Abstract
Typical plant aspartic protease zymogens comprise a characteristic and plant-specific insert (PSI). PSI domains can interact with membranes, and a role as a defensive weapon against pathogens has been proposed. However, the potential of PSIs as antimicrobial agents has not been fully investigated and explored yet due to problems in producing sufficient amounts of these domains in bacteria. Here, we report the development of an expression platform for the production of the PSI domain of cirsin in the generally regarded as safe (GRAS) yeast Kluyveromyces lactis. We successfully generated K. lactis transformants expressing and secreting significant amounts of correctly processed and glycosylated PSI, as well as its nonglycosylated mutant. A purification protocol with protein yields of ∼4.0 mg/liter was established for both wild-type and nonglycosylated PSIs, which represents the highest reported yield for a nontagged PSI domain. Subsequent bioactivity assays targeting phytopathogenic fungi indicated that the PSI of cirsin is produced in a biologically active form in K. lactis and provided clear evidence for its antifungal activity. This yeast expression system thereby emerges as a promising production platform for further exploring the biotechnological potential of these plant saposin-like proteins.
INTRODUCTION
Typical plant aspartic protease precursors comprise an internal domain of approximately 100 amino acids known as a plant-specific insert (PSI) that is not present in mammalian or microbial aspartic protease sequences. This PSI domain is structurally homologous to saposin-like proteins (SAPLIPs) (1–3), which are membrane-interacting proteins that have been linked with various physiological functions, such as lysosomal degradation of sphingolipids, antibacterial and antimicrobial activity, or regulation of the surface tension of pulmonary surfactant (4–6). Moreover, PSI domains were shown to interact with lipids. Their anticipated role in vacuolar sorting and their antipathogenic effects are suggested to be dependent on protein-membrane interactions (2, 7–9). Egas and coworkers demonstrated that the recombinant PSI from cardosin A is able to interact with phospholipid vesicles and to induce leakage of their contents in a pH- and lipid-dependent manner (8). Moreover, it was reported that recombinant PSI of Solanum tuberosum aspartic protease (StAP) is able to kill bacterial pathogens and to inhibit sporulation of phytopathogenic fungi through interaction with microbial membranes (9). Recently, Bryksa and coworkers (10) determined the structure of the isolated recombinant StAP PSI domain and demonstrated its fusogenic activity via pH-dependent membrane interactions. The two available crystal structures of PSI domains confirm their saposin fold, with phytepsin PSI displaying a “closed” fold in contrast to the “open” structure of StAP PSI (2, 10). Like other SAPLIPs (4), PSI domains contain six conserved cysteine residues engaged in three disulfide bonds, and a large majority also comprise a putative N-glycosylation site. Despite these conserved features, PSI domains are very diverse in their sequences, with amino acid identities ranging from 25 to 85%. This variety among PSIs in sequence and potential for posttranslational modification suggests that various mechanisms of membrane interaction and bioactivity/selectivity could apply, which is in line with what has been proposed for other SAPLIP family members with antimicrobial and/or cytolytic activity (4).
Fast-emerging resistance against conventional antibiotics, combined with the emergence of “superbugs,” has triggered an intensive search for alternative strategies to fight such resistant bacteria (11). Among the newly proposed antibiotics are antimicrobial peptides (AMPs), which might profit from high effectiveness, safety, and reduced levels of cross-resistance due to their mode of action (11–13). Structural similarities of PSI domains with SAPLIPs (2, 10), in combination with their ability to interact with and permeabilize phospholipid membranes (8, 10), resulting in antimicrobial activity (e.g., StAP PSI [9]), suggest a promising role for PSI domains as novel antibiotics.
So far, one of the major obstacles to further investigation of PSI domains and other AMPs (11) as effective alternatives to antibiotics is the high cost of production. To date, PSI domains have only been produced using prokaryotic systems, such as Escherichia coli, giving only very low yields of nonglycosylated material (8, 9). Increasing the production levels of such proteins has been a major challenge so far, with Bryksa and collaborators reporting their best results by expressing the StAP PSI fused to a solubility enhancement tag (10). These results illustrate the current limitations of expressing PSI domains in prokaryotic systems. They call for novel approaches to overcome this shortage in protein production in order to further explore the potential application of PSI domains as antimicrobial agents.
In this work, we report the development and optimization of a yeast (Kluyveromyces lactis)-based expression system for the production of the PSI domain of cirsin, an aspartic protease from Cirsium vulgare (14). We demonstrate that this generally regarded as safe (GRAS) yeast is an effective platform for producing good amounts of the cirsin PSI domain, in glycosylated as well as nonglycosylated forms. Furthermore, bioactivity assays run against different phytopathogenic fungi confirm that the cirsin PSI is produced in a biologically active form in K. lactis. These results provide evidence that the cirsin PSI acts as an antimicrobial agent with a potential role in pest control.
MATERIALS AND METHODS
Design of cirsin PSI constructs.
The sequence encoding the PSI domain of cirsin was amplified by PCR using the full-length precursor cDNA sequence (JN703462) as the template and the primers 5′-CTCGAGAAAAGAGTCATGAGCCAGCAATGC-3′ (forward) and 5′-AGATCTTTAATGATGATGATGATGATGACTGGGTAAGCGATCACAC-3′ (reverse). The forward primer introduced an XhoI restriction site (underlined) and a sequence encoding the Kex protease processing site (italics), in order to generate an in-frame fusion between the α mating factor (α-MF) secretion leader sequence present in the expression vector pKLAC1 (New England BioLabs [NEB]) and the PSI sequence. The reverse primer introduced a BglII restriction site (underlined), the sequence encoding a hexahistidine C-terminal tag (boldface), and a stop codon (italic). The resulting product of approximately 350 bp was cloned into the pGEM-T Easy vector (Promega), and plasmid DNA isolated from positive clones was analyzed by XhoI/BglII double digestion. The purified digestion product was then subcloned into the pKLAC1 expression vector (New England BioLabs) to obtain the construct α-MF_PSI(His)6.
The QuickChange site-directed mutagenesis kit (Stratagene) was used to generate a cirsin PSI glycosylation site mutant (N86S) in pKLAC1 using the following primers: 5′-GGATGCAAAACCAAATCAAACGATCTGAGACTGAAGATAACATAATC-3′ (forward primer) and 5′-GATTATGTTATCTTCAGTCTCAGATCGTTTGATTTGGTTTTGCATCC-3′ (reverse primer), where mutations are underlined [construct α-MF_PSIN86S(His)6]. Two additional PSI constructs were generated by site-directed mutagenesis (QuickChange site-directed mutagenesis kit), consisting only of the signal peptide (SP) of the α-MF secretion leader sequence in frame with PSI(His)6- and PSI(N86S)(His)6-encoding sequences, through deletion of the sequence corresponding to the propeptide of α-MF. The pKLAC1 constructs α-MF_PSI(His)6 and α-MF_PSI(N86S)(His)6 were used as templates in the deletion mutagenesis step using the primers 5′-CCGTTGTTATGGCTGCTCCACTCGAGGTCATGAGCCAGCAATGCAAAAC-3′ (forward primer) and 5′-GTTTTGCATTGCTGCCTCATGACCTCGAGTGGAGCAGCCATAACAACGG-3′ (reverse primer), yielding the pKLAC1 constructs SP_PSI(His)6 and SP_PSIN86S(His)6, respectively. A set of constructs comprising a tobacco etch virus (TEV) cleavage site upstream of the His tag was also generated to allow tag removal upon purification. The TEV recognition site was inserted by mutagenesis using the forward primer 5′-TGTGATCGCTTACCCAGTGAAAACTTGTATTTCCAAGGTCATCATCATCATCATCAT-3′ and the reverse primer 5′-ATGATGATGATGATGATGACCTTGGAAATACAAGTTTTCACTGGGTAAGCGATCACA-3′ (the TEV cleavage site is in boldface), yielding the pKLAC1 constructs SP_PSI_TEV(His)6 and SP_PSIN86S_TEV(His)6, respectively. E. coli strain TOP10F′ (Invitrogen) was used in all cloning steps. All constructs were confirmed by automated DNA sequencing.
K. lactis transformation and screening of transformants.
The K. lactis GG799 commercial strain (New England BioLabs) was used as the host for PSI expression. For preparation of K. lactis electrocompetent cells, GG799 cells were grown in YPGlu medium (1% yeast extract, 2% Bacto peptone [YP], 2% glucose) at 30°C, with shaking, until the culture reached an optical density at 600 nm (OD600) of 1. The cells were then pelleted at 1,000 × g for 5 min at 4°C and washed twice with cold sterile H2O. After centrifugation, the cells were washed and then resuspended in 1 M sorbitol (cold and sterile) and kept in ice until transformation. Before yeast transformation, expression cassettes were obtained by linearizing all pKLAC1-PSI constructs with SacII, according to the manufacturers' instructions (K. lactis Protein Expression kit; New England BioLabs). K. lactis competent cells were then incubated with purified DNA from each expression cassette (2 μg DNA) and transformed using a GenePulser Xcell electroporation system (Bio-Rad) under the following conditions: 1.5 kV, 25 μF, and 200 Ω. The cells were then allowed to recover by incubation for 30 min at 30°C with agitation in YPGlu medium and plated for selection in yeast carbon base (YCB) agar medium plates (3% 1 M sodium phosphate buffer, pH 7.0, 1.17% YCB medium supplied with the K. lactis Protein Expression kit [New England BioLabs], and 2% agar) containing 0.005 M acetamide for 4 days at 30°C. Transformants were screened by colony PCR using specific primers for the PSI sequence (5′-CAGTCATGAGCCAGCAATGC-3′ [forward primer] and 5′-GGGTAAGCGATCACACAGC-3′ [reverse primer]), followed by agarose gel electrophoresis. Small-scale expression assays were then performed to evaluate protein expression levels. For this screening, K. lactis transformants were grown in 10 ml YP medium containing 4% galactose (YPGal). At selected time points, culture medium samples were analyzed by Western blotting with an anti-His antibody.
Protein deglycosylation assays.
Culture medium enriched in the different forms of recombinant PSI—α-MF_PSI(His)6, α-MF_PSIN86S(His)6, and SP_PSI(His)6—were subjected to deglycosylation assays using two different commercial glycosidases, PNGase F and endo-β-N-acetylglucosaminidase H (endo H) (New England BioLabs), according to the manufacturers' instructions with minor modifications. Glycoprotein-denaturing buffer (0.5% SDS, 0.04 M dithiothreitol [DTT]) was combined with 36 μl of culture medium aliquots and heated at 100°C for 10 min. In the treatment with PNGase F, samples were further supplemented with 0.05 M sodium phosphate buffer, pH 7.5 (G7 reaction buffer; NEB), 1% NP-40, and 250 units of glycosidase, whereas for endo H treatment, 0.05 M sodium citrate buffer, pH 5.5 (G5 reaction buffer), and 250 units of endo H were added to the heat-denatured samples. The total reaction volumes were supplemented with MilliQ water to a final volume of 60 μl, and the mixture was incubated at 37°C for 5 h. Samples were then analyzed by Western blotting.
Production and purification of recombinant PSI fusion proteins.
Selected K. lactis recombinant clones were grown for protein purification by inoculating a starter culture (overnight growth at 30°C in 50 ml YP medium containing 2% glucose) into 300 ml YPGal medium (4% galactose) to an initial OD600 of 0.2 and were allowed to grow at 30°C for 24 or 72 h, depending on the expressed PSI construct. After expression, the following purification procedure was used for both glycosylated and nonglycosylated forms of recombinant cirsin PSI. Cells were harvested by centrifugation at 6,000 × g for 20 min, and the supernatant was filtered sequentially through 0.8- and 0.2-μm cellulose acetate filters (Whatman). The filtered supernatant was then precipitated with ammonium sulfate (90% saturation) at 4°C for 90 min and centrifuged at 10,000 × g (20 min). The protein pellet was resuspended in 0.02 M sodium phosphate buffer, pH 7.5, with 0.1 M NaCl, and high-molecular-weight protein aggregates and insoluble impurities were removed by ultracentrifugation at 144,000 × g for 20 min at 4°C. The resulting supernatant was applied to a HiLoad 26/60 Superdex 200 gel filtration column (GE Healthcare Life Sciences) connected to a fast protein liquid chromatography (FPLC) system (DuoFlow-Bio-Rad) and equilibrated in 0.02 M sodium phosphate buffer, pH 7.5, containing 0.1 M NaCl at a flow rate of 2.0 ml/min. The protein fractions were pooled and diluted to 0.02 M sodium phosphate buffer, pH 7.5, with 0.5 M sodium chloride and 0.01 M imidazole and applied to an Ni2+ affinity chromatography His Trap HP 1-ml column (GE Healthcare Life Sciences) connected to a low-pressure chromatographic system (BioLogic-Bio-Rad) previously equilibrated in the same buffer. The affinity chromatography was carried out at a flow rate of 0.75 ml/min, and the elution of proteins was performed in three sequential steps of increasing imidazole concentrations (0.05, 0.1, and 0.5 M imidazole) in the same buffer. Pooled fractions were further dialyzed overnight at 4°C against 0.02 M sodium phosphate buffer, pH 7.5, with 0.1 M NaCl. All the fractions collected were analyzed by SDS-PAGE and Western blotting. TEV cleavage assays were carried out by incubating purified nonglycosylated PSI with recombinant His-tagged TEV protease at a 1:10 molar ratio in the presence of 0.001 M EDTA and 3 mM glutathione-0.3 mM oxidized glutathione at 4°C overnight. A sample was diluted in 0.02 M sodium phosphate buffer, pH 7.5, containing 0.5 M NaCl, 0.01 M imidazole and applied to an Ni2+ affinity chromatography His Trap HP 1-ml column (GE Healthcare Life Sciences) for TEV protease removal. Flowthrough containing PSI was then dialyzed overnight at 4°C against 0.02 M sodium phosphate buffer, pH 7.5, with 0.1 M NaCl. Protein was quantified using a Thermo Scientific Pierce BCA Protein Assay Kit (Pierce) according to the manufacturers' instructions.
Analytical size exclusion chromatography.
The oligomeric status of purified recombinant nonglycosylated cirsin PSI was evaluated by analytical size exclusion chromatography under nondenaturing conditions on a Discovery Bio GFC 300 column (Supelco Analytical) connected to a high-performance liquid chromatography (HPLC) system (Prominence system; Shimadzu Corporation). The column was equilibrated in 0.02 M sodium phosphate, pH 7.5, containing 0.15 M NaCl at a flow rate of 1 ml/min and calibrated with Gel Filtration LMW and HMW calibration kits (GE Healthcare Life Sciences) according to the manufacturer's instructions. The molecular mass markers used for calibration were thyroglobulin (669 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), and RNase (13.7 kDa).
In vitro Kex cleavage assay.
α-MF_PSI(His)6 expression fractions (37 μl) were incubated overnight at 37°C with 1 μg Kex-2 (PeproTech) and 40 μl 0.2 M Tris-HCl buffer, pH 7.2, containing 0.002 M CaCl2. After incubation, samples were analyzed by Western blotting with an anti-His antibody.
Polyacrylamide gel electrophoresis and Western blotting.
Protein samples collected along with expression and purification were analyzed by SDS-PAGE using 12.5% polyacrylamide gels in a Bio-Rad Mini-Protean 3 Cell. Samples were treated with loading buffer (0.35 M Tris-HCl–0.28% SDS buffer, pH 6.8, 30% glycerol, 10% SDS, 0.6 M DTT, 0.012% bromophenol blue) at 95°C for 10 min before loading. The gels were stained with Coomassie brilliant blue R-250 (Sigma). For protein immunodetection, samples were separated by SDS-PAGE (12.5% gels) and transferred to a polyvinylidene difluoride membrane for immunoblotting (40 V overnight at 16°C). The membranes were blocked for 60 min with 5% nonfat dry milk in Tris-buffered saline (TBS) containing 0.1% Tween 20 and then incubated at room temperature for 60 min with His-tagged mouse antibody (GenScript; 1:10,000 dilution). After several washes with 0.5% nonfat dry milk plus 0.1% Tween 20 in TBS, the membranes were incubated at room temperature for 60 min with alkaline phosphatase-conjugated goat anti-mouse IgG plus IgM (GE Healthcare; 1:10,000 dilution). The membranes were washed again in TBS containing 0.1% Tween 20, and alkaline phosphatase activity was visualized by the enhanced chemifluorescence method using ECF substrate (GE Healthcare) on a Molecular Imager FX System (Bio-Rad).
Peptide mass fingerprinting by MALDI-TOF MS.
Electrophoretically homogeneous protein bands were excised from polyacrylamide gels, and in situ tryptic digestion was performed as follows. Samples were first reduced and carboamidomethylated with 10 mM DTT and 50 mM iodoacetamide and then were digested with 50 ng of trypsin at 37°C overnight. The tryptic digests were analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), and the spectra were acquired on a Bruker ultrafleXtrem spectrometer equipped with a Smartbeam-II laser in reflectron positive mode using a matrix of α-cyano-4-hydroxycinnamic acid (HCCA). The calibration standard mixture consisted of bradykinin (amino acids 1 to 7 [fragment 1-7]) (m/z 757.39916), angiotensin II (m/z 1,046.5418), angiotensin (m/z 1,296.6848), substance P (m/z 1,347.7354), bombesin (m/z 1,619.8223), renin substrate (m/z 1,758.93261), adrenocorticotropic hormone (ACTH) fragment 1-17 (m/z 2,093.0862), ATCH corticotropin-like intermediate lobe peptide (ACTH clip) fragment 18-39 (m/z 2,465.1983), and somatostatin 28 (m/z 3,147.4710). Theoretical mass maps of translated sequences were obtained with the software Sequence Editor 3.2 (Bruker Daltonics, Biotools 3.2), and the experimental and theoretical peptide matches were selected within a mass tolerance of 0.6 Da.
Antifungal assays.
Bioassay confrontation tests between K. lactis cells and fungi were conducted in petri dishes (10 cm in diameter) with YPGal medium at pH 5.0 containing 4% galactose and 1.5% agar. The antifungal activity of the SP_PSI(His)6 K. lactis transformant toward four different environmental phytopathogenic fungi was assessed: Botryosphaeria obtusa, Lewia infectoria, Alternaria alternata, and Drechslera biseptata. Untransformed K. lactis strain GG799 and a positive K. lactis transformant secreting green fluorescent protein (GFP) into the culture medium were used as negative controls. Yeast cells were inoculated near the edge of the plate and grown to a diameter of 1 cm; the fungal inoculum was placed on the opposite side of the plate at a constant distance from yeast cells in all assays. After 10 days of incubation at 30°C, confrontations were monitored and scored as a reduction in the growth of the fungus compared with cultures grown in the absence of yeast cells. Antifungal activity was expressed as a percentage of mycelial growth inhibition (MGI) as follows: MGI (%) = [(growth of the fungi in the absence of yeast − growth of the fungi in the presence of yeast)/(growth of fungi in the absence of yeast)] × 100. Assays were done in duplicate. The antifungal activity of purified recombinant nonglycosylated PSI [SP_PSIN86S(His)6] against A. alternata was also evaluated by a disk diffusion test. Sterile disks (Fluka) impregnated with purified PSI (0.47 mg/ml) were applied to potato dextrose broth agar (PDA) plates (1.5% agar) at two different pH values (pH 5.0 and 6.8). PSI-impregnated disks were placed near the edge of the plate, while the fungal inoculum was placed on the opposite side of the plate at a constant distance from the disk in all assays. After 7 days of incubation at 30°C, the growth-inhibitory effect was monitored and scored as a reduction in fungal growth compared with cultures grown in the presence of sample buffer (0.025 M sodium phosphate buffer, pH 7.5, containing 0.15 M NaCl). Antifungal activity was expressed as the percentage of mycelial growth inhibition as follows: MGI (%) = [(growth of the fungi in the presence of sample buffer − growth of the fungi in the presence of PSI)/(growth of fungi in the presence of sample buffer)] × 100. Commercial antifungal amphotericin B (0.25 mg/ml) was used as a positive control. Disk diffusion assays were performed in triplicate.
RESULTS
Cirsin PSI sequence analysis.
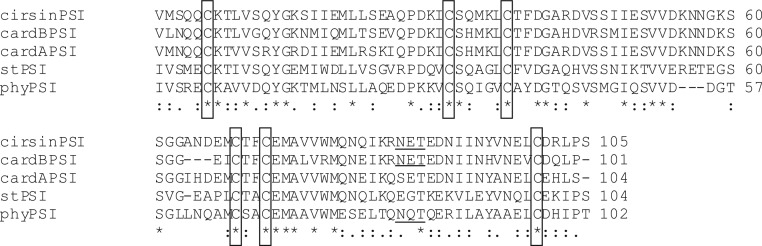
Cirsin, a plant aspartic protease from C. vulgare, was recently identified and characterized (14). Like many other typical plant aspartic proteases, it comprises a sequence with 105 amino acids typically named the PSI domain. The cirsin PSI domain bears a high degree of sequence conservation compared to other relevant plant PSI domains (Fig. 1). The highest identities were found with the PSI sequences of cardosin A and cardosin B (>80%) from Cynara cardunculus, whereas significantly lower sequence conservation was found for PSI domains from S. tuberosum AP (50%), and phytepsin (45%) from Hordeum vulgare. The cirsin PSI sequence contains the characteristic pattern of cysteine residues, as well as a putative glycosylation site. Although this glycosylation site is widely conserved and glycosylation might be relevant for proper location and/or functionality, no glycosylated PSI has ever been produced in heterologous hosts so far.
FIG 1.
Amino acid sequence alignment of the cirsin PSI domain with PSI domains from other typical plant aspartic proteases. Sequences retrieved from databases were automatically aligned using the CLUSTAL W algorithm. Conserved cysteines are boxed, identical residues are marked with asterisks, and gaps are indicated by dashes. Putative glycosylation sites were predicted using NetNGlyc 1.0, and the Asn-X-Thr sequons in the sequences are underlined. The PSI amino acid sequences were all obtained from the following reported sequences: cirsinPSI [cirsin, Cirsium vulgare (Savi) Ten.; GenBank accession no. JN703462], cardBPSI (cardosin B, Cynara cardunculus; GenBank accession no. CAB40349.1), cardAPSI (cardosin A, Cynara cardunculus; GenBank accession no. CAB40134.1), stPSI (Solanum tuberosum aspartic protease; GenBank accession no. AY672651), and phyPSI (phytepsin, Hordeum vulgarae; GenBank accession no. X56136). Colons indicate conservation between amino acids with strongly similar properties, and periods indicate conservation between amino acids with weakly similar properties.
Cirsin PSI fusion to the α-MF leader sequence is directed to secretion but not effectively processed in K. lactis.
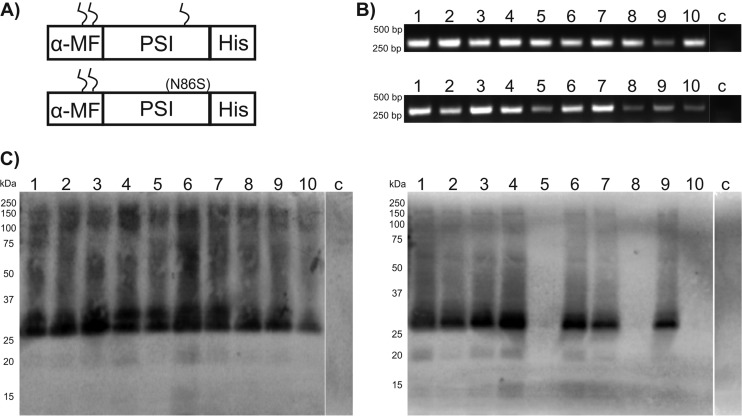
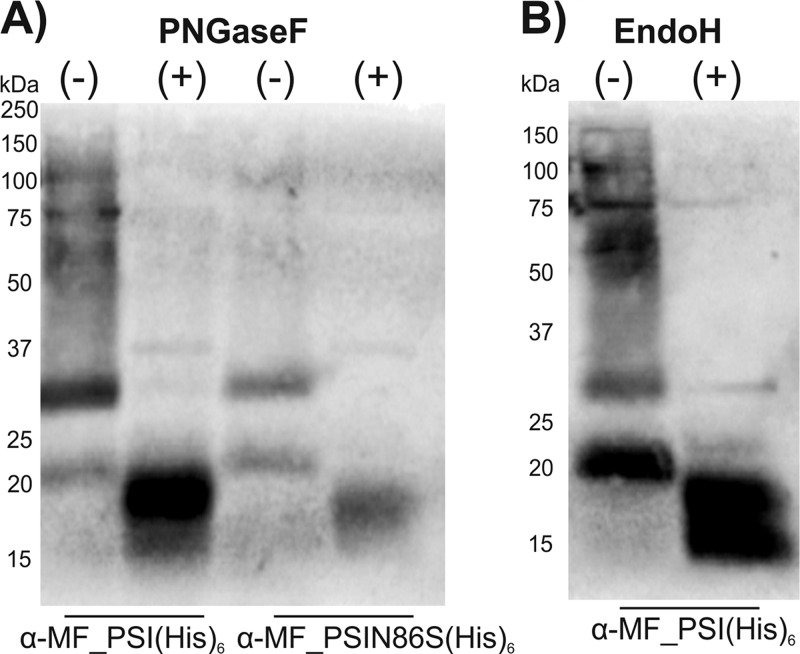
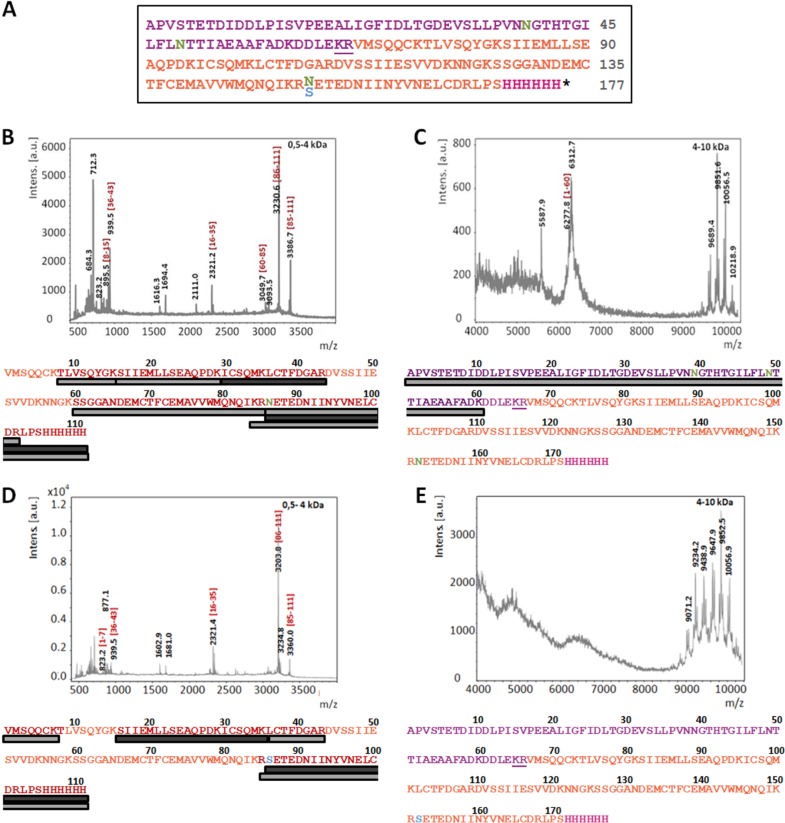
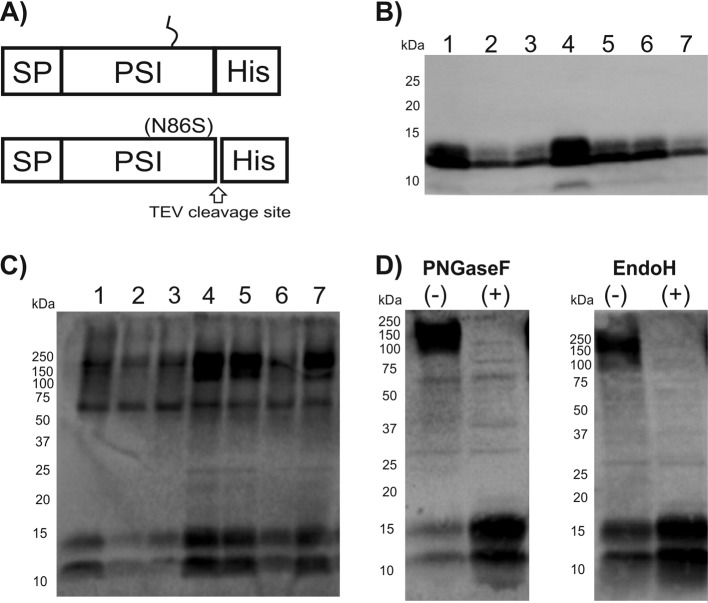
So far, recombinant PSI domains have been produced only in E. coli (8–10). The low yields reported to be obtained clearly indicate that this bacterial expression system is only suboptimally suited for expression of PSIs, as well as other AMPs (15, 16). In order to establish a more effective expression system for PSI domains (glycosylated and nonglycosylated) with improved production yields, we selected the GRAS yeast K. lactis as the tool of choice. Expression in this yeast strain is driven by a mutant form of the K. lactis LAC4 promoter (PLAC4-PBI) located in the vector pKLAC1 (NEB), which contains a fungal acetamidase gene for positive selection in K. lactis. Expression of acetamidase by yeast cells containing an integrated expression cassette allows growth on nitrogen-free minimal medium containing acetamide as the only nitrogen source (K. lactis Protein Expression kit; NEB). In our initial attempt, we intended to use the α-MF leader sequence present in pKLAC1 to drive protein transport and secretion through the yeast secretory pathway. The nucleotide sequence encoding cirsin PSI was modified with a C-terminal His tag and cloned into pKLAC1 downstream of the α-MF sequence, which includes a Kex-2 protease recognition site immediately upstream of the PSI N terminus [construct α-MF_PSI(His)6]. Since the cirsin PSI sequence includes a putative glycosylation site (NET motif), we wanted to address the relevance of this posttranslational modification to protein expression yields and bioactivity. Therefore, besides a clone for the PSI wild type (wt), we also generated a PSI mutant lacking the putative glycosylation site (Fig. 2A). Both expression cassettes, (i) α-MF_PSI(His)6 and (ii) α-MF_PSI(N86S)(His)6, were used to transform K. lactis strain GG799. Ten transformants were randomly selected for each construct, and integration of the expression cassette into the yeast genome was confirmed by PCR amplification (Fig. 2B). Assuming that protein expression levels might depend on the number of integrated cassettes, protein secretion was evaluated using small-scale expression assays, followed by Western blotting of culture supernatants with an anti-His antibody. Figure 2C illustrates expression profiles of K. lactis transformants for wt and mutant PSI(N86S) observed after 48 h of expression. Good levels of protein secretion were observed for all PSI wt constructs and 7 out of 10 transformants of the PSI(N86S) mutant. No protein expression was observed for the remaining three constructs. Although both forms of PSI appear to be effectively secreted into the culture medium, analysis of the immunostained bands for the PSI(N86S) mutant via electrophoretic mobility revealed significant accumulation of a protein of 25 kDa. This mass is clearly higher than the expected molecular mass for the nonglycosylated PSI(N86S) mutant (∼12.5 kDa). We ascribed the higher mass to additional posttranslational modifications present in the resulting protein, probably due to incomplete cleavage of the α-MF–PSI fusion proteins. Indeed, analysis of the α-MF leader sequence from K. lactis revealed the presence of two putative N-glycosylation sites, in line with previously described results for α-MF from Saccharomyces cerevisiae (17). To confirm this hypothesis, deglycosylation was performed for wt, as well as N86S-mutated, PSI by treating the respective culture supernatants with PNGase F, leading to proteins of ∼20 kDa (instead of 12.5 kDa) in both cases (Fig. 3A). These results clearly confirmed the presence of glycosylation in wt PSI, as well as the PSI(N86S) mutant. In parallel, deglycosylation of recombinant wt PSI with endo H was performed (Fig. 3B). The results clearly proved that the protein was sensitive to this glycosidase, as well, indicating that the smeared pattern observed may result from underglycosylated forms of PSI. To further confirm the presence of residual α-MF in the secreted protein, we performed peptide mass fingerprinting (PMF) on wt PSI and PSI(N86S). It was possible to identify the full-length α-MF–PSI(His)6 sequence (Fig. 4B and C), as well as nonglycosylated wt PSI, thereby indicating that a fraction of expressed protein is not glycosylated (Fig. 4B). A peak of 6,277.87 Da (Fig. 4C) indicated the presence of full-length nonglycosylated α-MF. From 9 kDa, a set of peaks with a difference of mass of 162 Da could be assigned to the α-MF domain heterogeneous glycosylation profile. The presence of the N86S mutation, as well as incomplete removal of the α-MF prodomain from the secreted protein, could be deduced from spectra shown in Fig. 4D and E. In summary, these spectra indicate that both recombinant PSI constructs (wt and N86S) were not correctly processed by Kex-2 before secretion.
FIG 2.
Cirsin PSI fused to the α-MF leader sequence is efficiently integrated into the K. lactis genome and secreted into culture medium. (A) Schematic representation of the α-MF_PSI(His)6 (top) and α-MF_PSI(N86S)(His)6 (bottom) fusion proteins. Both sequences comprise the α-MF preprodomain sequence and a hexahistidine tag. The two putative N-glycosylation sites in the α-MF leader sequence (NGT and NTT) are indicated. A putative N-glycosylation site (NET) is present in the α-MF_PSI(His)6 sequence, while in the construct α-MF_PSI(N86S)(His)6, this glycosylation site is mutated (N86S). (B) Confirmation by PCR of integration of the expression cassettes into the K. lactis genome. The numbers correspond to the selected clones and “c” to the amplification product using genomic DNA from untransformed K. lactis strain GG799 (negative control). (Top) α-MF_PSI(His)6. (Bottom) α-MF_PSI(N86S)(His)6. (C) Western blot analysis of recombinant cirsin PSI expression/secretion into culture media with an anti-His antibody. The numbers above the gels correspond to selected clones shown in panel B, and the lane marked with a c corresponds to the analysis of the culture medium of untransformed K. lactis GG799 cells (negative control). (Left) α-MF_PSI(His)6 transformants. (Right) α-MF_PSI(N86S)(His)6 transformants.
FIG 3.
Recombinant cirsin PSI fused to the α-MF leader sequence is secreted in a unprocessed form. (A) Western blot analysis of culture supernatants of selected transformants for α-MF_PSI(His)6 and α-MF_PSI(N86S)(His)6 treated with PNGase F using an anti-His antibody. (B) Western blot analysis of culture supernatant enriched in recombinant wt PSI [α-MF_PSI(His)6] subjected to deglycosylation with endo H using an anti-His antibody. +, presence, and −, absence of glycosidase.
FIG 4.
PMF of α-MF_PSI(His)6 and α-MF_PSI(N86S)(His)6. (A) Sequence considered in protein analysis. The sequences include the α-MF (purple), PSI domain (orange), XhoI site (LE), and Kex-2 cleavage site (underlined). The polyhistidine tail (dark pink), putative N-glycosylation sites (green), and N86S mutation (blue) are also indicated. (B and C) MALDI-TOF mass spectra of tryptic digests from purified α-MF-PSI(His)6 (∼25-kDa band); m/z values of prominent peaks are indicated. (D and E) MALDI-TOF mass spectra of tryptic digests from purified α-MF_PSI(N86S)(His)6 (∼25-kDa band); m/z values of prominent peaks are indicated. The spectra in panels C and E were acquired in a window of mass/charge ratios of 400 to 4,000 m/z; spectra of panel D and E were acquired in a window of 4,000 to 10,000 m/z. In each spectrum, bars under the sequences indicate the peak intensity (in greyscale) and sequence coverage of peptide fragments.
In order to evaluate the reason for incomplete processing of the two fusion proteins by Kex-2 in vivo, we performed in vitro assays with commercial Kex-2 protease on natural, as well as denatured, recombinant wt PSI. These in vitro assays showed cleavage of the fusion protein (data not shown), suggesting that the Kex-2 cleavage site is accessible to the enzyme, although cleavage appears to be compromised for protein in yeast. Incomplete processing of the α-MF prodomain can be caused either by hindered substrate recognition by Kex-2 in vivo, by low cleavage efficiency, or by insufficient amounts of Kex-2 present to handle the complete amount of overexpressed fusion protein. The necessary postprocessing for secreted PSI fusion proteins increases the number of production steps and costs, which led us to elaborate a novel production strategy.
The signal peptide of the α-MF domain is sufficient to achieve secretion of cirsin PSI.
In order to streamline production of recombinant cirsin PSI in K. lactis, we deleted the prodomain region of the α-MF leader sequence and kept only the SP of this leading peptide. As a consequence, recombinant PSI should be directed to the endoplasmic reticulum (ER) and secreted through the secretory pathway independent of the α-MF prodomain. In addition, we created a set of constructs with an inserted TEV cleavage site just upstream of the hexahistidine tag. The constructs were named SP_PSI(His)6, SP_PSI(N86S)(His)6, SP_PSI_TEV(His)6, and SP_PSI(N86S)_TEV(His)6. The corresponding expression cassettes were linearized and used to transform K. lactis strain GG799. A total of 7 clones of each construct were selected by colony PCR and analyzed for protein secretion at selected times of expression. The SP_PSI_TEV(His)6 clone produced only small amounts of protein and was discarded. The inclusion of the additional TEV cleavage site on the glycosylation mutant had only a small effect on protein expression levels, and therefore, we selected the SP_PSI(N86S)_TEV(His)6 and SP_PSI(His)6 constructs for further analysis (Fig. 5A). Western blot analysis of culture supernatants of SP_PSI(N86S)_TEV(His)6 and SP_PSI(His)6 transformants was performed with anti-His antibody after 24 h and 72 h of expression, respectively (Fig. 5B and C). Interestingly, the nonglycosylated mutant SP_PSI(N86S)_TEV(His)6 required only 24 h for accumulation of recombinant PSI in the culture medium. Moreover, and in contrast to the α-MF–PSI(N86S) clones, a protein in accordance with the calculated mass (∼12.5 kDa) of PSI was detected (cf. Fig. 3A and 5B). A second band with lower molecular mass was also observed. This most likely derives from a PSI with alternative N-terminal processing or degradation by exoproteinases.
FIG 5.
The signal peptide in the α-MF domain is sufficient to drive secretion of recombinant cirsin PSI. (A) Schematic representation of SP_PSI(His)6 (top) and SP_PSI(N86S)_TEV(His)6 (bottom) fusion proteins. Both constructs contain the signal peptide (SP) of the α-MF domain and a hexahistidine tag. The SP_PSI(His)6 putative glycosylation site (NET) is indicated; in SP_PSI(N86S)_TEV(His)6, this glycosylation site is mutated. The glycosylation mutant also contains a TEV protease cleavage site. (B and C) Analysis of recombinant PSI expression by Western blotting of culture supernatants with an anti-His antibody. (B) SP_PSI(N86S)_TEV(His)6. (C) SP_PSI(His)6. (D) Protein deglycosylation assays. The culture supernatant of a representative transformant for SP_PSI(His)6 was treated with PNGase F and endo H and analyzed by Western blotting with an anti-His antibody. +, presence, and −, absence of glycosidase.
Protein derived from the SP_PSI(His)6 construct showed a more homogeneous glycosylation pattern in the Western blot (Fig. 5C), confirming posttranslational modification of recombinant PSI before secretion. A second band at lower molecular mass emerging from nonglycosylated protein could also be detected, which is in line with previous reports describing the production of glycosylated, as well as nonglycosylated, protein in yeast (18–20). Glycosylated PSI from SP_PSI(His)6 was also treated with PNGase F and endo H. Immunoblots indicated that PSI deglycosylated with these two enzymes yielded protein running identically to the one found in the two lower-molecular-mass bands (Fig. 5D). This is strong evidence that these two bands indeed correspond to nonglycosylated forms with different N-terminal processing. The sensitivity of this glycoprotein to endo H is consistent with a high-mannose-type N-linked glycosylation.
In summary, these results corroborate the idea that the signal peptide of α-MF is sufficient to drive secretion and processing of glycosylated, as well as nonglycosylated, recombinant cirsin PSI in K. lactis.
Purification of glycosylated and nonglycosylated recombinant cirsin PSI.
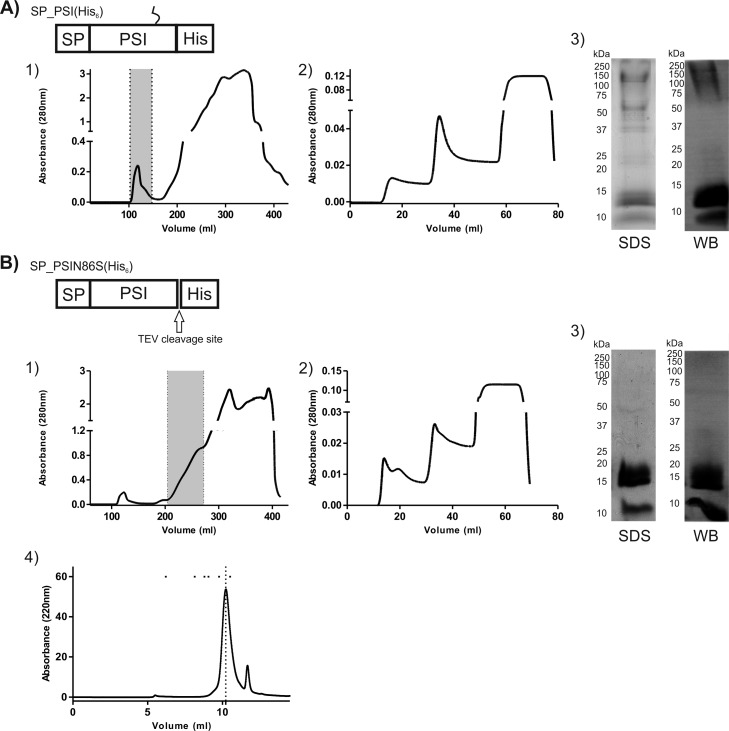
After establishing an efficient method for production of cirsin PSI in K. lactis, the purification protocol was optimized for glycosylated and nonglycosylated recombinant PSI. Transformants with higher expression levels were selected, and protein expression was performed in YPGal medium with 4% galactose for 24 h [SP_PSI(N86S)_TEV(His)6] or 72 h [SP_PSI(His)6]. For both constructs, the total soluble protein in the culture medium was concentrated by precipitation with ammonium sulfate before loading on a HiLoad 26/60 Superdex 200 column. This step was critical to remove medium components that could interfere with binding of the His-tagged protein to the column (immobilized metal ion affinity chromatography [IMAC]). The eluted samples were analyzed by Western blotting, and fractions enriched in recombinant PSI were pooled (Fig. 6A1 and B1). As expected, different elution volumes were obtained for proteins with different glycosylation states. The size exclusion eluates were then applied to a His Trap HP column (Fig. 6A2 and B2), and the His-tagged immunodetected fractions were pooled and dialyzed to remove imidazole. SDS-PAGE and Western blot analysis of purified fractions confirmed the effectiveness of the purification protocol (Fig. 6A3 and B3). Protein yields of 4.33 mg/liter and 4.67 mg/liter were obtained for glycosylated and nonglycosylated recombinant cirsin PSI, respectively. Interestingly, a small amount of nonglycosylated PSI could not be separated from the glycosylated material with this purification procedure. In order to assess the effect of the His tag on the purity and yield of the final PSI protein, we removed the His tag via TEV proteinase in the nonglycosylated PSIN86S mutant bearing a TEV cleavage site. After protein cleavage with TEV protease, the resulting mixture was applied to a His Trap HP column in order to remove the His tag and TEV protease, and the flowthrough comprising PSI was dialyzed and quantified. About 80% of recombinant PSI could be recovered in this way, confirming the effectiveness of the process (data not shown). Size exclusion chromatography under nondenaturing conditions and in the absence of a reducing agent revealed the vast majority of nonglycosylated cirsin PSI domain to be present as a dimer (Fig. 6B4).
FIG 6.
Purification of both glycosylated and nonglycosylated recombinant cirsin PSI. (A and B) Wild-type cirsin PSI [SP_PSI(His)6] and the glycosylation mutant [SP_PSI(N86S)_TEV_(His)6] were expressed for 24 and 72 h, respectively. The total protein content in the culture medium was concentrated, and recombinant proteins were purified by two chromatographic steps: (1) size exclusion chromatography (SEC) on a HiLoad 26/60 Superdex 200 column; (2) HisTrap affinity chromatography. Superdex 200 eluate fractions enriched in recombinant wt and glycosylation mutant PSI were pooled (marked with dotted lines) and further purified on a His Trap 1-ml column. (3) Purified fractions of both recombinant cirsin PSI forms were analyzed by SDS-PAGE and staining with Coomassie blue (SDS) and by Western blotting with an anti-His antibody (WB). (4) Analytical SEC of purified recombinant nonglycosylated PSI. The dotted line indicates the PSI elution volume. The dots indicate the elution volumes of molecular mass markers used for calibration (from left to right, thyroglobulin [669 kDa], aldolase [158 kDa], conalbumin [75 kDa], ovalbumin [43 kDa], carbonic anydrase [29 kDa], and RNase [13.7 kDa]).
Recombinant cirsin PSI displays antifungal activity.
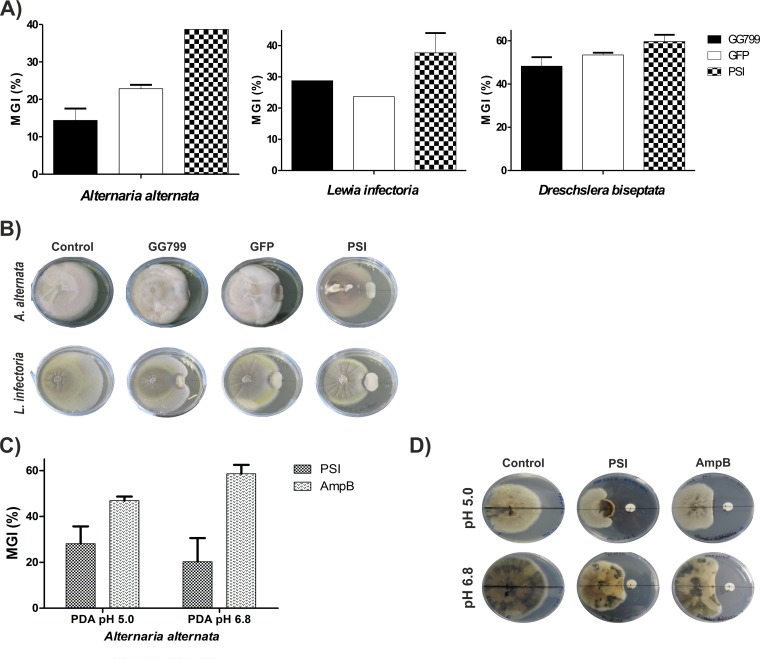
In order to evaluate the antimicrobial activity of recombinant cirsin PSI, we performed confrontation assays between the selected K. lactis transformants secreting wt PSI [SP_PSI(His)6] and four phytopathogenic fungi, B. obtusa, L. infectoria, A. alternata, and D. biseptata, with untransformed K. lactis cells (strain GG799) and a K. lactis transformant expressing GFP serving as negative controls. Cogrowth assays were carried out for 10 days at 30°C in YPGal medium at pH 5.0 supplemented with 4% galactose to promote PSI and GFP expression. After this incubation period, antifungal activity was visible as a zone of inhibited development of mycelial edges near the tested yeast colony. Antimicrobial activity was expressed as a percentage of mycelial growth inhibition (Fig. 7A). Surprisingly, K. lactis cells (untransformed or expressing GFP) were able to reduce the growth rates of three fungi (L. infectoria, A. alternata, and D. biseptata), inhibiting the growth rates of L. infectoria and A. alternata by around 20 to 25% and the growth of D. biseptata by ∼50%. No antifungal activity was detected against B. obtusa (data not shown). Although antifungal activity has been reported for other Kluyveromyces strains (Kluyveromyces phaffii and Kluyveromyces wickerhamii) (21–23), this protective activity of K. lactis observed against various plant pathogens has not been previously described. Despite this intrinsic antifungal activity, the total observed activity of K. lactis transformants expressing cirsin PSI was always enhanced (Fig. 7B). However, B. obtusa could not be stopped from growing by any of our K. lactis transformed strains. The presence of PSI decreased the growth of L. infectoria and D. biseptata by about 10% and was more pronounced for A. alternata (∼20%). These results provide evidence of antifungal activity of cirsin PSI, proving its expression in a biologically active form. The antifungal activity detected for recombinant cirsin PSI was also confirmed by a disk diffusion test using a purified sample of recombinant nonglycosylated cirsin PSI (0.47 mg/ml) against A. alternata. This test was performed at two different pHs (pH 5.0 and 6.8), with amphotericin B (0.25 mg/ml) serving as a positive control. Not surprisingly, the inhibitory activity of cirsin PSI on A. alternata appears to be potentiated under acidic conditions (Fig. 7C and D). These results are in line with data previously published by Egas et al. and Bryksa et al. (8, 10), who have reported increased interactions in a pH-dependent manner between PSI and phospholipid vesicles, leading to more pronounced vesicle leakage at low pH (pH 4.5).
FIG 7.
Recombinant cirsin PSI displays antifungal activity. (A) Confrontation assays between the selected K. lactis transformant secreting wt PSI [SP_PSI(His)6] and three phytopathogenic fungi (A. alternata, L. infectoria, and D. biseptata). Untransformed K. lactis (strain GG799) and a K. lactis transformant expressing GFP were used as negative controls. The antifungal activity is given by the percentage of mycelial growth inhibition {MGI = [(free growth of the fungi − growth of the fungi in the presence of yeast)/(free growth of the fungi)] × 100}. All assays were performed in duplicate. (B) Representative plates showing antifungal activity toward A. alternata and L. infectoria. (C and D) Disk diffusion test. Impregnated disks with a purified sample of recombinant nonglycosylated cirsin PSI (0.47 mg/ml) were assayed against the fungus A. alternata by evaluating the appearance of a clear zone of inhibition. The assay was performed in triplicate at two different pH values (pH 5.0 and 6.8) using the antifungal amphotericin B (0.25 mg/ml) as a positive control. (C) Antifungal activity is represented by the percentage of mycelial growth inhibition {MGI = [(growth of the fungi in the presence of sample buffer − growth of the fungi in the presence of PSI)/(growth of fungi in the presence of sample buffer)] × 100}. (D) Representative plates showing antifungal activity of PSI toward A. alternata. The error bars indicate standard deviations.
DISCUSSION
The cellular functions of PSI domains present in typical plant aspartic protease precursors are still not fully unraveled. However, several possible roles have been associated with these domains, including as a vacuolar sorting signal and/or as an antipathogen (2, 7–9). Most of the current knowledge on PSIs, like their membrane interaction properties, structural organization, and antimicrobial activity, was acquired in studies with PSI domains from phytepsin, cardosin A, and StAP (8–10, 24). However, the large number of PSI sequences available and the variety within these sequences clearly suggest that full characterization of all these plant domains has only started. An understanding of how this diversity influences PSI-membrane interactions and bioactivity and if/how this can be translated into biotechnological applications remains to be achieved. Further progress depends on additional in-depth studies with PSI domains, for which the development of more effective expression systems is required.
Here, we describe the development and optimization of heterologous production of the cirsin PSI domain in K. lactis. We have successfully generated K. lactis transformants expressing and secreting correctly folded and processed cirsin PSI. We demonstrate that cirsin PSI can be produced in a fully glycosylated form just as well as its nonglycosylated mutant, PSI(N86S). Successful results were obtained only after optimization of the fusion strategy between PSI and parts of the α-MF. The classical approach of fusing the cirsin PSI to the α-MF preprodomain so that the recombinant protein could be efficiently targeted to the ER and from there to the Golgi apparatus, where the α-MF prodomain would be cleaved off by Kex-2 endoprotease, yielded unprocessed PSI that still contained the α-MF prodomain. Incomplete protein processing was observed for glycosylated, as well as nonglycosylated, PSI with different experimental methods. Incomplete removal of the α-MF had already been observed when proforms of recombinant interleukin-1β and insulin precursors were expressed in S. cerevisiae (25) but has never been described for K. lactis. Fusion of the cirsin PSI with α-MF presignal alone was sufficient to produce correctly processed recombinant protein bearing high-mannose N-glycans and being effectively secreted into the culture medium. Correctly processed nonglycosylated PSI(N86S) mutant fused to α-MF presignal could also be obtained in reasonable amounts. This indicates that correct sorting happens regardless of whether PSI is glycosylated or not. Remarkably, the hyperglycosylation patterns of cirsin PSI fused to either the α-MF preprodomain or α-MF's presignal are different. Although both forms contain high-mannose-type N-glycans, fusion with the α-MF preprodomain results in a ladder of slightly underglycosylated forms of PSI that likely corresponds to the unprocessed fusion protein with heterogeneous glycosylation patterns. Incomplete cleavage of the α-MF leader sequence may be the consequence of limited Kex-2 processing in vivo or result from missorting of the α-MF–PSI fusion protein, thereby escaping Kex-2 processing in late Golgi compartments (26, 27).
The simplicity and effectiveness of the expression approach devised (fusion with only the signal peptide of α-MF) in K. lactis indicates a clear potential for expression of other PSI domains and proteins with similar properties. Moreover, this expression system proved its versatility by producing glycosylated, as well as nonglycosylated, cirsin PSI in good yields. Although the majority of PSI sequences of various species comprise a highly conserved N-glycosylation site, no expression of glycosylated PSI has been reported yet. Our results suggest that the expression system described here may be very useful in elucidating the role of glycosylation in PSI domains, although it must be kept in mind that plant-type N-glycans are different from their yeast counterparts (28). In addition, a robust purification protocol could be established for cirsin PSI in either glycosylated or nonglycosylated form. Our results confirm previous reports (10, 24) indicating that the tendency of nonglycosylated cirsin PSI to exist in dimeric form maybe due to exposed hydrophobic surface interactions.
The development of low-cost production of AMPs has received much attention in recent years (16). Some advances were achieved in E. coli to overcome AMP toxicity by generation of fusion constructs to sequester AMPs in inclusion bodies. Subsequent protein-refolding steps and cleavage with an external protease yielded the desired product (15, 16). The drawback of such an approach is multiple processing/purification steps, which take time and result in reduced protein yields and increased production costs. Expression in a eukaryotic system like yeast allows the introduction of posttranslational modifications and protein secretion into the culture medium (16). So far, the large majority of reports describing production of AMPs in yeast use Pichia pastoris as the expression host (16). We establish the yeast K. lactis as a promising alternative for production of these proteins, with the GRAS status of K. lactis providing the additional advantage of saving purification steps, leading to more cost-effective production processes (29, 30).
Plant diseases caused by viruses, bacteria, and fungi are responsible for significant losses or decreases in the quality and safety of agricultural products, accounting for worldwide losses of $30 to $50 billion/year (31). Although pest control is currently accomplished mainly with chemical pesticides, their use is increasingly restricted due to both health and environmental impacts (32). In fact, streptomycin- and oxytetracycline-resistant phytopathogens have already been found in plant crops (33). The huge economic impact of the deleterious effects of pathogens on crop cultures, together with the emergence and spread of superbugs, therefore, is triggering the development of alternative strategies to circumvent these problems (11). In this context, and given their plant origin and the reported activity of the StAP PSI domain against two potato pathogens, Phytophthora infestans and Fusarium solani (9), PSI domains emerge as interesting candidates in the development of alternative biocontrol tools. In this work, we provide clear evidence supporting the bioactivity of the cirsin PSI domain against plant fungal pathogens, like A. alternata, L. infectoria, and D. biseptata, compared with untransformed or GFP-expressing K. lactis cells. Unexpectedly, our results show that K. lactis itself also displays antifungal activity against these phytopathogens. The possibility that the observed killer activity of K. lactis may boost/mask cirsin PSI antifungal activity cannot be fully excluded. To our knowledge, antifungal activity of K. lactis has not been previously reported, and this finding deserves further attention. Earlier observed antifungal activity of nonglycosylated PSI domains (8, 10) could be confirmed with our nonglycosylated cirsin PSI. Full understanding of the observed pH-dependent antifungal activity awaits further investigations. Altogether, our studies confirm that cirsin PSI is produced in a biologically active form in K. lactis, and we provide evidence for its antiphytopathogenic activity.
In summary, we present a versatile K. lactis-based eukaryotic expression system as a platform for production of glycosylated, as well as nonglycosylated, PSI domains in their biologically active form. This work opens up new ways to effectively express these proteins, thereby providing tools to better understand the roles of PSI domains and their interaction with lipid vesicles. Further research in this direction will hopefully contribute to their future development as alternative biocontrol tools.
ACKNOWLEDGMENTS
We acknowledge the financial support of CONICET (PIP 0157) and the University of La Plata (Project X-613). L. Bakás is a member of the CICPBA Research Career Program; S. Vairo Cavalli is a member of the CONICET Research Career Program; D. Lufrano has been awarded a postdoctoral fellowship from CONICET.
The MALDI-TOF MS analyses were carried out in the Proteomics and Bioinformatics Facility of the Universitat Autònoma de Barcelona (SePBioEs-UAB). S.A.T. and SePBioEs are members of the ProteoRed-ISCIII network. We also thank Daniel Bur for his comments on the manuscript.
Footnotes
Published ahead of print 11 October 2013
REFERENCES
- 1.Guruprasad K, Tormakangas K, Kervinen J, Blundell TL. 1994. Comparative modelling of barley-grain aspartic proteinase: a structural rationale for observed hydrolytic specificity. FEBS Lett. 352:131–136. 10.1016/0014-5793(94)00935-X [DOI] [PubMed] [Google Scholar]
- 2.Kervinen J, Tobin GJ, Costa J, Waugh DS, Wlodawer A, Zdanov A. 1999. Crystal structure of plant aspartic proteinase prophytepsin: inactivation and vacuolar targeting. EMBO J. 18:3947–3955. 10.1093/emboj/18.14.3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simoes I, Faro C. 2004. Structure and function of plant aspartic proteinases. Eur. J. Biochem. 271:2067–2075. 10.1111/j.1432-1033.2004.04136.x [DOI] [PubMed] [Google Scholar]
- 4.Bruhn H. 2005. A short guided tour through functional and structural features of saposin-like proteins. Biochem. J. 389:249–257. 10.1042/BJ20050051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munford RS, Sheppard PO, O'Hara PJ. 1995. Saposin-like proteins (SAPLIP) carry out diverse functions on a common backbone structure. J. Lipid Res. 36:1653–1663 [PubMed] [Google Scholar]
- 6.Ryan MA, Akinbi HT, Serrano AG, Perez-Gil J, Wu H, McCormack FX, Weaver TE. 2006. Antimicrobial activity of native and synthetic surfactant protein B peptides. J. Immunol. 176:416–425 [DOI] [PubMed] [Google Scholar]
- 7.Duarte P, Pissarra J, Moore I. 2008. Processing and trafficking of a single isoform of the aspartic proteinase cardosin A on the vacuolar pathway. Planta 227:1255–1268. 10.1007/s00425-008-0697-1 [DOI] [PubMed] [Google Scholar]
- 8.Egas C, Lavoura N, Resende R, Brito RM, Pires E, de Lima MC, Faro C. 2000. The saposin-like domain of the plant aspartic proteinase precursor is a potent inducer of vesicle leakage. J. Biol. Chem. 275:38190–38196. 10.1074/jbc.M006093200 [DOI] [PubMed] [Google Scholar]
- 9.Munoz FF, Mendieta JR, Pagano MR, Paggi RA, Daleo GR, Guevara MG. 2010. The swaposin-like domain of potato aspartic protease (StAsp-PSI) exerts antimicrobial activity on plant and human pathogens. Peptides 31:777–785. 10.1016/j.peptides.2010.02.001 [DOI] [PubMed] [Google Scholar]
- 10.Bryksa BC, Bhaumik P, Magracheva E, De Moura DC, Kurylowicz M, Zdanov A, Dutcher JR, Wlodawer A, Yada RY. 2011. Structure and mechanism of the saposin-like domain of a plant aspartic protease. J. Biol. Chem. 286:28265–28275. 10.1074/jbc.M111.252619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parisien A, Allain B, Zhang J, Mandeville R, Lan CQ. 2008. Novel alternatives to antibiotics: bacteriophages, bacterial cell wall hydrolases, and antimicrobial peptides. J. Appl. Microbiol. 104:1–13. 10.1111/j.1365-2672.2007.03498.x [DOI] [PubMed] [Google Scholar]
- 12.Giuliani A, Pirri G, Nicoletto S. 2007. Antimicrobial peptides: an overview of a promising class of therapeutics. Cent. Eur. J. Biol. 2:1–33. 10.2478/s11535-007-0010-5 [DOI] [Google Scholar]
- 13.Yeaman MR, Yount NY. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55:27–55. 10.1124/pr.55.1.2 [DOI] [PubMed] [Google Scholar]
- 14.Lufrano D, Faro R, Castanheira P, Parisi G, Verissimo P, Vairo-Cavalli S, Simoes I, Faro C. 2012. Molecular cloning and characterization of procirsin, an active aspartic protease precursor from Cirsium vulgare (Asteraceae). Phytochemistry 81:7–18. 10.1016/j.phytochem.2012.05.028 [DOI] [PubMed] [Google Scholar]
- 15.Haught C, Davis GD, Subramanian R, Jackson KW, Harrison RG. 1998. Recombinant production and purification of novel antisense antimicrobial peptide in Escherichia coli. Biotechnol. Bioeng. 57:55–61. [DOI] [PubMed] [Google Scholar]
- 16.Parachin NS, Mulder KC, Viana AA, Dias SC, Franco OL. 2012. Expression systems for heterologous production of antimicrobial peptides. Peptides 38:446–456. 10.1016/j.peptides.2012.09.020 [DOI] [PubMed] [Google Scholar]
- 17.Caplan S, Green R, Rocco J, Kurjan J. 1991. Glycosylation and structure of the yeast MF alpha 1 alpha-factor precursor is important for efficient transport through the secretory pathway. J. Bacteriol. 173:627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy KP, Jr, Gagne P, Pazmany C, Moody MD. 1998. Expression of human interleukin-17 in Pichia pastoris: purification and characterization. Protein Expr. Purif. 12:208–214. 10.1006/prep.1997.0832 [DOI] [PubMed] [Google Scholar]
- 19.Saito A, Usui M, Song Y, Azakami H, Kato A. 2002. Secretion of glycosylated alpha-lactalbumin in yeast Pichia pastoris. J. Biochem. 132:77–82. 10.1093/oxfordjournals.jbchem.a003202 [DOI] [PubMed] [Google Scholar]
- 20.Yamada M, Inui K, Hamada D, Nakahira K, Yanagihara K, Sakai N, Nishigaki T, Ozono K, Yanagihara I. 2004. Analysis of recombinant human saposin A expressed by Pichia pastoris. Biochem. Biophys. Res. Commun. 318:588–593. 10.1016/j.bbrc.2004.04.069 [DOI] [PubMed] [Google Scholar]
- 21.Ciani M, Fatichenti F. 2001. Killer toxin of Kluyveromyces phaffii DBVPG 6076 as a biopreservative agent to control apiculate wine Yeasts. Appl. Environ. Microbiol. 67:3058–3063. 10.1128/AEM.67.7.3058-3063.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comitini F, De IJ, Pepe L, Mannazzu I, Ciani M. 2004. Pichia anomala and Kluyveromyces wickerhamii killer toxins as new tools against Dekkera/Brettanomyces spoilage yeasts. FEMS Microbiol. Lett. 238:235–240. 10.1111/j.1574-6968.2004.tb09761.x [DOI] [PubMed] [Google Scholar]
- 23.Rosini G. 1983. The occurrence of killer characters in yeasts. Can. J. Microbiol. 29:1462–1464. 10.1139/m83-224 [DOI] [PubMed] [Google Scholar]
- 24.Munoz F, Palomares-Jerez MF, Daleo G, Villalain J, Guevara MG. 2011. Cholesterol and membrane phospholipid compositions modulate the leakage capacity of the swaposin domain from a potato aspartic protease (StAsp-PSI). Biochim. Biophys. Acta 1811:1038–1044. 10.1016/j.bbalip.2011.08.013 [DOI] [PubMed] [Google Scholar]
- 25.Kjeldsen T. 2000. Yeast secretory expression of insulin precursors. Appl. Microbiol. Biotechnol. 54:277–286. 10.1007/s002530000402 [DOI] [PubMed] [Google Scholar]
- 26.Antebi A, Fink GR. 1992. The yeast Ca(2+)-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol. Biol. Cell 3:633–654. 10.1091/mbc.3.6.633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kjeldsen T, Brandt J, Andersen AS, Egel-Mitani M, Hach M, Pettersson AF, Vad K. 1996. A removable spacer peptide in an alpha-factor-leader/insulin precursor fusion protein improves processing and concomitant yield of the insulin precursor in Saccharomyces cerevisiae. Gene 170:107–112. 10.1016/0378-1119(95)00822-5 [DOI] [PubMed] [Google Scholar]
- 28.Rayon C, Lerouge P, Lc Faye. 1998. The protein N-glycosylation in plants. J. Exp. Botany 49:1463–1472 [Google Scholar]
- 29.Bonekamp F, Oosterom J. 1994. On the safety of Kluyveromyces lactis: a review. Appl. Microbiol. Biotechnol. 41:1–3. 10.1007/BF00166072 [DOI] [Google Scholar]
- 30.van Ooyen AJ, Dekker P, Huang M, Olsthoorn MM, Jacobs DI, Colussi PA, Taron CH. 2006. Heterologous protein production in the yeast Kluyveromyces lactis. FEMS Yeast Res. 6:381–392. 10.1111/j.1567-1364.2006.00049.x [DOI] [PubMed] [Google Scholar]
- 31.Montesinos E, Bardaji E. 2008. Synthetic antimicrobial peptides as agricultural pesticides for plant-disease control. Chem. Biodivers. 5:1225–1237. 10.1002/cbdv.200890111 [DOI] [PubMed] [Google Scholar]
- 32.Montesinos E. 2007. Antimicrobial peptides and plant disease control. FEMS Microbiol. Lett. 270:1–11. 10.1111/j.1574-6968.2007.00683.x [DOI] [PubMed] [Google Scholar]
- 33.Vidaver AK. 2002. Uses of antimicrobials in plant agriculture. Clin. Infect. Dis. 34(Suppl 3):S107–S110. 10.1086/340247 [DOI] [PubMed] [Google Scholar]