function to supply carbon for the de novo biosynthesis of fatty acids in CC1010 cells. We are now planning to demonstrate the contribution of photosynthesis through labeling experiments.
In the current study, supplementation of a high concentration of CO2 promoted TAG production in cells grown on MBM (Fig. 5A). This result was obtained under photoautotrophic conditions. C. reinhardtii can adapt to a broad range of external CO2 concentrations because of its inorganic carbon-concentrating system (53). Carbon flow through photosynthesis for TAG production might be supported by supplementation of a high concentration of CO2 under photoautotrophic conditions. In contrast, supplementation of CO2 did not promote TAG accumulation in cells grown on MBM-noN (Fig. 5B). Carbon assimilation is known to be balanced with nitrogen assimilation in cyanobacteria (49). The same is true for agricultural plants because fertilizers promote production as a whole. Nitrogen limitation in general shuts down carbon assimilation. Under nitrogen-deficient conditions with 5% CO2, the photosynthesis rate is most probably drastically limited by the balance with nitrogen metabolism, and this is the reason for lower TAG accumulation (Fig. 2). In view of the high variability in the accumulation of TAG and starch in various strains (44), the observed differences in TAG accumulation with different concentration of CO2 might be within the natural physiological variation caused by a subtle balance of carbon partitioning into TAG and starch.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by a grant-in-aid for Core Research for Evolutional Science and Technology (CREST) from the Japan Science and Technology Agency.
Footnotes
Published ahead of print 13 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00280-13.
REFERENCES
- 1.Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A. 2008. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54:621–639. 10.1111/j.1365-313X.2008.03492.x [DOI] [PubMed] [Google Scholar]
- 2.Spoehr HA, Milner HW. 1949. The chemical composition of Chlorella; effect of environmental conditions. Plant Physiol. 24:120–149. 10.1104/pp.24.1.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris EH. 2001. Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52:363–406. 10.1146/annurev.arplant.52.1.363 [DOI] [PubMed] [Google Scholar]
- 4.Merchant SS, Kropat J, Liu B, Shaw J, Warakanont J. 2012. TAG, you're it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr. Opin. Biotechnol. 23:352–363. 10.1016/j.copbio.2011.12.001 [DOI] [PubMed] [Google Scholar]
- 5.Harris EH. 2009. The sexual cycle, p 119–157 In Harris EH. (ed), The Chlamydomonas sourcebook, 2nd ed, vol 1 Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 6.Yamano T, Iguchi H, Fukuzawa H. 2013. Rapid transformation of Chlamydomonas reinhardtii without cell-wall removal. J. Biosci. Bioeng. 115:691–694. 10.1016/j.jbiosc.2012.12.020 [DOI] [PubMed] [Google Scholar]
- 7.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Cardol P, Cerutti H, Chanfreau G, Chen CL, Cognat V, Croft MT, Dent Dutcher RS, Fernandez E, Fukuzawa H, Gonzalez-Ballester D, Gonzalez-Halphen D, Hallmann A, Hanikenne M, Hippler M, Inwood W, Jabbari K, Kalanon M, Kuras R, Lefebvre PA, Lemaire SD, Lobanov AV, Lohr M, Manuell A, Meier I, Mets L, Mittag M, Mittelmeier T, et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318:245–250. 10.1126/science.1143609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riekhof WR, Sears B, Benning C. 2005. Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: discovery of the betain lipid synthase BTA1Cr. Eukaryot. Cell 4:242–252. 10.1128/EC.4.2.242-252.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giroud C, Gerber A, Eichenberger W. 1988. Lipids of Chlamydomonas reinhardtii. Analysis of molecular species and intracellular site(s) of biosynthesis. Plant Cell Physiol. 29:587–595 [Google Scholar]
- 10.Eichenberger W. 1976. Lipids of Chlamydomonas reinhardi under different growth conditions. Phytochemistry 15:459–463. 10.1016/S0031-9422(00)88947-5 [DOI] [Google Scholar]
- 11.Sato N. 1988. Dual role of methionine in the biosynthesis of diacylglyceryltrimethylhomoserine in Chlamydomonas reinhardtii. Plant Physiol. 86:931–934. 10.1104/pp.86.3.931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato N. 1989. Modulation of lipid and fatty acid content by carbon dioxide in Chlamydomonas reinhardtii. Plant Sci. 61:17–21. 10.1016/0168-9452(89)90113-1 [DOI] [Google Scholar]
- 13.Sato N. 1992. Betain lipids. Bot. Mag. 105:185–197. 10.1007/BF02489414 [DOI] [Google Scholar]
- 14.Eichenberger W. 1982. Distribution of diacylglyceryl-O-4′-N,N,N-trimethyl)-homoserine in different algae. Plant Sci. Lett. 24:91–95. 10.1016/0304-4211(82)90012-8 [DOI] [Google Scholar]
- 15.Klug RM, Benning C. 2001. Two enzymes of diacylglyceryl-O-4′-(N,N,N-trimethyl)homoserine biosynthesis are encoded by btaA and btaB in the purple bacterium Rhodobacter sphaeroides. Proc. Natl. Acad. Sci. U. S. A. 98:5910–5915. 10.1073/pnas.101037998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Proschold T, Harris EH, Colemen AW. 2005. Portrait of a species: Chlamydomonas reinhardtii. Genetics 170:1601–1610. 10.1534/genetics.105.044503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris EH. 2009. The genus Chlamydomonas, p 1–24 In Harris EH. (ed), The Chlamydomonas sourcebook, 2nd ed, vol 1 Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 18.Fan J, Andre C, Xu C. 2011. A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett. 585:1985–1991. 10.1016/j.febslet.2011.05.018 [DOI] [PubMed] [Google Scholar]
- 19.James GO, Hocart CH, Hillier W, Chen H, Kordbacheh F, Price GD, Djordjevic MA. 2011. Fatty acid profiling of Chlamydomonas reinhardtii under nitrogen deprivation. Bioresour. Technol. 102:3343–3351. 10.1016/j.biortech.2010.11.051 [DOI] [PubMed] [Google Scholar]
- 20.Moellering ER, Benning C. 2010. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell 9:97–106. 10.1128/EC.00203-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U. 2009. Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell 8:1856–1868. 10.1128/EC.00272-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Work VH, Radakovits R, Jinkerson RE, Meuser JE, Elliott LG, Vinyard DJ, Laurens LM, Dismukes GC, Posewitz MC. 2010. Increased lipid accumulation in the Chlamydomonas reinhardtii sta7-10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains. Eukaryot. Cell 9:1251–1261. 10.1128/EC.00075-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giroud C, Eichenberger W. 1989. Lipids of Chlamydomonas reinhardtii. Incorporation of [14C]acetate, [14C]palmitate and [14C]oleate into different lipids and evidence for lipid-linked desaturation of fatty acids. Plant Cell Physiol. 30:121–128 [Google Scholar]
- 24.Goodson C, Roth R, Wang ZT, Goodenough U. 2011. Structural correlates of cytoplasmic and chloroplast lipid body synthesis in Chlamydomonas reinhardtii and stimulation of lipid body production with acetate boost. Eukaryot. Cell 10:1592–1606. 10.1128/EC.05242-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato N. 2012. Scientific elan vital: entropy deficit or inhomogeneity as a unified concept of driving forces of life in hierarchical biosphere driven by photosynthesis. Entropy 14:233–251. 10.3390/e14020233 [DOI] [Google Scholar]
- 26.Watanabe A. 1960. List of algal strains in collection at the Institute of Applied Microbiology, University of Tokyo. J. Gen. Appl. Microbiol. 6:283–292. 10.2323/jgam.6.283 [DOI] [Google Scholar]
- 27.Porra RJ, Thompson WA, Kriedemann PE. 1989. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975:384–394. 10.1016/S0005-2728(89)80347-0 [DOI] [Google Scholar]
- 28.Ishikawa M, Fujiwara M, Sonoike K, Sato N. 2009. Orthogenomics of photosynthetic organisms: bioinformatic and experimental analysis of chloroplast proteins of endosymbiont origin in Arabidopsis and their counterparts in Synechocystis. Plant Cell Physiol. 50:773–788. 10.1093/pcp/pcp027 [DOI] [PubMed] [Google Scholar]
- 29.Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917. 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- 30.Sato N, Furuya M. 1983. Isolation and identification of diacylglyceryl-O-4′-(N,N,N-trimethyl)-homoserine from the fern Adiantum capillusveneris L. Plant Cell Physiol. 24:1113–1120 [Google Scholar]
- 31.Andersson B, Holman R. 1974. Pyrrolidides for mass spectrometric determination of the position of the double bond in monounsaturated fatty acids. Lipids 9:185–190. 10.1007/BF02532690 [DOI] [PubMed] [Google Scholar]
- 32.Capella P, Zorzut CM. 1968. Determination of double bond position in monounsaturated fatty acids esters by mass spectrometry of their trimethylsilyloxy derivatives. Anal. Chem. 40:1458–1463. 10.1021/ac60266a013 [DOI] [Google Scholar]
- 33.Janssen G, Parmentier G. 1978. Determination of double bond positions in fatty acids with conjugated double bonds. Biomed. Mass Spectrom. 5:439–443. 10.1002/bms.1200050704 [DOI] [PubMed] [Google Scholar]
- 34.Janssen G, Parmentier G, Verhulst A, Eyssen H. 1985. Location of the double bond positions in microbial isomerization and hydrogenation products of α- and γ-linolenic acids. Biomed. Mass Spectrom. 3:134–138 [Google Scholar]
- 35.Castruita M, Casero C, Karpowicz SJ, Kropat J, Vieler A, Hsieh SI, Yan W, Cokus S, Loo JA, Benning C, Pellegrini M, Merchant SS. 2011. Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 32:1273–1292. 10.1105/tpc.111.084400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urzica EI, Vieler A, Hong-Hermesdorf A, Page MD, Casero D, Gallaher SD, Kropat J, Pellegrini M, Benning C, Merchant SS. 2013. Remodeling of membrane lipids in iron-starved Chlamydomonas. J. Biol. Chem. 288:30246–30258. 10.1074/jbc.M113.490425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X, Moellering ER, Liu B, Johnny C, Fedewa M, Sears BB, Kuo M-H, Benning C. 2012. A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 24:4670–4686. 10.1105/tpc.112.105106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riekhof WR, Benning C. 2009. Glycerolipid biosynthesis, p 41–68 In Stern DB. (ed), The Chlamydomonas sourcebook, 2nd ed, vol 2 Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 39.Sato N, Fujiwara S, Kawaguchi A, Tsuzuki M. 1997. Cloning of a gene for chloroplast ω6 desaturase of a green algae, Chlamydomonas reinhardtii. J. Biochem. 122:1224–1232. 10.1093/oxfordjournals.jbchem.a021885 [DOI] [PubMed] [Google Scholar]
- 40.Kajikawa M, Yamamoto KT, Kohzu Y, Shoji S, Matsui K, Tanaka Y, Sakai Y, Fukuzawa H. 2006. A front-end desaturase from Chlamydomonas reinhardtii produces pinolenic and coniferonic acids by ω13 desaturation in methylotrophic yeast and tobacco. Plant Cell Physiol. 47:64–73. 10.1093/pcp/pci224 [DOI] [PubMed] [Google Scholar]
- 41.Zauner S, Jochum W, Bigorowski T, Benning C. 2012. A cytochrome b5-containing plastid-located fatty acid desaturase from Chlamydomonas reinhardtii. Eukaryot. Cell 11:856–863. 10.1128/EC.00079-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen HM, Cuine S, Beyly-Adriano A, Legeret B, Billon E, Auroy P, Beisson F, Peltier G, Li-Beisson Y. 2013. The green microalga Chlamydomonas reinhardtii has a single ω-3 fatty acid desaturase which localizes to the chloroplast and impacts both plastidic and extraplastidic membrane lipids. Plant Physiol. 163:914–928. 10.1104/pp.113.223941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Msanne J, Xu D, Konda AR, Casas-Mollano JA, Awada T, Cahoon EB, Cerutti H. 2012. Metabolic and gene expression changes triggered by nitrogen deprivation in the photoautotrophically grown microalgae Chlamydomonas reinhardtii and Coccomyxa sp. C-169. Phytochemistry 75:50–59. 10.1016/j.phytochem.2011.12.007 [DOI] [PubMed] [Google Scholar]
- 44.Siaut M, Cuine S, Cagnon C, Fessler B, Nguyen M, Carrier P, Beyly A, Beisson F, Triantaphylides C, Li-Beisson Y, Peltier G. 2011. Oil accumulation in the model green algae Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 11:7. 10.1186/1472-6750-11-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cakmak T, Angun P, Ozkan AD, Cakmak Z, Olmez TT, Tekinay T. 2012. Nitrogen and sulfur deprivation differentiate lipid accumulation targets of Chlamydomonas reinhardtii. Bioengineered 3:343–346. 10.4161/bioe.21427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemschemeier A, Casero D, Liu B, Benning C, Pellegrini M, Happe T, Merchant SS. 2013. Copper response regulator1-dependent and -independent responses of the Chlamydomonas reinhardtii transcriptome to dark anoxia. Plant Cell 25:3186–3211. 10.1105/tpc.113.115741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kropat J, Hong-Hermesdorf A, Casero D, Ent P, Castruita M, Pellegrini M, Merchant SS, Malasarn D. 2011. A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 66:770–780. 10.1111/j.1365-313X.2011.04537.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mason CB, Manuel LJ, Moroney JV. 1990. A new chloroplast protein is induced by growth on low CO2 in Chlamydomonas reinhardtii. Plant Physiol. 93:833–836. 10.1104/pp.93.2.833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turpin DH. 1991. Effects of inorganic N availability on algal photosynthesis and carbon metabolism. J. Phycol. 27:14–20. 10.1111/j.0022-3646.1991.00014.x [DOI] [Google Scholar]
- 50.Plumley FG, Schmidt GW. 1989. Nitrogen-dependent regulation of photosynthetic gene expression. Proc. Natl. Acad. Sci. U. S. A. 86:2678–2682. 10.1073/pnas.86.8.2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Philipps G, Happe T, Hemschemeier A. 2012. Nitrogen deprivation results in photosynthetic hydrogen production in Chlamydomonas reinhardtii. Planta 235:729–745. 10.1007/s00425-011-1537-2 [DOI] [PubMed] [Google Scholar]
- 52.Draber W, Trebst A, Harth E. 1970. On a new inhibitor of photosynthetic electron-transport in isolated chloroplasts. Z. Naturforsch. B 25:1157–1159 [DOI] [PubMed] [Google Scholar]
- 53.Badger MR, Kaplan A, Berry JA. 1980. Internal inorganic carbon pool of Chlamydomonas reinhardtii: evidence for a carbon dioxide-concentrating mechanism. Plant Physiol. 66:407–413. 10.1104/pp.66.3.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.