Abstract
Lignocellulosic biomass is digested in nature by the synergistic activities of enzymes with complementary properties, and understanding synergistic interactions will improve the efficiency of industrial biomass use for sustainable fuels and chemicals. Cel9A and Cel48A from a model bacterium, Thermobifida fusca (TfCel9A and TfCel48A, respectively), are two cellulases with different properties and have previously been shown to synergize well with each other. TfCel9A is a processive endocellulase with relatively high activity on crystalline cellulose. TfCel48A is a reducing end-directed exocellulase with very low activity on crystalline cellulose. Neither enzyme fits its respective role in the classical synergism model of enzymatic cellulose digestion. Using the results of time course, endpoint, and sequential addition activity assays, we propose a model of synergistic cooperation between the two cellulases. TfCel9A is most effective on fresh bacterial cellulose with a presumably uniform surface at the molecular level. Its processive activity likely erodes the surface and thus reduces its own activity. TfCel48A is able to hydrolyze the TfCel9A-modified substrate efficiently and replenish the uniform surface required by TfCel9A, creating a feedback mechanism. The model of synergistic interactions is comparable to an earlier proposed model for Trichoderma reesei Cel7A and Cel7B, but the roles of endo- and exocellulases are reversed, a finding which suggests that bacteria and fungi may have evolved different approaches to efficient biomass degradation.
INTRODUCTION
Fuels and chemicals derived from lignocellulosic biomass have the potential to displace a significant fraction of petroleum-based products in the near future. One of the key challenges in the utilization of biomass for such applications is the cost associated with enzymatic hydrolysis of cellulose to fermentable sugars. Consequently, mechanistic studies of cellulases and other biomass-degrading enzymes will play an important role in the advancement of biomass use for the production of renewable low-carbon-footprint fuels and chemicals.
In nature, most biomass is degraded by the complementary action of multiple enzymes produced by cellulolytic microorganisms. Cellulose constitutes a major component of plant cell walls and is composed of anhydrous glucose units covalently linked via β-1,4 glycosidic bonds to form long polymer chains. Hydrogen bonding between individual chains forms a recalcitrant heterogeneous material of variable crystallinity that is relatively resistant to enzymatic hydrolysis. Cellulases belong to different glycoside hydrolase (GH) families and are commonly divided into two major classes. Exocellulases typically contain an active site inside a tunnel and thus initiate hydrolysis at cellulose chain ends. Endocellulases usually contain an active site in an open cleft and are able to initiate hydrolysis anywhere along the cellulose chain. The processivity of cellulases is defined as the average number of consecutive cleavages that a cellulase will carry out before its dissociation from the chain. Due to technical limitations, the processivity of cellulases acting on bulk cellulose is usually reported as a relative value. For exocellulases, this is commonly a ratio of cellobiose (G2) to cellotriose (G3) and/or glucose (G1) (1–3), while for endocellulases, it is usually the ratio of soluble and insoluble reducing ends after digestion. Exocellulases are believed to be more processive than endocellulases, in particular, as judged by the soluble-to-insoluble-reducing-end postdigestion ratios (4). There is a subclass of processive endocellulases, the most studied of which is Cel9A from Thermobifida fusca (TfCel9A).
Synergistic interactions between different cellulases can strongly enhance the rate and extent of enzymatic cellulose hydrolysis. Since Reese et al. first documented synergism between different cellulase fractions in 1950 (5), a lot of work has been carried out to understand the mechanisms by which these enzymes act on their substrates. In 1979, Wood and McCrae proposed what became the classical endocellulase-exocellulase model of enzymatic cellulose hydrolysis (6). According to the model, endocellulases attack the amorphous fractions of cellulose, creating more chain ends for exocellulase attack. In turn, exocellulase activity exposes new amorphous regions within the bulk substrate and thus stimulates additional endocellulase activity. Support for this model has been extensively demonstrated in the literature, but it is also likely that the model is incomplete (7–9). Synergistic interactions have also been demonstrated for different types of exocellulases that preferentially attack either the reducing or the nonreducing end of the cellulose chain (10, 11). More recently, synergism between cellulases and auxiliary activity (AA) proteins from the AA10 (formerly CBM33) and AA9 (formerly GH61) families has been demonstrated (12–14). In addition, removal of obstacles and the release of unproductively bound exocellulases appear to be important components of effective synergistic cellulase mixtures (9, 15, 16). Continued studies of synergistic interactions in cellulose hydrolysis can therefore contribute to the development of models more complete than those that are presently available.
Here we report data for synergistic interactions between Cel9A and Cel48A, two cellulases secreted by the model cellulolytic bacterium Thermobifida fusca (TfCel9A and TfCel48A, respectively). TfCel9A is a processive endocellulase which contains two cellulose binding modules, CBM2 and CBM3c (17), and is relatively effective as a single cellulase in the digestion of crystalline cellulose (4). CBM2 is attached via a flexible linker (which includes a fibronectin III domain) to CBM3c. CBM3c is, in turn, connected almost rigidly to the GH9 catalytic domain (18) and is believed to be responsible for the processivity of TfCel9A and ability of TfCel9A to digest crystalline cellulose effectively (17, 18). TfCel48A is a reducing end-directed exocellulase with very poor activity on crystalline cellulose (19). Despite its low activity, TfCel48A makes up about a third of total secreted cellulases when T. fusca is grown on cellulose (20), and it has been shown to interact synergistically with other T. fusca cellulases, including TfCel9A (4, 19). Given that TfCel9A is processive and is the most effective T. fusca cellulase on crystalline substrates (4), it is unlikely to serve the typical role assigned to endocellulases in the endosynergism-exosynergism model. In addition, the very low activity of TfCel48A on crystalline cellulose also makes it incompatible with the presumed role of exocellulases in the classical model of enzymatic cellulose digestion. Hence, it is likely that TfCel48A and TfCel9A employ a distinct mechanism of synergism, and here we propose a model for their cooperation.
MATERIALS AND METHODS
Substrates and enzymes.
Bacterial cellulose (BC) was produced by Acetobacter aceti subsp. xylinus (“Acetobacter xylinum”) and was obtained as a gift from Monsanto. BC cake was washed three times with deionized (DI) water by centrifugation and resuspended in DI water with 0.04% sodium azide (Sigma-Aldrich). The concentration was determined as dry weight per volume. Recombinant TfCel9A and TfCel48A were expressed in Escherichia coli BL21 cells and purified as previously described in references 17 and 21, respectively.
Time course activity assays.
All reactions were conducted in triplicate in Eppendorf 2-ml Protein LoBind plastic tubes. One milligram substrate was combined with 83 nM TfCel9A and/or 750 nM TfCel48A in 0.6 ml 50 mM sodium acetate buffer, pH 5.5. Only buffer and substrate were combined for use as negative controls. Upon mixing, the BC reaction mixtures were immediately placed in a 50°C water bath. Samples were removed in triplicate at the given time points and placed on dry ice to stop the reaction. Frozen samples were later placed in a boiling bath for 10 min in order to denature the enzyme. It was verified experimentally that boiling does not alter the soluble sugar profiles detected by high-performance liquid chromatography (HPLC). The remaining substrate was removed using Corning Spin-X centrifuge tube filters, and the soluble sugar concentrations were measured using a Shimadzu HPLC system fitted with a Bio-Rad Aminex HPX-87P analytical column and a refractive index detector. The mobile phase was Milli-Q water at a flow rate of 0.6 ml/min. Sample injection was performed by an autosampler installed on the instrument.
BC hydrolysis assays of different concentrations of TfCel48A with a constant concentration of TfCel9A.
Reaction mixtures were prepared in the same manner as described above for the time course assays. TfCel9A (8.3 nM) was combined with 0 to 158 nM TfCel48A, and the mixture was incubated with 1 mg BC for 19 h. Samples were then processed for HPLC in the same manner as described above.
Sequential BC addition assays.
Sequential BC addition assays were conducted under the same conditions used for the time course assays. Enzymes and substrate were incubated for 20 h and divided into three groups. One group was boiled for 5 min to denature the enzyme and was then processed for HPLC analysis, as described above. To the other two groups, either 1 mg fresh BC or buffer was added and the reaction mixtures were incubated for another 20 h. The reaction mixtures were then boiled and processed for HPLC.
Sequential enzyme addition assays.
Sequential enzyme addition assays were conducted under the same conditions used for the time course assays. For endpoint sequential assays, enzymes and substrate were incubated for 20 h and boiled for 5 min. The corresponding synergistic partner was then added, and the reaction mixtures were incubated for 2.25 h. A comparison between the total amount of soluble sugar produced by enzymes on fresh and pretreated substrates was made. In the case of substrate pretreatment, the total amount of sugar produced before the addition of the synergistic partner was determined experimentally and subtracted from the total amount of soluble sugar present after digestion by the freshly added enzyme. For time course sequential assays, reaction mixtures were initially prepared by mixing 83 nM TfCel9A with 1 mg BC. Triplicate samples were collected at the intervals indicated below and frozen on dry ice to stop the reaction. After overnight incubation, all remaining reaction mixtures were boiled for 5 min and divided into two groups. TfCel9A (83 nM) was added to one group, and TfCel48A (750 nM) was added to the other group. Samples from both groups were removed in triplicate at the indicated intervals. After overnight incubation, the remaining reaction mixtures containing TfCel48A were boiled for 5 min and 83 nM TfCel9A was added. Samples were removed in triplicate at the time intervals indicated below. Controls were run to ensure that boiling of the reaction mixtures was sufficient to stop all additional activity when they were incubated at 50°C. It was also verified experimentally that boiling does not alter the HPLC profiles of the soluble sugars or affect the reactivity of BC.
Data analysis.
HPLC data were processed with the OriginPro (version 8) program (OriginLab Corporation). Product identities and concentrations were determined by Gaussian peak fitting, using standard solutions with known concentrations of soluble cellooligosaccharides for reference. Soluble sugar concentrations at time zero were subtracted from all of the subsequently obtained concentrations. The soluble sugar produced upon initial mixing of the enzyme and the substrate is primarily due to the burst activity, as described previously (22, 23), whereas the model used here is concerned with the digestion of the more recalcitrant portions of cellulose. The A- and b-parameter values of the time course profiles (where A is the total activity of the added enzyme, and b is the hydrolysis power factor that quantifies the dependence of activity on time) were determined using the nonlinear least-squares fit of equation 1.
Kinetic model of cellulose hydrolysis.
We recently developed a kinetic model for mechanistic studies of enzymatic cellulose hydrolysis (21). The model relies on two parameters to quantify cellulose digestion by individual cellulases and their mixtures over time. It is based on a pseudo-zero-order Michaelis-Menten kinetic scheme but replaces an activity constant with a time (digestion)-dependent activity coefficient:
| (1) |
where X is percent digestion, and t is time. The values of A and b were determined by fitting equation 1 to the time course data for cellulose digestion. Parameter A is a product of specific activity and the productively bound cellulase concentration, neither of which is directly measurable for cellulases acting on bulk substrates. At low enzyme loads, the value of A is strongly dependent on the total added enzyme concentration and follows the Arrhenius relationship with respect to temperature. Parameter b is an intrinsic constant for a given cellulase on a given substrate and is independent of the total amount of added enzyme at low enzyme-to-substrate ratios. The theoretical limits of b are 0, in which case no products are formed, and 1, in which case the activity is constant over time, as is the case in classical kinetics. Thus, the value of b is indicative of the cellulase's ability to overcome substrate recalcitrance, and it can be used to better understand synergistic interactions between biomass-active enzymes. For example, we showed that the addition of a T. fusca AA10 enzyme E7 to TfCel48A acting on BC increased the b value from 0.34 to 0.65 (21), which is consistent with the presumed ability of AA10 enzymes to disrupt crystalline regions of cellulose and chitin (14, 24).
RESULTS
Time course data for BC hydrolysis by TfCel9A and TfCel48A, alone and together, are shown in Fig. 1. The very low activity of TfCel48A on crystalline cellulose is reflected by its low A and b values: 0.77 and 0.34, respectively. On the other hand, the relatively high TfCel9A A and b values of 3.35 and 0.56, respectively, reflect its ability to hydrolyze BC well. When the two enzymes are combined together at the same loads used in individual assays, the b value of the mixture is 0.55, which is the same as that of TfCel9A alone, and the A value is increased by ca. 260% compared to the sum of the A values measured for the two cellulases alone. Equation 1 is typically applicable only for the initial range of digestion, which can vary strongly for different enzymes (21). It is likely that the observed drop-off is due to the contribution of additional factors other than substrate recalcitrance to the decline of the cellulose digestion rate (e.g., surface erosion and obstacle formation [9, 16, 25]). Importantly, while the drop-off point is below 2% digestion for TfCel48A and ca. 12% for TfCel9A, it increases to ca. 35% when the two enzymes are combined.
FIG 1.
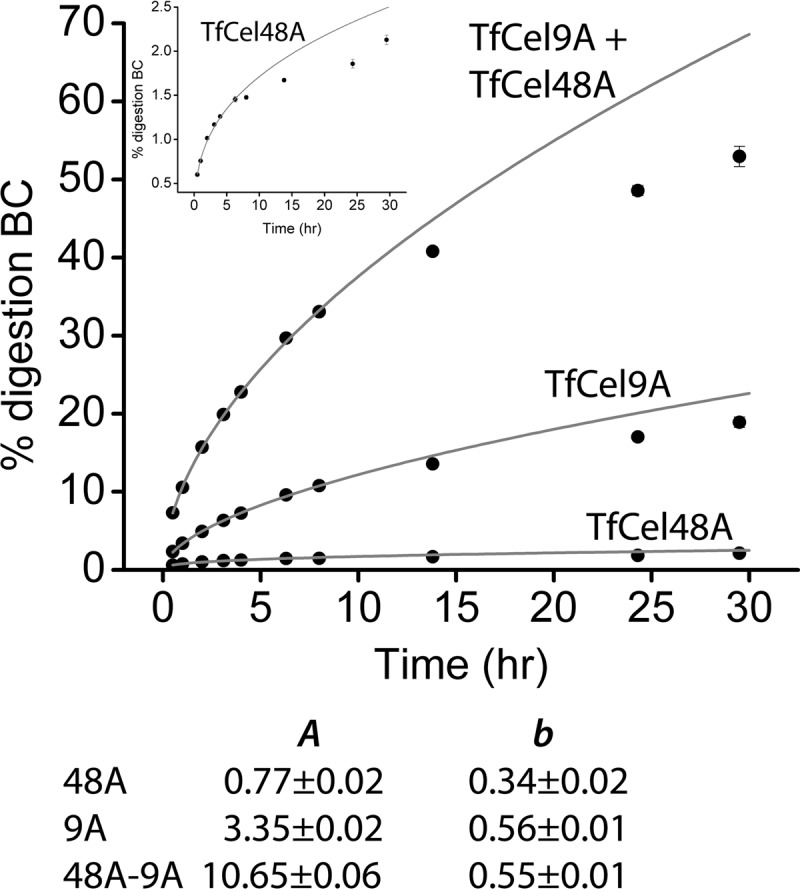
Time course BC hydrolysis assays of 750 nM TfCel48A, 83 nM TfCel9A, and their mixture. (Inset) Magnification of the TfCel48A time course. All data points were obtained in triplicate, and the error bars represent 1 standard deviation. The table lists the A- and b-parameter values obtained by fitting the time course data to equation 1.
The fact that the b-parameter value of the TfCel48A-TfCel9A mixture is the same as that of TfCel9A while the A-parameter value is significantly increased suggests that the two enzymes acting together are able to digest the BC much faster than either one acting alone, but they are not more effective at overcoming substrate recalcitrance per se. The BC hydrolysis rate increase for the mixture may be the result of the higher activity of either or both enzymes. As a processive enzyme, TfCel9A initially produces cellotetraose (G4), which is later cleaved in solution (by TfCel9A and other cellulases that may be present) to cellotriose (G3), cellobiose (G2), and glucose (G1) (Fig. 2a). On the other hand, TfCel48A produces mostly G2 and G3 and only a trace amount of G4 (Fig. 2b), which it is able to hydrolyze only slowly. Therefore, most of the G4 present during the initial stages of BC digestion by the TfCel48A-TfCel9A mixture is produced by TfCel9A and can be used to compare the activity of TfCel9A alone and in the mixture. As shown in Fig. 3a, the G4 concentration profile during the initial 14 h of BC digestion is roughly the same for TfCel9A alone and TfCel9A in combination with TfCel48A. This strongly suggests that the rate of BC hydrolysis by TfCel9A is not significantly increased in the presence of TfCel48A and implies that most of the rate increase observed for the two enzymes acting together is due to additional TfCel48A activity. Further support for this proposition is shown in Fig. 3b, which demonstrates a strong dependence of the total soluble sugar produced on an increasing amount of TfCel48A added to a constant amount of TfCel9A in endpoint assays of BC hydrolysis. Interestingly, even though the total amount of soluble sugar produced is strongly dependent on TfCel48A concentrations even at high TfCel48A/TfCel9A ratios, the specific activity of the total added enzyme is the highest at a ratio of 0.5. This suggests that the productively bound fraction of TfCel48A decreases as its ratio to TfCel9A increases.
FIG 2.
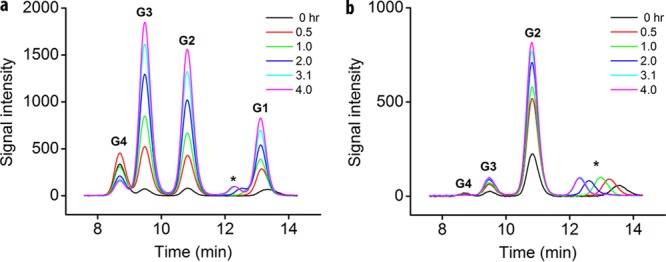
Sample HPLC profiles from time courses of 750 nM TfCel48A and 83 nM TfCel9A on 1 mg BC. (a) TfCel9A initially produces mostly cellotetraose (G4), which is later hydrolyzed in solution to cellotriose (G3), cellobiose (G2), and glucose (G1). (b) TfCel48A produces mostly G3 and G2 at a constant ratio (which is indicative of its processivity) and a trace amount of G4. G4 is only slowly cleaved by TfCel48A. The data for the zero time point were collected by mixing the samples along with all other samples and placing them in boiling water to denature the enzymes at the same time that other samples were placed in a 50°C bath. * indicates an artifact peak always observed in the HPLC system employed here.
FIG 3.
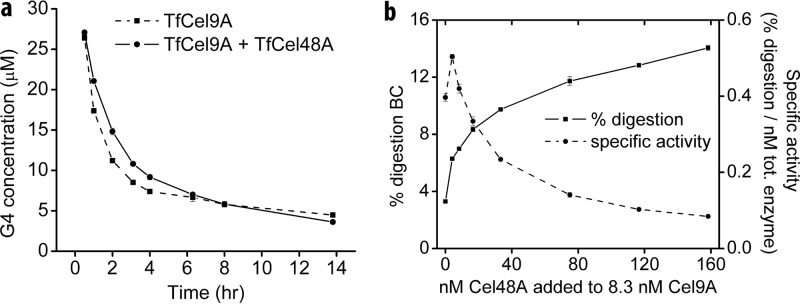
TfCel9A and TfCel48A relative activity in the synergistic mixture. (a) Cellotetraose (G4) concentration profile for BC hydrolysis by TfCel9A alone and the TfCel9A-TfCel48A (1:9 ratio) mixture; (b) extent of digestion and specific activity as a function of the TfCel48A concentration increase with a constant concentration of TfCel9A in BC hydrolysis 19-h endpoint assays.
The main factors that may contribute to the decline in the rate of enzymatic cellulose digestion at low enzyme loads are inherent substrate recalcitrance, morphological substrate changes, and enzyme deactivation (due to poor thermostability and/or unproductive irreversible binding). To determine whether the decline in the rate of BC digestion by TfCel9A and TfCel48A was caused primarily by substrate or enzyme-related changes, we carried out sequential BC addition endpoint assays, the results of which are shown in Fig. 4. After BC hydrolysis assay mixtures with TfCel48A or TfCel9A alone were incubated overnight, the same amount of fresh BC or buffer was added to the reaction mixtures and the mixtures were incubated for the same amount of time. For both TfCel48A and TfCel9A, the addition of fresh BC resulted in activity much higher than that achieved with addition of buffer only. Incubation of TfCel9A with fresh and partially digested BC resulted in the formation of the same amount of additional product as that formed in the initial digestion, while continued digestion of the original BC with fresh buffer produced only half as much additional product. This suggests that TfCel9A activity is primarily limited by the substrate-related factors and that the enzyme was not deactivated over the 2-day period during which the assays were run. Similarly, TfCel48A was able to hydrolyze fresh BC much better than the partially digested BC after overnight incubation, indicating that its activity is also primarily limited by substrate-related factors. However, the activity of TfCel48A on fresh BC resulted in only 70% additional product formation after the same incubation time, indicating that some fraction of TfCel48A is deactivated during the initial digestion. Presumably, its deactivation is primarily due to irreversible unproductive binding to the substrate, as this enzyme is generally very stable over time and multiple freeze-thaw cycles.
FIG 4.
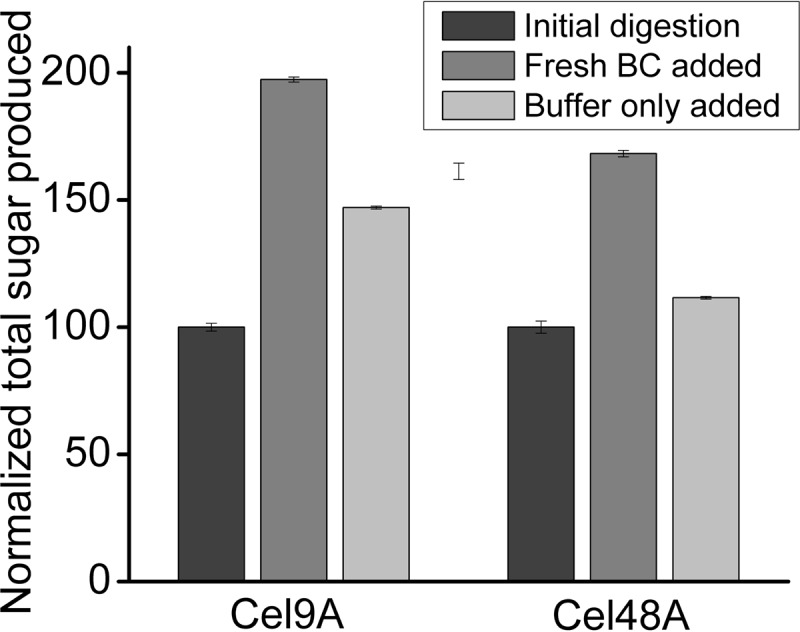
Sequential BC addition in endpoint assays. The initial 20-h BC (1 mg) digestion was followed by the addition of fresh BC (1 mg) or buffer to the reaction mixtures. The reaction mixtures were then incubated for another 20 h, and the amount of soluble sugar measured in the reaction mixtures is presented (normalized such that the value of 100 represents the amount of sugar produced after the first overnight incubation with the enzyme). Both enzymes showed enhanced activity when fresh BC was added. The double amount of soluble sugar produced by TfCel9A after the second addition of BC implies that the enzyme was largely dissociated from the original BC fraction, while in the case of TfCel48A, some enzyme may be deactivated by irreversible unproductive binding to the original substrate.
Sequential addition of synergistically acting cellulases can provide insight into the roles played by synergistic partners in cooperative substrate digestion (9, 26, 27). For this reason, we tested the effect of substrate pretreatment by TfCel48A and TfCel9A on each other's activities (Fig. 5a). Substrate was incubated with either enzyme overnight, and the reaction mixtures were then boiled to denature the enzymes. The synergistic partner was then added, the reaction mixtures were incubated for 2.25 h, and the results were compared to those of assays in which the reaction mixtures were prepared with fresh BC. The results indicate that predigestion of BC with TfCel9A enhances TfCel48A overnight endpoint activity by ca. 100%. On the other hand, TfCel48A predigestion of BC did not enhance the activity of TfCel9A under the tested conditions. These observations are consistent with other published data that show enhanced exocellulase activity following substrate pretreatment with an endocellulase, but not vice versa (27).
FIG 5.
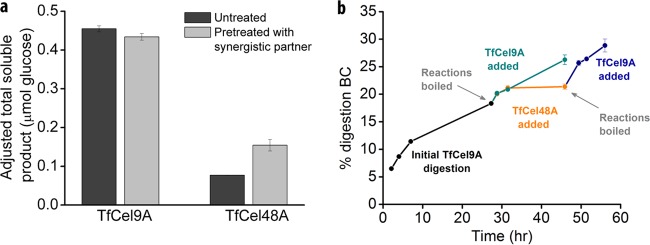
Effect of sequential additions of TfCel48A and TfCel9A on each other's activities. (a) BC was pretreated with either TfCel48A or TfCel9A for 20 h, and the reaction mixtures were boiled to denature the enzyme. The synergistic partner was then added, and the reaction mixtures were incubated for 2.25 h. Comparisons of activity on untreated and enzyme-pretreated substrate are provided. For the pretreated reaction mixtures, the provided value was adjusted by subtracting the amount of soluble sugar present in the reaction mixture measured after the pretreatment from the total amount of sugar measured at the end of the assay. (b) Initial TfCel9A (83 nM) digestion was followed by the addition of TfCel9A (83 nM) with or without an intermediate incubation with TfCel48A (750 nM). The intermediate addition of TfCel48A enhanced the activity of the freshly added TfCel9A, as evidenced by the steeper percent digestion profile following incubation with TfCel48A.
To further understand the role of TfCel48A in its interaction with TfCel9A, we carried out more sequential enzyme addition assays such that the initial digestion with TfCel9A was followed up either with fresh addition of TfCel9A or with addition of TfCel48A followed by TfCel9A (Fig. 5b). The goal was to see whether the intermediate addition of TfCel48A would enhance the activity of the newly added TfCel9A, and this was indeed the case. After overnight incubation with TfCel9A and boiling of the reaction mixtures to denature the enzyme, addition of fresh TfCel9A led to an increase in the extent of BC digestion by less than 2.5 percentage points in 4 h. However, when fresh TfCel9A was added following intermediate overnight incubation with TfCel48A, the extent of BC digestion increased by more than 4 percentage points in 4 h, indicating that TfCel9A activity was enhanced by ca. 60% after the intermediate addition of TfCel48A.
DISCUSSION
TfCel48A and TfCel9A are important components of the T. fusca biomass-degrading arsenal, and their properties make them incompatible with the classical endocellulase-exocellulase synergism model. To understand the potentially distinct mode of cooperation between these enzymes, we carried out endpoint, time course, and sequential addition BC hydrolysis assays, the results of which allow the following suppositions to be made. When combined, the two enzymes are able to hydrolyze BC much faster than either one alone, but their ability to overcome BC recalcitrance is the same as that of TfCel9A acting alone (Fig. 1). The decline in the hydrolysis rate of individual enzyme assays is primarily caused by substrate-related factors for both cellulases, although it also appears that a fraction of TfCel48A is deactivated by irreversible binding when the enzyme is acting alone (Fig. 4). Most of the synergistic product formation was due to the activity of TfCel48A, and the rate of BC hydrolysis by TfCel9A was not apparently enhanced in the presence of TfCel48A (Fig. 3). Pretreatment of fresh BC by TfCel9A enhanced the ability of TfCel48A to hydrolyze the same substrate, but the reverse was not true. However, TfCel48A hydrolysis of BC predigested by TfCel9A stimulated further hydrolysis by TfCel9A (Fig. 5).
Based on these observations, we propose a model of synergism, as follows (illustrated in Fig. 6). TfCel9A appears to be best adapted for the hydrolysis of fresh BC. CBM3c is necessary for the ability of TfCel9A to hydrolyze BC (17), and its relatively flat binding surface may function best on a uniform substrate surface (18, 28). According to Monte Carlo simulations (16) and high-resolution imaging of cellulose before and after digestion (25, 29), cellulose hydrolysis by processive cellulases creates an eroded surface, which may no longer be optimal for TfCel9A binding. BC surface erosion may, therefore, explain why TfCel9A activity drops off more rapidly above ca. 12% substrate digestion. On the other hand, the very low activity of TfCel48A on BC (Fig. 1) suggests that it cannot effectively access the individual chains in this primarily crystalline substrate (30). The eroded BC surface formed by TfCel9A activity may provide a much more accessible substrate for TfCel48A, as it creates new chain ends and less tightly bound chains. TfCel48A would preferentially hydrolyze looser chains, replenishing the smooth surface required for efficient TfCel9A activity. Such a feedback mechanism would allow the continuous optimized activity by both cellulases, and this is reflected by their high total activity on BC as well as the equation 1 drop-off point of digestion well above that observed for either enzyme alone.
FIG 6.
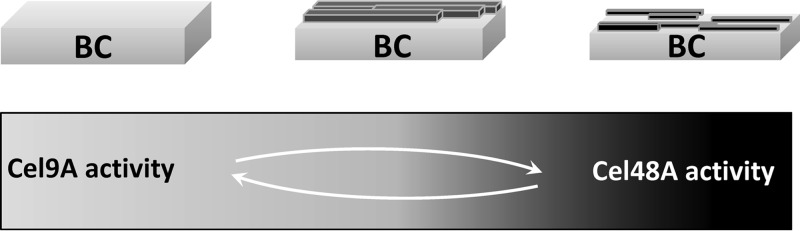
Proposed model of synergistic cooperation between TfCel9A and TfCel48A. TfCel9A appears to be best adapted for the hydrolysis of fresh BC. Its processive mode of action creates an eroded surface, which may no longer be optimal for its continued binding to the substrate. TfCel48A cannot effectively access the individual chains in crystalline substrates, but the eroded BC surface formed by TfCel9A activity provides a more accessible substrate for TfCel48A. TfCel48A preferentially hydrolyzes looser chains, replenishing the smooth surface required for efficient TfCel9A activity.
A comparable model of synergism was proposed by Valjamae et al. (9) for Trichoderma reesei enzymes Cel7A (TrCel7A; CBH I, a processive exocellulase) and Cel7B (TrCel7B; EG I, a nonprocessive endocellulase). In their study of BC hydrolysis by the two cellulases, the authors concluded that the processive action of TrCel7A creates an eroded surface with obstacles that reduce the activity of the enzyme. The modified surface, however, is efficiently hydrolyzed by TrCel7B and allows sustained TrCel7A activity. TfCel9A and TfCel48A, therefore, appear to play similar roles as TrCel7A and TrCel7B, respectively, in the model proposed in the above-described study. It is important to note, however, that the roles of the endo- and exocellulases are reversed in the two models, which suggests that T. fusca and T. reesei may have evolved somewhat different approaches to biomass digestion. If this is indeed the case, it would be interesting to determine whether such differences apply more generally to cellulolytic bacteria and fungi.
The model of synergism between TfCel48A and TfCel9A proposed here was initiated from the time course data shown in Fig. 1 by comparing equation 1 parameter values obtained for the individual enzymes and their mixture. Additional assays were then carried out to further develop and verify the TfCel48A-TfCel9A synergism model. We believe that this study provides additional support for the utility of our recently developed kinetic model for the mechanistic studies of enzymatic cellulose digestion.
ACKNOWLEDGMENT
This work was supported by the BioEnergy Science Center, a U.S. Department of Energy (DOE) center supported by the Office of Biological and Environmental Research in the DOE Office of Science.
Footnotes
Published ahead of print 25 October 2013
REFERENCES
- 1.Medve J, Karlsson J, Lee D, Tjerneld F. 1998. Hydrolysis of microcrystalline cellulose by cellobiohydrolase I and endoglucanase II from Trichoderma reesei: adsorption, sugar production pattern, and synergism of the enzymes. Biotechnol. Bioeng. 59:621–634 [PubMed] [Google Scholar]
- 2.von Ossowski I, Stahlberg J, Koivula A, Piens K, Becker D, Boer H, Harle R, Harris M, Divne C, Mahdi S, Zhao YX, Driguez H, Claeyssens M, Sinnott ML, Teeri TT. 2003. Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Ce17A. A comparison with Phanerochaete chrysosporium Cel7D. J. Mol. Biol. 333:817–829. 10.1016/S0022-2836(03)00881-7 [DOI] [PubMed] [Google Scholar]
- 3.Vuong TV, Wilson DB. 2009. Processivity, synergism, and substrate specificity of Thermobifida fusca Cel6B. Appl. Environ. Microbiol. 75:6655–6661. 10.1128/AEM.01260-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irwin DC, Spezio M, Walker LP, Wilson DB. 1993. Activity studies of eight purified cellulases: specificity, synergism, and binding domain effects. Biotechnol. Bioeng. 42:1002–1013. 10.1002/bit.260420811 [DOI] [PubMed] [Google Scholar]
- 5.Reese ET, Siu RG, Levinson HS. 1950. The biological degradation of soluble cellulose derivatives and its relationship to the mechanism of cellulose hydrolysis. J. Bacteriol. 59:485–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood TM, McCrae SI. 1979. Synergism between enzymes involved in the solubilization of native cellulose, p 181–209 In Advances in chemistry. American Chemical Society, Washington, DC [Google Scholar]
- 7.Fox JM, Jess P, Jambusaria RB, Moo GM, Liphardt J, Clark DS, Blanch HW. 2013. A single-molecule analysis reveals morphological targets for cellulase synergy. Nat. Chem. Biol. 9:356–361. 10.1038/nchembio.1227 [DOI] [PubMed] [Google Scholar]
- 8.Kostylev M, Wilson DB. 2012. Synergistic interactions in cellulose hydrolysis. Biofuels 3:61–70. 10.4155/bfs.11.150 [DOI] [Google Scholar]
- 9.Valjamae P, Sild V, Nutt A, Pettersson G, Johansson G. 1999. Acid hydrolysis of bacterial cellulose reveals different modes of synergistic action between cellobiohydrolase I and endoglucanase I. Eur. J. Biochem. 266:327–334. 10.1046/j.1432-1327.1999.00853.x [DOI] [PubMed] [Google Scholar]
- 10.Barr BK, Hsieh YL, Ganem B, Wilson DB. 1996. Identification of two functionally different classes of exocellulases. Biochemistry 35:586–592. 10.1021/bi9520388 [DOI] [PubMed] [Google Scholar]
- 11.Fagerstam L, Pettersson L. 1980. The 1.4-β-glucan cellobiohydrolases of Trichoderma reesei QM 9414A new type of cellulolytic synergism. FEBS Lett. 119:97–100. 10.1016/0014-5793(80)81006-4 [DOI] [Google Scholar]
- 12.Harris PV, Welner D, McFarland KC, Re E, Navarro Poulsen JC, Brown K, Salbo R, Ding H, Vlasenko E, Merino S, Xu F, Cherry J, Larsen S, Lo Leggio L. 2010. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry 49:3305–3316. 10.1021/bi100009p [DOI] [PubMed] [Google Scholar]
- 13.Moser F, Irwin D, Chen S, Wilson DB. 2008. Regulation and characterization of Thermobifida fusca carbohydrate-binding module proteins E7 and E8. Biotechnol. Bioeng. 100:1066–1077. 10.1002/bit.21856 [DOI] [PubMed] [Google Scholar]
- 14.Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sorlie M, Eijsink VG. 2010. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330:219–222. 10.1126/science.1192231 [DOI] [PubMed] [Google Scholar]
- 15.Igarashi K, Uchihashi T, Koivula A, Wada M, Kimura S, Okamoto T, Penttila M, Ando T, Samejima M. 2011. Traffic jams reduce hydrolytic efficiency of cellulase on cellulose surface. Science 333:1279–1282. 10.1126/science.1208386 [DOI] [PubMed] [Google Scholar]
- 16.Valjamae P, Sild V, Pettersson G, Johansson G. 1998. The initial kinetics of hydrolysis by cellobiohydrolases I and II is consistent with a cellulose surface-erosion model. Eur. J. Biochem. 253:469–475. 10.1046/j.1432-1327.1998.2530469.x [DOI] [PubMed] [Google Scholar]
- 17.Irwin D, Shin DH, Zhang S, Barr BK, Sakon J, Karplus PA, Wilson DB. 1998. Roles of the catalytic domain and two cellulose binding domains of Thermomonospora fusca E4 in cellulose hydrolysis. J. Bacteriol. 180:1709–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakon J, Irwin D, Wilson DB, Karplus PA. 1997. Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat. Struct. Biol. 4:810–818. 10.1038/nsb1097-810 [DOI] [PubMed] [Google Scholar]
- 19.Irwin DC, Zhang S, Wilson DB. 2000. Cloning, expression and characterization of a family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur. J. Biochem. 267:4988–4997. 10.1046/j.1432-1327.2000.01546.x [DOI] [PubMed] [Google Scholar]
- 20.Spiridonov NA, Wilson DB. 1998. Regulation of biosynthesis of individual cellulases in Thermomonospora fusca. J. Bacteriol. 180:3529–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kostylev M, Wilson D. 2013. Two-parameter kinetic model based on a time-dependent activity coefficient accurately describes enzymatic cellulose digestion. Biochemistry 52:5656–5664. 10.1021/bi400358v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cruys-Bagger N, Elmerdahl J, Praestgaard E, Tatsumi H, Spodsberg N, Borch K, Westh P. 2012. Pre-steady-state kinetics for hydrolysis of insoluble cellulose by cellobiohydrolase Cel7A. J. Biol. Chem. 287:18451–18458. 10.1074/jbc.M111.334946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Praestgaard E, Elmerdahl J, Murphy L, Nymand S, McFarland KC, Borch K, Westh P. 2011. A kinetic model for the burst phase of processive cellulases. FEBS J. 278:1547–1560. 10.1111/j.1742-4658.2011.08078.x [DOI] [PubMed] [Google Scholar]
- 24.Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VG. 2012. Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 5:45. 10.1186/1754-6834-5-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganner T, Bubner P, Eibinger M, Mayrhofer C, Plank H, Nidetzky B. 2012. Dissecting and reconstructing synergism: in situ visualization of cooperativity among cellulases. J. Biol. Chem. 287:43215–43222. 10.1074/jbc.M112.419952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeoh T, Wilson DB, Walker LP. 2006. Effect of cellulase mole fraction and cellulose recalcitrance on synergism in cellulose hydrolysis and binding. Biotechnol. Prog. 22:270–277. 10.1021/bp050266f [DOI] [PubMed] [Google Scholar]
- 27.Nidetzky B, Steiner W, Claeyssens M. 1994. Cellulose hydrolysis by the cellulases from Trichoderma reesei: adsorptions of two cellobiohydrolases, two endocellulases and their core proteins on filter paper and their relation to hydrolysis. Biochem. J. 303(Pt 3):817–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira OV, Freitas LCG, Straatsma TP, Lins RD. 2009. Interaction between the CBM of Cel9A from Thermobifida fusca and cellulose fibers. J. Mol. Recognit. 22:38–45. 10.1002/jmr.925 [DOI] [PubMed] [Google Scholar]
- 29.Chanzy H, Henrissat B, Vuong R, Schulein M. 1983. The action of 1,4-beta-d-glucan cellobiohydrolase on Valonia cellulose microcrystals. An electron microscopy study. FEBS Lett. 153:113–118 [Google Scholar]
- 30.Park S, Baker JO, Himmel ME, Parilla PA, Johnson DK. 2010. Cellulose crystallinity index: measurement techniques and their impact on interpreting cellulase performance. Biotechnol. Biofuels 3:10. 10.1186/1754-6834-3-10 [DOI] [PMC free article] [PubMed] [Google Scholar]


