Abstract
In the Netherlands, extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli bacteria are highly prevalent in poultry, and chicken meat has been implicated as a source of ESBL-producing E. coli present in the human population. The current study describes the isolation of ESBL-producing E. coli from house flies and blow flies caught at two poultry farms, offering a potential alternative route of transmission of ESBL-producing E. coli from poultry to humans. Overall, 87 flies were analyzed in 19 pools. ESBL-producing E. coli bacteria were detected in two fly pools (10.5%): a pool of three blow flies from a broiler farm and a pool of eight house flies from a laying-hen farm. From each positive fly pool, six isolates were characterized and compared with isolates obtained from manure (n = 53) sampled at both farms and rinse water (n = 10) from the broiler farm. Among six fly isolates from the broiler farm, four different types were detected with respect to phylogenetic group, sequence type (ST), and ESBL genotype: A0/ST3519/SHV-12, A1/ST10/SHV-12, A1/ST58/SHV-12, and B1/ST448/CTX-M-1. These types, as well as six additional types, were also present in manure and/or rinse water at the same farm. At the laying-hen farm, all fly and manure isolates were identical, carrying blaTEM-52 in an A1/ST48 genetic background. The data imply that flies acquire ESBL-producing E. coli at poultry farms, warranting further evaluation of the contribution of flies to dissemination of ESBL-producing E. coli in the community.
INTRODUCTION
Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae are increasing in prevalence worldwide (1, 2). ESBLs confer resistance to most beta-lactam antibiotics, including 3rd- and 4th-generation cephalosporins, which severely limits treatment possibilities for infections caused by these bacteria. Often, options for treatment are further restricted by the multiresistant nature of ESBL-producing bacteria, which has led to increased use of, and increasing prevalence of Enterobacteriaceae resistant to, last-resort antibiotics, such as carbapenems (3). Although initially ESBL production was mainly observed in hospital infections caused by Klebsiella pneumoniae, today it is also frequently associated with community-acquired infections, mostly urinary tract infections caused by Escherichia coli (4, 5), as well as commensal E. coli strains isolated from humans and food-producing animals (6–8).
Not only can dissemination of ESBL-producing E. coli in the community be facilitated by direct contact with human or animal carriers, but also, the presence of the bacterium in environmental compartments, such as surface water (9–12) and wildlife (13), suggests that the environment should also be considered in this regard. In the Netherlands, ESBL-producing E. coli is highly prevalent in poultry: in 2009, ESBL-producing (and/or AmpC-producing) E. coli bacteria were detected on 100% (n = 26) of Dutch broiler farms studied (14). Because of the high prevalence of ESBL-producing E. coli on Dutch retail chicken meat and the overlap between ESBL genotypes from chicken meat and clinical E. coli isolates (15, 16), chicken meat has been suggested as a source of ESBL-producing E. coli in the Netherlands.
The present study was aimed at assessing a potential alternative, indirect route of transmission of ESBL-producing E. coli from poultry to humans, namely, through flies. These insects have been recognized as transmitters of infectious diseases for some time (17). They move between feces and carcasses and food meant for human consumption. Bacteria acquired from filth can be transmitted to food, either via the fly exterior, e.g., body surface and mouth parts, or with feces and vomit that is produced during feeding (18–23). In an experimental setting, flies were shown to transmit Campylobacter between chickens (24), and several studies have demonstrated similar pathogenic E. coli and Klebsiella strains on flies and in humans in hospital settings, as well as in small rural communities, suggesting their potential as transmission vehicles (25–27). Even though flies generally stay close to their breeding source, they may also move over considerable distances (28, 29). The current study demonstrates the dissemination of ESBL-producing E. coli from laying hens and broilers to houseflies and blow flies, indicating a possible role for flies in the dissemination of ESBL-producing E. coli from poultry to the general public.
MATERIALS AND METHODS
Sampling of flies.
A broiler farm, with a capacity of 90,000 broiler chickens distributed over four poultry houses, and a laying-hen farm, with a capacity of 30,000 free-range chickens, were visited in September and October 2011, respectively. Broilers are commonly kept for a period of 5 to 7 weeks, after which the entire flock is transported to the slaughterhouse and the poultry houses are cleaned before the next flock is introduced. The broiler farm was therefore visited twice, once while a flock of broilers 38 days of age were present and once after the poultry houses had been emptied of the flock and were being cleaned, for the purpose of evaluating the presence of ESBL-producing E. coli on the farm after cleaning. Laying hens are kept for well over a year, and therefore, the laying-hen farm was visited once, at which time the hens were 52 weeks old. During all visits, flies were collected using nontoxic sticky flypaper and harvested within 24 h after placement. When the number of flies stuck on the flypaper was considered low compared to the number of flies flying around the farm, they were also collected using a fly swatter and stored in sterile containers. At the broiler farm, 10 flies were caught when broilers were present and 27 when the poultry houses were being cleaned; at the laying-hen farm, 51 flies were collected. The 87 flies were analyzed in 19 separate pools, each consisting of 1 to 8 flies that were identical with respect to collection location and fly species (see Table 3).
TABLE 3.
Characteristics of fly samples
| Fly scientific name | Fly common name |
nb |
|||||
|---|---|---|---|---|---|---|---|
| Broiler farm |
Laying-hen farm |
||||||
| Broilers present |
At cleaning |
||||||
| Flies | Pools | Flies | Pools | Flies | Pools | ||
| Musca domestica | Common housefly | 9 | 2 (3–6) | 45 | 6 (5–8) | ||
| Fannnia canicularis | Lesser housefly | 9 | 4 (1–4) | 11 | 2 (4–7) | ||
| Lucilia spp.a | Blow fly | 1 | 1 | 6 | 2 (3) | ||
| Stomoxys calcitrans | Stable fly | 6 | 2 (2–4) | ||||
| Total | 10 | 5 | 26 | 6 | 51 | 8 | |
Flies were not identified to species level.
Indicated in parentheses are the minimum and maximum number of flies per pool.
Sampling of manure and rinse water.
At both farms, poultry manure of varying degrees of freshness was sampled. During the first visit to the broiler farm, while the broilers were present, 10 manure samples were collected: 8 samples of fresh manure (“fresh” was defined as still soft and warm) from poultry houses, 2 samples per house, and 1 sample of semifresh manure (“semifresh” was defined as generally still identifiable as individual droppings but no longer soft and warm) from a small flock of laying hens kept as a hobby on the premises near the private house. During cleaning of the poultry houses, when there were no broilers present, two manure samples were collected from a 2-day-old dung heap. At the laying-hen farm, also, 10 manure samples were collected: 2 samples of fresh manure from the poultry house, 7 semifresh samples (3 from manure belts inside the poultry house, 2 from the free-range area, 1 from the premises but outside the official free-range area, and 1 from soil beneath the manure transport belt outside the poultry house), and 1 sample of dried (“dusty”) manure from a manure storage container. The samples consisted of five pooled individual droppings, with the exception of the samples from the dung heap and storage container, where 20 to 50 g were collected. All manure samples were placed in sterile containers. At the broiler farm, 0.5 liter of rinse water was sampled from wastewater pits that filled up during cleaning of the poultry houses. The samples were transported in cool boxes containing ice packs and stored at 4°C.
Isolation of ESBL-producing E. coli.
All samples were analyzed within 24 h after sampling. Fly pools were collected in 24 to 33 ml phosphate-buffered saline (Biotrading, Mijdrecht, the Netherlands) with 0.5% Tween 20 and transferred to sterile filter bags (Interscience, St Nom La Bretêche, France) using sterile pairs of tweezers. The flies in bags were thoroughly crushed using thumb and forefinger (from the outside of the bag) and then homogenized using a Stomacher400 (Seward, Worthing, United Kingdom) at 230 rpm. Of these homogenates, 100 μl was streaked on ChromID ESBL medium (bioMérieux, Boxtel, the Netherlands). For manure, 10 g was diluted 10 times in buffered peptone water (BPW), followed by homogenization using a Pulsifier (Microgen Bioproducts Ltd., Camberley, United Kingdom). These 10−1 homogenates were again diluted 10 times (10−2 dilution) using peptone saline (Biotrading), and 100 μl (each) of 10−1 and 10−2 dilutions were streaked on ChromID ESBL medium. Wastewater samples were filtered through membrane filters with a pore size of 0.45 μm (Millipore, Amsterdam, the Netherlands) in volumes ranging from 0.1 μl to 1 ml. Filters were placed on ChromID ESBL medium. For fly pool and manure samples, additionally, 10 ml of homogenates was preenriched in BPW either supplemented or not with 1 μg/ml cefotaxime (BPW-CTX) and streaked on ChromID ESBL medium. After the first five pools of flies had been analyzed (together containing 10 flies caught at the broiler farm while broilers were present) (see Table 3), the 14 fly sample pools collected at later time points (i.e., 77 flies caught during cleaning at the broiler farm and at the laying-hen farm) were additionally analyzed for the presence of total E. coli, using Tryptone Bile X-glucuronide (TBX) agar (Bio-Rad, Veenendaal, the Netherlands).
All cultures were incubated for 4 to 5 h at 37°C, followed by 16 to 20 h at 44°C to increase selectivity for E. coli (30). In manure and wastewater samples, ESBL-producing E. coli bacteria were quantified, and 95% confidence intervals (CI) were calculated using Mathematica 9.0.1 (Wolfram Research, Oxfordshire, United Kingdom).
Confirmation of ESBL production and identification of ESBL genes.
Isolates were tested for ESBL production using disk diffusion following CLSI guidelines (31). Using Sensi-Disc test discs (BD, Breda, the Netherlands), zone diameters were determined for cefotaxime (30 μg/ml), cefotaxime (30 μg/ml) plus clavulanic acid (10 μg/ml), ceftazidime (30 μg/ml), ceftazidime plus clavulanic acid (10 μg/ml), and cefoxitin (30 μg/ml). ESBL-producing isolates were defined as strains resistant to cefotaxime (zone diameter, ≤22 mm) and/or ceftazidime (zone diameter, ≤17 mm) and with a reduction in zone diameter of ≥5 mm with the disks containing clavulanic acid (31). Phenotypically confirmed ESBL-producing E. coli isolates were analyzed for the presence of genes encoding CTX-M group 1, CTX-M group 2, and CTX-M group 9 ESBLs and of blaOXA, blaSHV, and blaTEM genes by multiplex PCRs using primers described by Dallenne et al. (32). Material from a single colony was suspended in Tris-EDTA buffer (pH 8.0; Sigma-Aldrich, Zwijndrecht, the Netherlands), and the cells were lysed at 70°C for 5 min. DNA extracts were stored at −20°C. For amplification, 3 μl of DNA extract was mixed with 10 pmol of each primer and 12.5 μl Qiagen Multiplex PCR mix (Qiagen, Venlo, the Netherlands) in a final volume of 25 μl. Amplification conditions were as described by Dallenne et al. PCR products were analyzed on agarose gels. PCR products of the expected size were treated with ExoSap-It (GE Healthcare, Hoevelaken, the Netherlands) and sequenced using the same primers used to generate the PCR products and a BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Bleiswijk, the Netherlands). The sequences obtained were compared with ESBL gene sequences in the GenBank database and on the Lahey website (http://www.lahey.org/studies).
Phylogenetic typing.
ESBL-producing isolates were allotted to phylogenetic group A, B1, B2, or D by PCR targeted to the chuA and yjaA genes and the TspE4.C2 DNA fragment, using primers described by Clermont et al. (33). For amplification, 1.5 μl of 10-times-diluted DNA extract (the same extract used for ESBL genotyping) was mixed with 5 pmol of each primer and 12.5 μl iQ Supermix (Bio-Rad, Veenendaal, the Netherlands) in a final volume of 25 μl. The amplification conditions were as follows: 5 min at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 60°C (TspE4.C2) or 62°C (chuA and yjaA), 30s at 72°C, and a final elongation step of 10 min at 72°C. Strains were subgrouped according to the method of Escobar-Paramo et al. (34): subgroup A0, chuA, yjaA, andTspE4.C2 negative; subgroup A1, chuA negative, yjaA+, TspE4.C2 negative; group B1, chuA negative, yjaA+ or negative, TspE4.C2+; subgroup B22, chuA+, yjaA+, TspE4.C2 negative; subgroup B23, chuA+, yjaA+, TspE4.C2+; subgroup D1, chuA+, yjaA negative, TspE4.C2 negative; subgroup D2, chuA+, yjaA negative, TspE4.C2+.
MLST.
For multilocus sequence typing (MLST) of ESBL-producing E. coli isolates, seven housekeeping genes were amplified (adk, fumC, gyrB, icd, mdh, purA, and recA), as described by Wirth et al. (35). Primer sequences were obtained from the E. coli MLST database website (http://mlst.ucc.ie/mlst/dbs/Ecoli). For amplification, 2 μl of 10-times-diluted DNA extract was mixed with 200 pmol of each primer, 1× PCR buffer (Invitrogen, Bleiswijk, the Netherlands), 2.5 mM MgCl2 (Invitrogen), 200 μM deoxynucleoside triphosphate (dNTP) mixture (Invitrogen), and 1.25 U Taq polymerase (Invitrogen) in a final volume of 50 μl. The amplification conditions were as follows: 5 min at 95°C, followed by 35 cycles of 30s at 95°C, 30s at 60°C (adk, icd, mdh, purA, and recA) or 30s at 64°C (fumC and gyrB), 45s at 72°C, and a final elongation step of 10 min at 72°C. The PCR products were analyzed on agarose gels, and PCR products of the expected size were treated with ExoSap-It (GE Healthcare, Hoevelaken, the Netherlands), followed by sequencing with the same primers used to generate PCR products using a BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Bleiswijk, the Netherlands). Sequences were imported into the E. coli MLST database website (http://mlst.ucc.ie/mlst/dbs/Ecoli) to determine MLST types.
RESULTS
Prevalence of ESBL-producing E. coli in manure and rinse water.
At the broiler farm, ESBL-producing E. coli bacteria were detected in 82% of manure samples and all rinse water samples (Table 1). ESBL-producing E. coli bacteria were found in fresh manure from three of four poultry houses, as well as in 2-day-old dung heap manure samples. Additionally, they were detected in manure from hobby laying hens that were kept on the premises. In the dung heap samples, ESBL-producing E. coli bacteria were detected only after preenrichment. In these samples, concentrations were at least 0.1 CFU/g and most likely below 173 CFU/g, where the upper limit is represented by the 97.5% CI obtained from the negative direct cultures. The average ESBL-producing E. coli concentrations in quantifiable manure samples were 5.3 × 105 CFU/g (range, 1.5 × 103 to 2.4 × 106 CFU/g). In rinse water sampled from five wastewater pits when poultry houses were being cleaned, ESBL-producing E. coli was detected with an average concentration of 2.5 × 107 CFU/liter (range, 3.9 × 106 to 5.8 × 107 CFU/liter).
TABLE 1.
Concentrations of ESBL-producing E. coli bacteria in manure and rinse water at a Dutch broiler farm
| Sampling location | Freshness of manure | Concnd | 95% CI |
|---|---|---|---|
| Manure | |||
| Poultry house 1 (A) | Fresh | 4.5 × 105 | 3.6 × 105–5.4 × 105 |
| Poultry house 1 (B) | Fresh | 1.5 × 103 | 1.1 × 103–2.1 × 103 |
| Poultry house 2 (A) | Fresh | 2.0 × 105 | 1.9 × 105–2.2 × 105 |
| Poultry house 2 (B) | Fresh | 2.4 × 106 | 2.2 × 106–2.7 × 106 |
| Poultry house 3 (A) | Fresh | <0.1a | 0–86 |
| Poultry house 3 (B) | Fresh | <0.1a | 0–86 |
| Poultry house 4 (A) | Fresh | 2.7 × 104 | 2.5 × 104–3.0 × 104 |
| Poultry house 4 (B) | Fresh | 4.2 × 104 | 3.4 × 104–5.1 × 104 |
| Hobby laying hens | Mixed | 2.0 × 104 | 1.5 × 105–2.6 × 104 |
| Dung heap (A)c | 2 days old | ≥0.1b | 0–173 |
| Dung heap (B)c | 2 days old | ≥0.1b | 0–173 |
| Rinse waterc | |||
| Wastewater reservoir 1 | NAe | 2.6 × 107 | 2.0 × 107–3.4 × 107 |
| Wastewater reservoir 2 | NA | 5.8 × 107 | 4.8 × 107–6.9 × 107 |
| Wastewater reservoir 3 | NA | 3.9 × 106 | 3.1 × 106–4.7 × 106 |
| Wastewater reservoir 4 | NA | 1.8 × 107 | 1.2 × 107–2.4 × 107 |
| Assembly pit | NA | 2.0 × 107 | 1.4 × 107–2.6 × 107 |
Negative in direct culture and after enrichment.
Positive after enrichment only.
Sampled at cleaning. The 95% CI are based on the results of direct culture.
Concentrations are CFU/g for manure and CFU/liter for rinse water.
NA, not applicable.
At the laying-hen farm, ESBL-producing E. coli bacteria were detected in 8 of 10 (80%) manure samples (Table 2). ESBL-producing E. coli bacteria were found both inside the poultry house and outside, in the official free range but also at other places on the premises. Positive samples included one of two fresh manure samples and all samples of mixed freshness. No ESBL-producing E. coli bacteria were detected in dried manure stored in containers. In one of the samples, ESBL-producing E. coli bacteria were detected only after preenrichment. In this case, the concentration of ESBL-producing E. coli bacteria could not be established accurately but was at least 0.1 CFU/g and most likely below 87 CFU/g, where the upper limit is represented by the 97.5% CFI obtained from the negative direct cultures. The average ESBL-producing E. coli concentration in quantifiable samples was 2.8 × 103 CFU/g (range, 45 to 9.3 × 103 CFU/g).
TABLE 2.
Concentrations of ESBL-producing E. coli in manure at a Dutch laying-hen farm
| Sampling location | Freshness of manure | Concn (CFU/g) | 95% CI |
|---|---|---|---|
| Poultry house (A) | Fresh | 45 | 2.6–100 |
| Poultry house (B) | Fresh | <0.1a | 0–87 |
| Transport belt 1 | Mixed | 2.7 × 103 | 2.1 × 103–3.5 × 103 |
| Transport belt 2 | Mixed | 3.4 × 102 | 1.2 × 102–7.3 × 102 |
| Transport belt 3 | Mixed | 3.8 × 103 | 3.0 × 103–4.6 × 103 |
| Free range (A) | Mixed | 9.3 × 103 | 8.1 × 103–1.1 × 104 |
| Free range (B) | Mixed | 3.1 × 103 | 2.5 × 103–3.9 × 103 |
| Premises | Mixed | 4.1 × 102 | 2.0 × 102–7.4 × 102 |
| Soil beneath transport belt | Mixed | ≥0.1b | 0–87 |
| Storage container | Dry | <0.1a | 0–87 |
Negative in direct culture and after enrichment.
Positive after enrichment only. The 95% CI are based on the results of direct culture.
Prevalence of ESBL-producing E. coli in/on flies.
Overall, 87 flies were caught at the poultry farms: 54 house flies (Musca domestica), 20 lesser house flies ( Fannia canicularis), 6 stable flies (Stomoxys calcitrans), and 7 blow flies (Lucilia spp.) (Table 3). The flies were analyzed in 19 pools, each consisting of one to eight flies that were identical with respect to collection location and fly species. ESBL-producing E. coli bacteria were detected in two pools (10.5%): a pool of three blow flies from the broiler farm caught during cleaning and a pool of eight house flies from the laying-hen farm. In comparison, total E. coli bacteria were detected in 12 of the 14 (85.7%) fly pools that were also analyzed for total E. coli. E. coli bacteria were detected in 8/8 housefly pools, 2/2 blow fly pools, 1/2 lesser house fly pools, and 1/2 stable fly pools. Two fly pools did not contain detectable levels of E. coli: one fly pool consisting of four F. canicularis flies from the broiler farm and one fly pool consisting of two S. calcitrans flies from the laying-hen farm. The 17 ESBL-producing E. coli-negative pools together consisted of 77 flies, meaning that at least 88.5% (77/87) of all flies analyzed did not carry detectable levels of ESBL-producing E. coli bacteria. The total numbers of ESBL-producing E. coli bacteria in the positive fly homogenates were 2.5 × 104 CFU and 1.2 × 103 CFU for the blow flies and house flies, respectively. Given the relatively low overall prevalence of ESBL-producing E. coli on flies, it is most likely that each positive pool contained only one ESBL-producing E. coli-positive fly, meaning that the indicated numbers of ESBL-producing E. coli were derived from one fly each. However, the possibility that multiple ESBL-producing E. coli-carrying flies were present in the positive pools cannot be excluded.
ESBL genes in ESBL-producing E. coli isolates.
Overall, 113 suspected ESBL-producing E. coli isolates were obtained: at the broiler farm, 40 from broiler manure, 5 from hobby laying-hen manure, 25 from rinse water, and 6 from the positive blow fly pool; at the laying-hen farm, 31 from laying-hen manure and 6 from the positive housefly pool. All isolates were confirmed ESBL producers, based on the effect of clavulanic acid on cefotaxime resistance alone (single effect; n = 8) or that of clavulanic acid on cefotaxime and ceftazidime resistance (double effect; n = 105). Of note, even though all isolates had at least reduced susceptibility to cefotaxime (zone diameter, ≤25 mm), all single-effect isolates and 10 of the double-effect isolates did not appear resistant to ceftazidime (zone diameter, >20 mm in the absence of clavulanic acid). The eight single-effect isolates were all derived from the broiler farm: one from manure from one of the poultry houses, five from dung heap manure, and two from the fly sample.
All 12 fly isolates (six from the broiler farm fly pool and six from the laying-hen farm fly pool), all 31 manure isolates from the laying-hen farm, and a random selection of 24 manure and 10 wastewater isolates from the broiler farm were characterized with respect to ESBL genotype. Four of six (67%) fly isolates from the broiler farm carried blaSHV-12, and two (33%) carried blaCTX-M-1 (Fig. 1). The same ESBL genes were detected in manure and rinse water isolates from the same farm, with similar relative distributions (23/34 blaSHV-12 versus 6/34 blaCTX-M-1). Five manure isolates, all obtained from one dung heap sample, carried blaTEM-52. The eight single-effect isolates were all identified as blaCTX-M-1-carrying isolates. At the laying-hen farm, all six fly isolates and all 31 manure isolates carried blaTEM-52.
FIG 1.
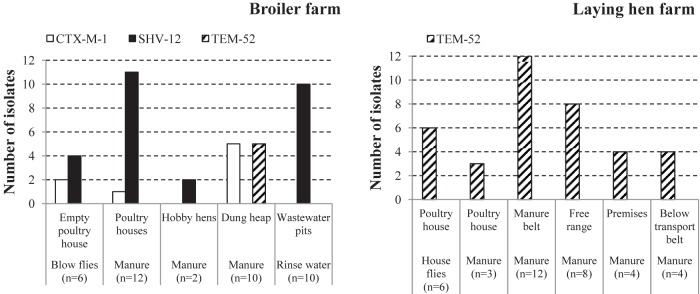
ESBL type distribution in ESBL-producing E. coli isolates from a broiler farm and a laying-hen farm. The bars represent the numbers of isolates with genes encoding CTX-M-1, SHV-12, and TEM-52 in flies, manure, and rinse water. The manure isolates are grouped according to sampling location within the respective farms.
Phylogenetic typing and MLST of ESBL-producing E. coli.
Combining ESBL genotypes and phylogenetic profiles of isolates, six different ESBL-producing E. coli types were recognized at the broiler farm: A0/SHV-12, A1/SHV-12, B11/SHV-12, D2/SHV-12, B11/CTX-M-1, and B22/TEM-52 (Fig. 2). The six ESBL-producing E. coli isolates obtained from the blow fly pool represented four different ESBL-producing E. coli types. All of these types were also present in at least one of the other sample types (broiler manure, hobby laying-hen manure, dung heap manure, and/or wastewater). At the laying-hen farm, housefly and manure isolates all shared the same type: A1/TEM-52.
FIG 2.
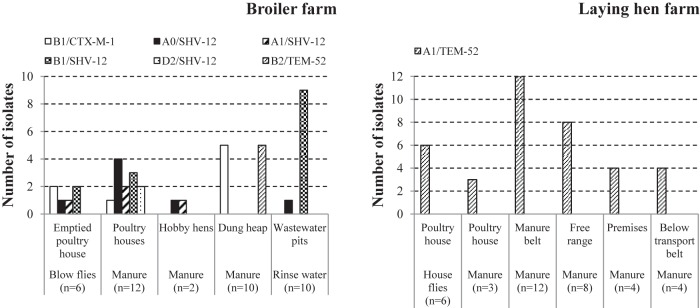
Phylogenetic group/ESBL type variants among ESBL-producing E. coli isolates from a broiler farm and a laying-hen farm. The bars represent the numbers of isolates with the indicated combinations of phylogenetic groups and ESBL types in flies, manure, and rinse water. The manure isolates are grouped according to sampling location within the respective farms.
A selection of 25 ESBL-producing E. coli isolates from the broiler farm (including 6 isolates from the blow fly pool) and 11 ESBL-producing E. coli isolates from the laying-hen farm (including 3 isolates from the house fly pool) were further characterized using MLST. From each matrix-location combination indicated on the x axes in Fig. 1 and 2, at least one phylogenetic group/ESBL genotype variant was selected, so that all phylogenetic group-ESBL genotype combinations were represented. Ten different ESBL-producing E. coli sequence types (STs) were recognized at the broiler farm (Table 4). Among these, two new sequence types were identified. Each sequence type was always observed in combination with one specific ESBL genotype. In the fly pool, four different sequence type-ESBL genotype combinations were identified, and all of them were also present in manure and/or wastewater (Table 4). All A1/TEM-52 isolates from the laying hen farm (3 isolates from flies, 1 from manure in the poultry house, 3 from manure on the manure belt, 2 from manure of free-range chickens, and 2 from manure collected at other sites on the premises) had the same sequence type, ST48.
TABLE 4.
ESBL-producing E. coli sequence types/ESBL genotypes at broiler and laying-hen farmsa
| Phylogenetic group/ESBL gene | Sequence type | No. of isolates originating from matrix |
Total | ||||
|---|---|---|---|---|---|---|---|
| Flies | Manure |
Rinse water | |||||
| Poultry house | Dung heap | Hobby hens | |||||
| Broiler farm | |||||||
| B11/CTX-M-1 | Total | 2 | 1 | 1 | NA | NA | 4 |
| ST 448 | 2 | 1 | 1 | NA | NA | 4 | |
| A0/SHV-12 | Total | 1 | 2 | NA | 1 | 1 | 5 |
| ST 746 | 0 | 0 | NA | 1 | 0 | 1 | |
| ST 3519 | 1 | 2 | NA | 0 | 1 | 4 | |
| A1/SHV-12 | Total | 1 | 2 | NA | 1 | NA | 4 |
| ST 10 | 1 | 2 | NA | 1 | NA | 4 | |
| B11/SHV-12 | Total | 2 | 1 | NA | NA | 7 | 10 |
| ST 58 | 2 | 1 | NA | NA | 3 | 6 | |
| ST 155 | 0 | 0 | NA | NA | 2 | 2 | |
| ST 2079 | 0 | 0 | NA | NA | 1 | 1 | |
| New | 0 | 0 | NA | NA | 1 | 1 | |
| D2/SHV-12 | Total | NA | 1 | NA | NA | NA | 1 |
| ST 420 | NA | 1 | NA | NA | NA | 1 | |
| B22/TEM-52 | Total | NA | NA | 1 | NA | NA | 1 |
| New | NA | NA | 1 | NA | NA | 1 | |
| Grand total | 6 | 7 | 2 | 2 | 8 | 25 | |
From each matrix/location combination indicated on the x axes of Fig. 1 and 2, at least one representative of each phylogenetic group/ESBL genotype variant was selected for MLST. Indicated in the table are, for each phylogenetic group/ESBL gene variant and each sample type, the total number of isolates tested in MLST analysis (Total) and the number of isolates with the indicated sequence type. NA, not applicable (the specific phylogenetic group/ESBL gene variants were not observed in the indicated sample type).
DISCUSSION
At the broiler farm, three different ESBL genes, blaCTX-M-1, blaSHV-12, and blaTEM-52, were circulating, associated with 10 different E. coli sequence types, all during one production round. In contrast, at the laying-hen farm only one ESBL gene, blaTEM-52, and one sequence type were detected. In the Netherlands, the total amount of antimicrobials used in broilers is approximately 20 times higher than that in laying hens, i.e., during the first half of 2013, 20 daily doses per animal year (dd/ay) were registered for conventionally held broilers compared to 0.7 to 1.2 dd/ay for free-range and battery laying hens, respectively (Dutch Product Boards of Livestock, Meat, and Eggs, personal communication). Possibly, the more restricted use of antimicrobials in laying hens and the high turnover of flocks in broiler farms could explain the difference in diversity in ESBL-producing E. coli between farm types. However, only one farm of each type was included in the current study, and more farms need to be analyzed to establish whether this difference in diversity is indeed a feature specific to the two types of poultry farms. The three ESBL genes detected at the farms are the most commonly observed genes on poultry meat in the Netherlands and are, with the exception of blaSHV-12, equally common in humans (15). Several of the E. coli sequence types found on poultry farms, i.e., ST10, ST48, ST58, and ST448, have been previously detected in human and animal clinical isolates obtained in the Netherlands or the neighboring countries Belgium and Germany, albeit generally associated with ESBL genes other than those observed in the current study (15, 16, 36–40). At both types of farms, flies carrying ESBL-producing E. coli were caught, and all fly isolates had genotypes also present in manure and rinse water, strongly implying the broilers and laying hens were the sources.
In 2012, the Netherlands had 2,140 poultry farms with 95 million chickens (Statistics Netherlands [CBS] [http://www.statline.cbs.nl]) in a country with an area of 41,256 km2. In conventionally kept broilers, the prevalence of ESBL-producing E. coli is very high, with 100% of farms positive (37), and 77 to 94% of all retail chicken meat is contaminated with ESBL-producing E. coli bacteria (15, 16). The prevalence in laying hens has not been published yet, but preliminary data suggest 100% positivity in conventionally kept laying hens, as well, albeit at lower concentrations than in broilers (unpublished observations). Assuming that farm flies may travel beyond farm premises, they could facilitate the spread of ESBL-producing E. coli from farms to the general public by contaminating food meant for human consumption. House flies have been reported to travel up to 30 km, although maximum distances of 0.5 to 4 km appear to be more common (28, 29). Monitoring the travel behavior of marked house flies on two mixed poultry and dairy farms over 3 days, Lysyk and Axtell (41) observed that the majority of house flies stayed in animal housings in which they were released (66% to 85%) or moved between poultry and dairy housings (7% to 24%), which were approximately 100 m apart on both farms. A minority of flies (4% to 6%) moved from the animal housings to surrounding pastures and fields and could be retrieved up to 250 m from the release sites (41). Even when flies travel only relatively short distances from farms, for instance, 200 m, ESBL-producing E. coli may still be disseminated to people living or pursuing recreation in agricultural areas.
Assuming that both positive fly pools contained one ESBL-producing E. coli strain each, 2.8% and 2.0% of the flies at the broiler and laying-hen farms, respectively, were carriers. In contrast, total E. coli bacteria were detected in the vast majority (83% and 88%) of fly samples, implying that the chances of flies becoming contaminated with ESBL-producing E. coli largely depended on the ESBL-producing E. coli/total E. coli ratio in the food animals at the farm or in their manure. In fresh manure, this ratio can vary from 1:200 to 1:2 × 105 between poultry farms (unpublished). Overall, 1 of 54 (1.9%) house flies and 1 of 7 (14%) blow flies were carriers of ESBL-producing E. coli bacteria. Two recently published studies, one performed at a horseback-riding center in the Czech Republic and the other on a cattle farm in Japan, describe the presence of ESBL-producing E. coli on flies, with at least some identity homology with variants isolated from animals and/or stables (42, 43). In the Czech study, ESBL-producing E. coli bacteria were isolated from 18% of unspecified fly species, and in the Japanese study, cephalosporin-resistant E. coli bacteria were detected in 14.3 and 10.3% of houseflies and false stable flies, respectively. The detection of ESBL-producing E. coli on house flies (current and Japanese studies), blow flies (current study), and false stable flies (Japanese study) and of total E. coli on stable flies and lesser houseflies (current study) demonstrates that at least these five fly species may act as ESBL-producing E. coli carriers.
As well as in poultry, ESBL-producing E. coli bacteria are commonly observed in other food animals in the Netherlands, especially veal calves and slaughter pigs (8). The risk for the general public to be exposed to ESBL-producing bacteria from flies originating from farms can be estimated using quantitative risk assessment (QMRA) (44, 45). For that purpose, data are required on the prevalence and concentrations of ESBL-producing E. coli on flies, on the survival potential of E. coli in or on flies, and on the ecology and behavior of the particular fly species that are identified as possible carriers of ESBL-producing E. coli. Examples of this type of information are the number of flies that live on farms, their life spans and feeding and breeding properties (which determine both the chance of flies acquiring E. coli and the possibility of transmitting these bacteria to food), the percentage of flies that move from the farms, and the distances they travel. Such analysis will provide insight into the overall contribution of flies to the spread of ESBL-producing E. coli from food animals to the community and allow comparison with the contributions of other possible transmission routes, such as direct contact with human and animal carriers or consumption of contaminated food. Mapping the relative roles of all potential transmission routes, including transmission through various fly species, is necessary to purposefully reduce the spread of community-associated ESBL-producing E. coli.
ACKNOWLEDGMENTS
This work was supported by the Netherlands Food and Consumer Product Safety Authority.
We thank Henk J. M. Aarts, Wilma F. Jacobs-Reitsma, and Rozemarijn Q. J. van der Plaats for their contributions to this study.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 25 October 2013
REFERENCES
- 1.Cantón R, Novais A, Valverde A, Machado E, Peixe L, Baquero F, Coque TM. 2008. Prevalence and spread of extended-spectrum ß-lactamase-producing Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 14(Suppl. 1):144–153. 10.1111/j.1469-0691.2007.01850.x [DOI] [PubMed] [Google Scholar]
- 2.Castanheira M, Mendes RE, Rhomberg PR, Jones RN. 2008. Rapid emergence of blaCTX-M among Enterobacteriaceae in U.S. Medical Centers: molecular evaluation from the MYSTIC Program (2007). Microb. Drug Resist. 14:211–216. 10.1089/mdr.2008.0827 [DOI] [PubMed] [Google Scholar]
- 3.Cantón R, Akova M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Miriagou V, Naas T, Rossolini GM, Samuelsen O, Seifert H, Woodford N, Nordmann P. 2012. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 18:413–431. 10.1111/j.1469-0691.2012.03821.x [DOI] [PubMed] [Google Scholar]
- 4.Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, Ayala J, Coque TM, Kern-Zdanowicz I, Luzzaro F, Poirel L, Woodford N. 2007. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59:165–174. 10.1093/jac/dkl483 [DOI] [PubMed] [Google Scholar]
- 5.Paterson DL, Bonomo RA. 2005. Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657–686. 10.1128/CMR.18.4.657-686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huijbers PM, de Kraker M, Graat EA, van Hoek AH, van Santen MG, de Jong MC, van Duijkeren E, de Greeff SC. 2013. Prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in humans living in municipalities with high and low broiler density. Clin. Microbiol. Infect. 19:E256–E259. 10.1111/1469-0691.12150 [DOI] [PubMed] [Google Scholar]
- 7.Trott D. 2013. Beta-lactam resistance in gram-negative pathogens isolated from animals. Curr. Pharm. Des. 19:239–249. 10.2174/138161213804070339 [DOI] [PubMed] [Google Scholar]
- 8.Mevius DJ, Koene MGJ, Wit B, van Pelt W, Bondt N. 2012. Monitoring of antimicrobial resistance and antibiotic usage in animals in the Netherlands in 2010/2011. Nethmap_MARAN report http://www.uu.nl/SiteCollectionImages/Fac_DGK/Nieuwsplaatjes/Nieuws/2012/NethmapMaran_Web.pdf
- 9.Blaak H, van Rooyen SR, Schuijt MS, Docters van Leeuwen AE, van den Berg HHJL, Lodder-Verschoor F, Italiaander R, Schets FM, de Roda Husman AM. 2011. Prevalence of antibiotic resistant bacteria in the rivers Meuse, Rhine and New Meuse. RIVM report 703719071 National Institute for Public Health and the Environment, Bilthoven, The Netherlands [Google Scholar]
- 10.Chen H, Shu W, Chang X, Chen JA, Guo Y, Tan Y. 2010. The profile of antibiotics resistance and integrons of extended-spectrum beta-lactamase producing thermotolerant coliforms isolated from the Yangtze River basin in Chongqing. Environ. Pollut. 158:2459–2464. 10.1016/j.envpol.2010.03.023 [DOI] [PubMed] [Google Scholar]
- 11.Dhanji H, Murphy NM, Akhigbe C, Doumith M, Hope R, Livermore DM, Woodford N. 2011. Isolation of fluoroquinolone-resistant O25b:H4-ST131 Escherichia coli with CTX-M-14 extended-spectrum ß-lactamase from UK river water. J. Antimicrob. Chemother. 66:512–516. 10.1093/jac/dkq472 [DOI] [PubMed] [Google Scholar]
- 12.Hong H, Chun J, Lee Y. 2004. Detection of extended-spectrum β-lactamase-producing, multidrug-resistant environmental isolates of Escherichia coli that bind to human bladder cells. Microb. Drug Resist. 10:184–189. 10.1089/1076629041310145 [DOI] [PubMed] [Google Scholar]
- 13.Guenther S, Ewers C, Wieler LH. 2011. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front. Microbiol. 2:246. 10.3389/fmicb.2011.00246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dierikx C, van der Goot J, Fabri T, van Essen-Zandbergen A, Smith H, Mevius D. 2013. Extended-spectrum-beta-lactamase- and AmpC-beta-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J. Antimicrob. Chemother. 68:60–67. 10.1093/jac/dks349 [DOI] [PubMed] [Google Scholar]
- 15.Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, Heck M, Savelkoul P, Vandenbroucke-Grauls C, van der Zwaluw K, Huijsdens X, Kluytmans J. 2011. Extended-spectrum beta-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg. Infect. Dis. 17:1216–1222. 10.3201/eid1707.110209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, Voets GM, van den Munckhof MP, van Essen-Zandbergen A, Platteel T, Fluit AC, van de Sande-Bruinsma N, Scharinga J, Bonten MJ, Mevius DJ. 2011. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 17:873–880. 10.1111/j.1469-0691.2011.03497.x [DOI] [PubMed] [Google Scholar]
- 17.Greenberg B. 1973. Flies and disease, vol II: biology and disease transmission. Princeton University Press, Princeton, NJ [Google Scholar]
- 18.De Jesus AJ, Olsen AR, Bryce JR, Whiting RC. 2004. Quantitative contamination and transfer of Escherichia coli from foods by houseflies, Musca domestica L. (Diptera: Muscidae). Int. J. Food Microbiol. 93:259–262. 10.1016/j.ijfoodmicro.2003.12.003 [DOI] [PubMed] [Google Scholar]
- 19.Forster M, Klimpel S, Mehlhorn H, Sievert K, Messler S, Pfeffer K. 2007. Pilot study on synanthropic flies (e.g. Musca, Sarcophaga, Calliphora, Fannia, Lucilia, Stomoxys) as vectors of pathogenic microorganisms. Parasitol. Res. 101:243–246. 10.1007/s00436-007-0522-y [DOI] [PubMed] [Google Scholar]
- 20.Forster M, Sievert K, Messler S, Klimpel S, Pfeffer K. 2009. Comprehensive study on the occurrence and distribution of pathogenic microorganisms carried by synanthropic flies caught at different rural locations in Germany. J. Med. Entomol. 46:1164–1166. 10.1603/033.046.0526 [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi M, Sasaki T, Saito N, Tamura K, Suzuki K, Watanabe H, Agui N. 1999. Houseflies: not simple mechanical vectors of enterohemorrhagic Escherichia coli O157:H7. Am. J. Trop. Med. Hyg. 61:625–629 [DOI] [PubMed] [Google Scholar]
- 22.Sasaki T, Kobayashi M, Agui N. 2000. Epidemiological potential of excretion and regurgitation by Musca domestica (Diptera: Muscidae) in the dissemination of Escherichia coli O157: H7 to food. J. Med. Entomol. 37:945–949. 10.1603/0022-2585-37.6.945 [DOI] [PubMed] [Google Scholar]
- 23.Pava-Ripoll M, Pearson RE, Miller AK, Ziobro GC. 2012. Prevalence and relative risk of Cronobacter spp., Salmonella spp., and Listeria monocytogenes associated with the body surfaces and guts of individual filth flies. Appl. Environ. Microbiol. 78:7891–7902. 10.1128/AEM.02195-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shane SM, Montrose MS, Harrington KS. 1985. Transmission of Campylobacter jejuni by the housefly (Musca domestica). Avian Dis. 29:384–391. 10.2307/1590499 [DOI] [PubMed] [Google Scholar]
- 25.Echeverria P, Harrison BA, Tirapat C, McFarland A. 1983. Flies as a source of enteric pathogens in a rural village in Thailand. Appl. Environ. Microbiol. 46:32–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fotedar R, Banerjee U, Samantray JC, Shriniwas 1992. Vector potential of hospital houseflies with special reference to Klebsiella species. Epidemiol. Infect. 109:143–147 [PMC free article] [PubMed] [Google Scholar]
- 27.Moriya K, Fujibayashi T, Yoshihara T, Matsuda A, Sumi N, Umezaki N, Kurahashi H, Agui N, Wada A, Watanabe H. 1999. Verotoxin-producing Escherichia coli O157:H7 carried by the housefly in Japan. Med. Vet. Entomol. 13:214–216. 10.1046/j.1365-2915.1999.00161.x [DOI] [PubMed] [Google Scholar]
- 28.Murvosh CM, Taggard CW. 1966. Ecological studies of the housefly. Ann. Entomol Soc. Am. 59:534–547 [DOI] [PubMed] [Google Scholar]
- 29.Nazni WA, Luke H, Wan Rozita WM, Abdullah AG, Sa'diyah I, Azahari AH, Zamree I, Tan SB, Lee HL, Sofian MA. 2005. Determination of the flight range and dispersal of the house fly, Musca domestica (L.) using mark release recapture technique. Trop. Biomed. 22:53–61 [PubMed] [Google Scholar]
- 30.Anonymous. 2001. International standard. ISO 16649-2. Microbiology of food and animal feeding stuffs: horizontal method for the enumeration of beta-glucuronidase-positive Escherichia coli. Part 2. Colony count technique at 44°C using 5-bromo-4-chloro-3-indolyl beta-d-glucuronide. International Organization for Standardization, Geneva, Switzerland [Google Scholar]
- 31.Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement M100-S20 CLSI, Wayne, PA [Google Scholar]
- 32.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- 33.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558. 10.1128/AEM.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Escobar-Paramo P, Le Menac'h A, Le Gall T, Amorin C, Gouriou S, Picard B, Skurnik D, Denamur E. 2006. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ. Microbiol. 8:1975–1984. 10.1111/j.1462-2920.2006.01077.x [DOI] [PubMed] [Google Scholar]
- 35.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deplano A, Denis O, Rodriguez-Villalobos H, De Ryck R, Struelens MJ, Hallin M. 2011. Controlled performance evaluation of the DiversiLab repetitive-sequence-based genotyping system for typing multidrug-resistant health care-associated bacterial pathogens. J. Clin. Microbiol. 49:3616–3620. 10.1128/JCM.00528-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dierikx CM, van Duijkeren E, Schoormans AH, van Essen-Zandbergen A, Veldman K, Kant A, Huijsdens XW, van der Zwaluw K, Wagenaar JA, Mevius DJ. 2012. Occurrence and characteristics of extended-spectrum-beta-lactamase- and AmpC-producing clinical isolates derived from companion animals and horses. J. Antimicrob. Chemother. 67:1368–1374. 10.1093/jac/dks049 [DOI] [PubMed] [Google Scholar]
- 38.Reuland EA, Overdevest IT, Al Naiemi N, Kalpoe JS, Rijnsburger MC, Raadsen SA, Ligtenberg-Burgman I, van der Zwaluw KW, Heck M, Savelkoul PH, Kluytmans JA, Vandenbroucke-Grauls CM. 2013. High prevalence of ESBL-producing Enterobacteriaceae carriage in Dutch community patients with gastrointestinal complaints. Clin. Microbiol. Infect. 19:542–549. 10.1111/j.1469-0691.2012.03947.x [DOI] [PubMed] [Google Scholar]
- 39.Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, Claeys G, Lontie M, Van Meensel B, Herman L, Haesebrouck F, Butaye P. 2010. Characterization of extended-spectrum beta-lactamases produced by Escherichia coli isolated from hospitalized and nonhospitalized patients: emergence of CTX-M-15-producing strains causing urinary tract infections. Microb. Drug Resist. 16:129–134. 10.1089/mdr.2009.0132 [DOI] [PubMed] [Google Scholar]
- 40.Schink AK, Kadlec K, Kaspar H, Mankertz J, Schwarz S. 2013. Analysis of extended-spectrum-beta-lactamase-producing Escherichia coli isolates collected in the GERM-Vet monitoring programme. J. Antimicrob. Chemother. 68:1741–1749. 10.1093/jac/dkt123 [DOI] [PubMed] [Google Scholar]
- 41.Lysyk TJ, Axtell RC. 1986. Movement and distribution of house flies (Diptera: Muscidae) between habitats in two livestock farms. J. Econ. Entomol. 79:993–998 [DOI] [PubMed] [Google Scholar]
- 42.Dolejska M, Duskova E, Rybarikova J, Janoszowska D, Roubalova E, Dibdakova K, Maceckova G, Kohoutova L, Literak I, Smola J, Cizek A. 2011. Plasmids carrying blaCTX-M-1 and qnr genes in Escherichia coli isolates from an equine clinic and a horseback riding centre. J. Antimicrob. Chemother. 66:757–764. 10.1093/jac/dkq500 [DOI] [PubMed] [Google Scholar]
- 43.Usui M, Iwasa T, Fukuda A, Sato T, Okubo T, Tamura Y. 2013. The role of flies in spreading the extended-spectrum beta-lactamase gene from cattle. Microb. Drug Resist. 19:415–420. 10.1089/mdr.2012.0251 [DOI] [PubMed] [Google Scholar]
- 44.Snary EL, Kelly LA, Davison HC, Teale CJ, Wooldridge M. 2004. Antimicrobial resistance: a microbial risk assessment perspective. J. Antimicrob. Chemother. 53:906–917. 10.1093/jac/dkh182 [DOI] [PubMed] [Google Scholar]
- 45.Heller J, Kelly L, Reid SW, Mellor DJ. 2010. Qualitative risk assessment of the acquisition of Meticillin-resistant staphylococcus aureus in pet dogs. Risk Anal. 30:458–472. 10.1111/j.1539-6924.2009.01342.x [DOI] [PubMed] [Google Scholar]


