Abstract
To assess the host specificity of Enterocytozoon bieneusi and to track the sources of E. bieneusi contamination, we genotyped E. bieneusi in wildlife and stormwater from the watershed of New York City's source water, using ribosomal internal transcribed spacer (ITS)-based PCR and sequence analyses. A total of 255 specimens from 23 species of wild mammals and 67 samples from stormwater were analyzed. Seventy-four (29.0%) of the wildlife specimens and 39 (58.2%) of the stormwater samples from streams were PCR positive. Altogether, 20 E. bieneusi genotypes were found, including 8 known genotypes and 12 new ones. Sixteen and five of the genotypes were seen in animals and stormwater from the watershed, respectively, with WL4 being the most common genotype in both animals (35 samples) and stormwater (23 samples). The 20 E. bieneusi genotypes belonged to five genogroups (groups 1, 3, 4, and 7 and an outlier), with only 23/113 (20.4%) E. bieneusi-positive samples belonging to zoonotic genogroup 1 and 3/20 genotypes ever being detected in humans. The two genogroups previously considered host specific, groups 3 and 4, were both detected in multiple groups of mammals. Thus, with the exception of the type IV, Peru11, and D genotypes, which were detected in only 7, 5, and 2 animals, respectively, most E. bieneusi strains in most wildlife samples and all stormwater samples in the watershed had no known public health significance, as these types have not previously been detected in humans. The role of different species of wild mammals in the contribution of E. bieneusi contamination in stormwater was supported by determinations of host-adapted Cryptosporidium species/genotypes in the same water samples. Data from this study indicate that the host specificity of E. bieneusi group 3 is broader than originally thought, and wildlife is the main source of E. bieneusi in stormwater in the watershed.
INTRODUCTION
Microsporidiosis is a significant cause of diarrhea, especially in children and immunocompromised persons. Although over a dozen microsporidian parasites have been found in humans, Enterocytozoon bieneusi is the species responsible for >90% of human microsporidian infections (1–3). It is also commonly found in animals, especially mammals and some birds (4). Although animals have been generally considered a major source of human E. bieneusi infection, this is based largely on results of genetic characterizations of parasites from humans and some animals (3, 4).
Water potentially plays an important role in the transmission of microsporidiosis. Epidemiological and environmental studies have frequently identified drinking of untreated water as a potential risk factor for microsporidiosis in humans (5, 6). A putative waterborne outbreak of intestinal microsporidiosis caused by E. bieneusi and Encephalitozoon intestinalis was reported in France (7), although the validity of the claim was disputed (8). Spores of E. bieneusi have been identified in surface water at high frequencies (9–14).
Sequence analysis of the ribosomal internal transcribed spacer (ITS) has been used extensively in molecular epidemiologic studies of E. bieneusi infection in humans and assessment of the human-infective potential of E. bieneusi in animals (3, 4). Over 100 E. bieneusi genotypes have been identified, belonging to at least six phylogenetically distinct genogroups (groups 1 to 5 and the so-called outlier in dogs) (15–17). Host adaptation is apparent among E. bieneusi genotypes (18–20), with almost all human isolates and some animal isolates belonging to genotypes in group 1 (3, 4). Other genogroups contain mostly animal-adapted genotypes, including group 2 genotypes in cattle, group 3 genotypes in muskrats, group 4 genotypes in raccoons, group 5 genotypes in primates, and the outlier in dogs (15–17). Sequence characterizations of other genes support the ITS classification of genogroups, as PCR primers based on nucleotide sequences of group 1 largely cannot amplify DNA from other E. bieneusi genogroups (21). Some additional genogroups, such as groups 6 and 7, have also been identified recently (22, 23), although the host specificity of them is less clear.
Determination of host adaptation of E. bieneusi genotypes, however, was based largely on ITS sequence characterizations of specimens from only a few domestic and wild animal species (18, 19, 24, 25). Although the bovine-adapted nature of group 2 genotypes was confirmed in subsequent studies (26–29), the host specificity of other genogroups remains to be determined. Indeed, group 4 genotypes, originally shown to be common in raccoons in the United States (18), have recently been found in wastewater in Tunisia, where raccoons do not exist, indicating that some other animals are probably also natural hosts of this group of genotypes (30). More studies on the distribution of E. bieneusi genotypes in wildlife are needed before we have a better understanding of host adaptation in E. bieneusi.
Despite the zoonotic potential of E. bieneusi in wildlife, few studies have examined the distribution of E. bieneusi genotypes in wild mammals in drinking source watersheds. In a study of fecal specimens from 239 muskrats, 85 beavers, 67 foxes, 55 raccoons, and 19 otters in watersheds in the Chesapeake Bay area, 17 E. bieneusi genotypes (WL1 to WL17) were found among 59 PCR-positive specimens. Three E. bieneusi genogroups were found, with groups 1, 3, and 4 seen in 43 animals (from 13 beavers, 11 muskrats, 9 foxes, 8 raccoons, and 2 otters), 9 animals (all from muskrats), and 7 animals (all from raccoons), respectively (18). Likewise, only a few studies have assessed the public health potential of E. bieneusi in water by using genotyping tools, which showed the presence of group 1 genotypes in a small number of samples genotyped (13, 14).
In this study, E. bieneusi in fecal specimens from wildlife living in the watershed of the New York City (NYC) water supply system was detected and genotyped by PCR and sequence analysis of the ITS. The host range of each genotype was used to assess the extent of host adaptation in E. bieneusi. Data on the host range of E. bieneusi genotypes were used to infer the source and potential public health significance of E. bieneusi in storm runoff within the watershed. The latter interpretation was also aided by data on the distribution of host-adapted Cryptosporidium species/genotypes present in the water samples. Data from this study indicated that most E. bieneusi isolates in wild animals and all isolates from water in the watershed had not been found in humans, although the host range of some of the so-called host-adapted genogroups was broader than previously believed.
MATERIALS AND METHODS
Animal specimens.
Fecal specimens were collected between September 2005 and July 2007 from 255 wild mammals in the New York City Department of Environmental Protection (NYCDEP) watershed. They were a part of a previous study on the distribution of Cryptosporidium species and genotypes in wildlife (31). The collection areas included reservoir buffer lands (forest, woodland, transition edge, and mixed meadow/shrub), areas along stream corridors, and maintained surfaces (lawns and parks). The collection of animal fecal specimens was described in detail previously (31). All animals were identified by species (except Peromyscus) and age, with most animals being adults. Samples were initially collected by NYCDEP staff and then shipped in coolers to the laboratory at the Centers for Disease Control and Prevention (CDC) for pathogen detection by PCR.
Stormwater samples.
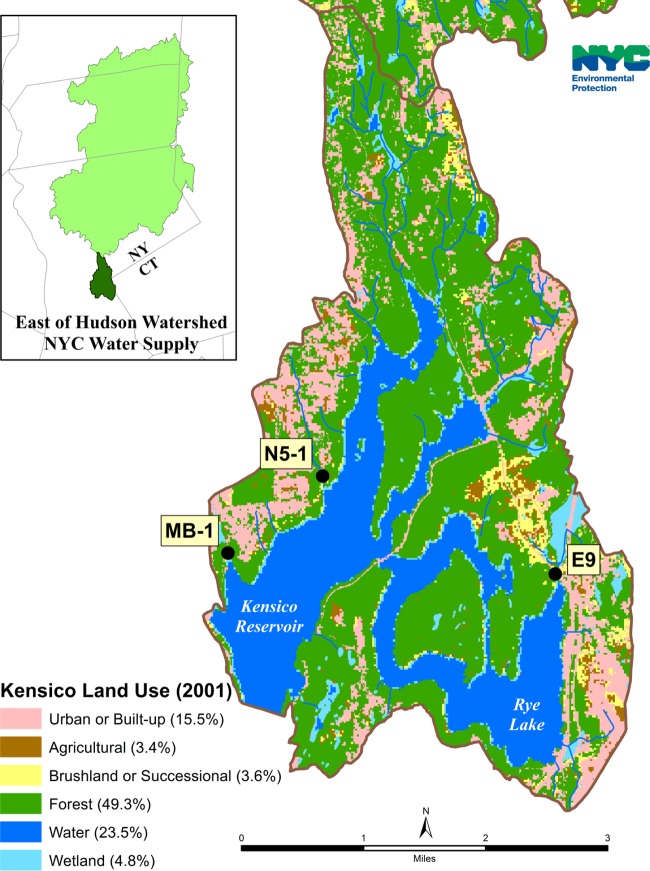
Sixty-seven stormwater samples were collected during July 2002 to October 2003 from three sites within the NYCDEP Kensico watershed in Valhalla, NY, including the E9, Malcolm Brook, and N5 basins. They were mostly a part of a previous study on the distribution and public health potential of Cryptosporidium species and genotypes in stormwater within the New York City drinking source water supply (32). The N5 stream basin is 298 acres, is located on the west side of the watershed, and consists almost entirely of residential lots (91%). The Malcolm Brook basin is 95 acres, is close to the N5 site, and has large wooded areas (39% of the total area) and corporate parks (22%), in addition to relatively high-density suburban residential lots (34%) with public sewer systems. The E9 stream basin, in contrast, is located on the east side of the watershed, is 321 acres, and drains a large wetland area (18.4% of the total area). In addition, it has large wooded areas (56.5%) and some development areas (17.6%) (Fig. 1).
FIG 1.
Sampling sites in the Kensico watershed of the New York City Department of Environmental Protection (NYCDEP).
Stormwater samples were collected with preset autosamplers (Isco 6700; Isco, Inc., Lincoln, NE), which were set to collect 20-liter composite samples over a predicted intensity of the storm event. The samples were filtered through an Envirochek HV filter (Pall Gelman Laboratory, Ann Arbor, MI) in the NYCDEP laboratory according to procedures described in U.S. Environmental Protection Agency Method 1623 (33). Material on the filter was eluted, and a portion of the concentrated pellet from each sample was transported to the CDC laboratory for molecular analysis. The Malcolm Brook and N5 basin sites and stormwater sample collection were described in detail previously (32). All stormwater samples were collected in stream channels prior to entry into the reservoir; therefore, they were not yet subjected to reservoir residence time, treatment with chlorine, or UV disinfection.
DNA extraction.
After washing the specimens twice with distilled water by centrifugation, genomic DNA was extracted from 0.2 ml of fecal specimens or 0.5 ml of stormwater concentrates by using the FastDNA Spin kit for soil (MP Biomedicals, Santa Ana, CA) and eluted in 100 μl of reagent-grade water, as described previously (34). The extracted DNAs from both fecal specimens and stormwater samples were used for PCR detection of E. bieneusi, whereas those from stormwater samples were further analyzed for Cryptosporidium by PCR. To neutralize residual PCR inhibitors in the extracted DNA, 400 ng/μl of nonacetylated bovine serum albumin (Sigma-Aldrich, St. Louis, MO) was used for primary PCR for both E. bieneusi and Cryptosporidium detection. In a previous study on the distribution of Cryptosporidium genotypes in these and other stormwater samples (32), DNA was extracted by using a QIAamp DNA minikit from immunomagnetic separation-purified oocysts present in a separate portion of the water concentrates.
E. bieneusi detection and genotyping.
A nested PCR analysis of a ∼392-bp fragment of the rRNA gene containing the entire internal transcribed spacer (ITS) was used to detect E. bieneusi, using a dog-specific E. bieneusi genotype (PtEbIX) as the positive control (18). The primers used included AL4037 (5′-GATGGTCATAGGGATGAAGAGCTT-3′) and AL4039 (5′-ACGGATCCAAGTGATCCTGTATT-3′) for primary PCR and AL4038 (5′-AGGGATGAAGAGCTTCGGCTCTG-3′) and AL4040 (5′-AGTGATCCTGTATTAGGGATATT-3′) for secondary PCR. Each animal specimen was analyzed twice by using 2 μl of extracted DNA per PCR, whereas each stormwater sample was analyzed once by using of 3 μl of extracted DNA per PCR. Positive PCR products were sequenced to determine E. bieneusi genotypes. E. bieneusi genotypes were named according to an established nomenclature system (35). The genotypes with ITS sequences identical to those in the GenBank database were considered known genotypes. In contrast, genotypes that produced ITS sequences with any single-nucleotide substitutions, deletions, or insertions and were confirmed by DNA sequencing of at least two PCR products were considered new genotypes.
Cryptosporidium detection and genotyping.
To aid in the interpretation of the source of E. bieneusi genotypes in stormwater samples, we also genotyped Cryptosporidium present in the same extracted DNAs by nested PCR amplification of an approximately 830-bp fragment of the small-subunit (SSU) rRNA gene, as Cryptosporidium spp. are known to have more strict host specificity. The primers used include AL1687 (5′-TTCTAGAGCTAATACATGCG-3′) and AL3417 (5′-CCCATTTCCTTCGAAACAGGA-3′) for primary PCR and AL3032 (5′-GGAAGGGTTGTATTTATTAGATAAAG-3′) and AL4871 (5′-CTCATAAGGTGCTGAAGGAGTA-3′) for secondary PCR (32). DNA of Cryptosporidium serpentis was used as the positive control. The Cryptosporidium species and genotypes present were differentiated by restriction fragment length polymorphism (RFLP) analysis of the secondary PCR products using the restriction enzymes SspI and VspI (32). All positive PCR products were sequenced to confirm the genotype identification. To detect the occurrence of mixed Cryptosporidium species and genotypes, each water sample was analyzed six times by the PCR-RFLP technique, using 0.5, 1.0, 1.5, 2.0, 2.5, and 3.0 μl of DNA per PCR.
Sequence analysis.
After purification using Montage PCR filters (Millipore, Bedford, MA), the secondary PCR products were sequenced in both directions with secondary PCR primers by using the ABI BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) and an ABI3130 genetic analyzer (Applied Biosystems). Nucleotide sequences obtained were aligned with reference sequences downloaded from the GenBank database by using ClustalX (http://www.clustal.org/) to determine E. bieneusi genotypes and Cryptosporidium species or genotypes. For ITS sequences of E. bieneusi, to confirm the genogroup designation and assess the genetic relationship among genotypes, a neighbor-joining tree was constructed by using Mega 5.2 (http://www.megasoftware.net/), based on genetic distances calculated by the Kimura two-parameter model. A sequence (GenBank accession no. DQ885585) from the dog-specific genotype (PtEbIX) was used as the outgroup. The reliability of various clusters was evaluated by the bootstrap method with 1,000 replicates. The topology of the tree was supported by a maximum parsimony analysis of the same sequence alignment, with all alignment sites taken into consideration.
Nucleotide sequence accession numbers.
Unique E. bieneusi ITS sequences obtained from wildlife and stormwater during the study were deposited in the GenBank database under accession numbers KF591677 to KF591688.
RESULTS
Prevalence of E. bieneusi in wild mammals.
E. bieneusi was detected in 74 (29.0%) of the 255 wild mammals sampled (Table 1). These animals belonged to 23 species in six orders. Most of the positive animals (64/74) were rodents and carnivores, while only 10 were other mammals. Rodents, however, accounted for over one-half (142/255) of the sampled animals and had an E. bieneusi infection rate (38/142; 26.8%) similar to the average infection rate. Within rodents, members of the squirrel family had the highest infection rate (21/49; 42.9%), while the remaining 17 positive rodents were all from the Cricetidae (deer mice and voles). Among two other groups of animals commonly examined, carnivores had an infection rate (26/42; 61.9%) much higher than the average, whereas ruminants (white-tailed deer) had an infection rate (6/49; 12.2%) lower than the average. The small numbers of insectivores (5 animals), lagomorphs (8), and marsupials (9) sampled did not allow an accurate determination of E. bieneusi infection rates; only 3/8 eastern cottontails and 1/9 Virginia opossums sampled were positive (Table 1).
TABLE 1.
Prevalence of Enterocytozoon bieneusi genotypes in wild mammals in the NYCDEP watershed
| Animal | No. of samples | No. of positive samples | Infection rate (%) | Genotype(s) (no. of samples positive) | Genogroup(s) (no. of samples positive) |
|---|---|---|---|---|---|
| Rodents | 142 | 38 | 26.8 | ||
| Sciuridae | 49 | 21 | 42.9 | ||
| Sciurus carolinensis (eastern gray squirrel) | 34 | 11 | 32.4 | Type IV (3), WL4 (5), WW6 (2), PtEbV + WL21 (1) | 1 (4), 3 (5), 4 (2) |
| Sciurus vulgaris (red squirrel) | 2 | 0 | 0 | ||
| Glaucomys volans (southern flying squirrel) | 1 | 0 | 0 | ||
| Tamias striatus (eastern chipmunk) | 7 | 5 | 71.4 | Type IV (1), WL4 (3), WL23 (1) | 1 (1), 3 (4) |
| Marmota monax (woodchuck) | 5 | 5 | 100.0 | Type IV + WL20 (1), WL4 (2), WL22 (1), WW6 (1) | 1 (1), 3 (3), 4 (1) |
| Castoridae | 16 | 0 | 0 | ||
| Castor canadensis (beaver) | 16 | 0 | 0 | ||
| Muridae | 1 | 0 | 0 | ||
| Rattus norvegicus (Norway rat) | 1 | 0 | 0 | ||
| Zapodidae | 1 | 0 | 0 | ||
| Napaeozapus insignis (woodland jumping mouse) | 1 | 0 | 0 | ||
| Cricetidae | 71 | 17 | 23.9 | ||
| Peromyscus sp. (deer mouse) | 55 | 13 | 23.6 | WL4 (10), WL23 (2), WL25 (1) | 3 (13) |
| Myodes gapperi (boreal red-backed vole) | 5 | 1 | 20.0 | WL20 + WL21(1) | 1 (1) |
| Microtus pennsylvanicus (meadow vole) | 10 | 3 | 30.0 | Peru11 (1), Peru11 + type IV (1), WL21 + unknown (1) | 1 (3) |
| Ondatrini zibethicus (muskrat) | 1 | 0 | 0 | ||
| Erethizontidae | 4 | 0 | 0 | ||
| Erethizon dorsatum (porcupine) | 4 | 0 | 0 | ||
| Carnivores | 42 | 26 | 61.9 | ||
| Mustela vison (mink) | 4 | 0 | 0 | ||
| Mustela erminea (ermine) | 1 | 1 | 100.0 | WL4 (1) | 3 (1) |
| Procyon lotor (raccoon) | 22 | 18 | 81.8 | WL4 (8), WW6 (7), Peru11 (1), WL26 (1), WL24 (1) | 3 (8), 4 (8), 1 (1), 7 (1) |
| Ursus americanus (black bear) | 5 | 2 | 40.0 | Type IV (1), WL4 (1) | 1 (1), 3 (1) |
| Lontra canadensis (river otter) | 8 | 5 | 62.5 | D (2), WL4 (2), WL2 (1) | 1 (2), 3 (2), 4 (1) |
| Mephitis mephitis (striped skunk) | 2 | 0 | 0 | ||
| Insectivores | 5 | 0 | 0 | ||
| Blarina brevicauda (northern short-tailed shrew) | 5 | 0 | 0 | ||
| Lagomorpha | 8 | 3 | 37.5 | ||
| Sylvilagus floridanus (eastern cottontail) | 8 | 3 | 37.5 | Peru11 (1), Peru11 + WL4 (1), mixed (1) | 1 (1), 1 + 3 (1), unknown (1) |
| Ruminants | 49 | 6 | 12.2 | ||
| Odocoileus virginianus (white-tailed deer) | 49 | 6 | 12.2 | WL18 (2), WL19 (2), WL4 (2) | 1 (4), 3 (2) |
| Marsupials | 9 | 1 | 11.1 | ||
| Didelphis virginiana (Virginia opossum) | 9 | 1 | 11.1 | Mixed (1) | Unknown (1) |
| Totala | 255 | 74 | 29.0 |
There were 16 total genotypes detected and 4 total genogroups detected.
E. bieneusi genotypes in wildlife.
Altogether, 16 E. bieneusi genotypes were found among the 74 PCR-positive specimens, including 7 known genotypes (D, Peru11, PtEbV, type IV, WL2, WL4, and WW6) and 9 new ones (named WL18 to WL26 in this study). WL4 was the most common genotype (35), followed by WW6 (10), type IV (7), and Peru11 (5). Other genotypes were detected in only 1 to 3 animals (Table 1). Seven specimens had mixed genotypes, as reflected by discordant genotype results in replicate PCRs (five samples) or underlying signals in sequence trace files, which prevented determination of genotypes (two samples).
E. bieneusi genotypes in stormwater samples.
Thirty-nine (58.2%) of the 67 stormwater samples analyzed were PCR positive for E. bieneusi, including 3 of 4 (75.0%) samples from the E9 basin, 3 of 15 (20.0%) samples from Malcolm Brook, and 33 of 48 (68.8%) samples from the N5 basin. These isolates belonged to five genotypes, including two known genotypes (WL4 and WL6) and three new genotypes (called SW1, SW2, and SW3). WL4 was the most common genotype, being seen in 23 samples. This was followed by WL6, being seen in five samples. In contrast, the three new genotypes were each seen in only 1 to 2 samples. Seven samples had mixed genotypes, as reflected by the underlying signals in sequence trace files. There were apparent differences in the occurrences and genotype distributions of E. bieneusi among the three sampling sites. The N5 and E9 basins had a higher occurrence of E. bieneusi than Malcolm Brook, with PCR positivities of 68.8%, 75.0%, and 20.0%, respectively. WL4 was also the dominant E. bieneusi genotype in the N5 and E9 basins but was absent in Malcolm Brook (Table 2).
TABLE 2.
Enterocytozoon bieneusi and Cryptosporidium genotypes in stormwater samples from the NYCDEP watershed
| Site | No. of samples |
E. bieneusi |
Cryptosporidium |
Possible major animal source(s) for pathogens in water | |||||
|---|---|---|---|---|---|---|---|---|---|
| No. (%) of positive samples | Genotypes (no. of samples positive) | Genogroups (no. of samples positive) | Major known host(s) | No. (%) of positive samples | Genotypesa (no. of replicate PCR-positive samples) | Known hosts | |||
| E9 basin | 4 | 3 (75.0) | WL4 (2), mixed (1) | 3 (2), unknown (1) | Deer mice, muskrats, raccoons, squirrel family | 3 (75.0) | W7 (4), W10 (2), W11 (2), W16 (1) | Muskrats, birds, snakes | Muskrats |
| Malcolm Brook | 15 | 3 (20.0) | SW1 (1), SW3 (1), mixed (1) | 1 (1), outlier (1), unknown (1) | Unknown | 6 (40.0) | W4 (10), W10 (2), W7 (1) | Squirrel family, birds | Squirrel family |
| N5 basin | 48 | 33 (68.8) | SW1 (1), SW2 (1), WL4 (21), WL6 (5), mixed (5) | 1 (2), 3 (26), unknown (5) | Deer mice, muskrats, raccoons, squirrel family | 35 (72.9) | W4 (23), W1 (15), W13 (14), W10 (8), W7 (7), W19 (5), W16 (3), W17 (2), W8 (2), W12 (2), W15 (2), W5 (1), W11 (1) | Squirrel family, raccoons | Squirrel family, raccoons, muskrats |
| Totalb | 67 | 39 (58.2) | 44 (65.7) | ||||||
The abbreviations of Cryptosporidium genotypes found in water are as follows: W1, deer mouse genotype III; W4, C. ubiquitum; W5, shrew genotype; W7, muskrat genotype I; W8, opossum genotype II; W10, C. baileyi; W11, snake genotype; W12, unknown genotype; W13, skunk genotype; W15, vole genotype; W16, muskrat genotype II; W17, chipmunk genotype I; W19, rat genotype IV.
There were 5 total genotypes and 2 total genogroups for E. bieneusi and 13 total Cryptosporidium genotypes detected.
Phylogenetic relationship of E. bieneusi isolates.
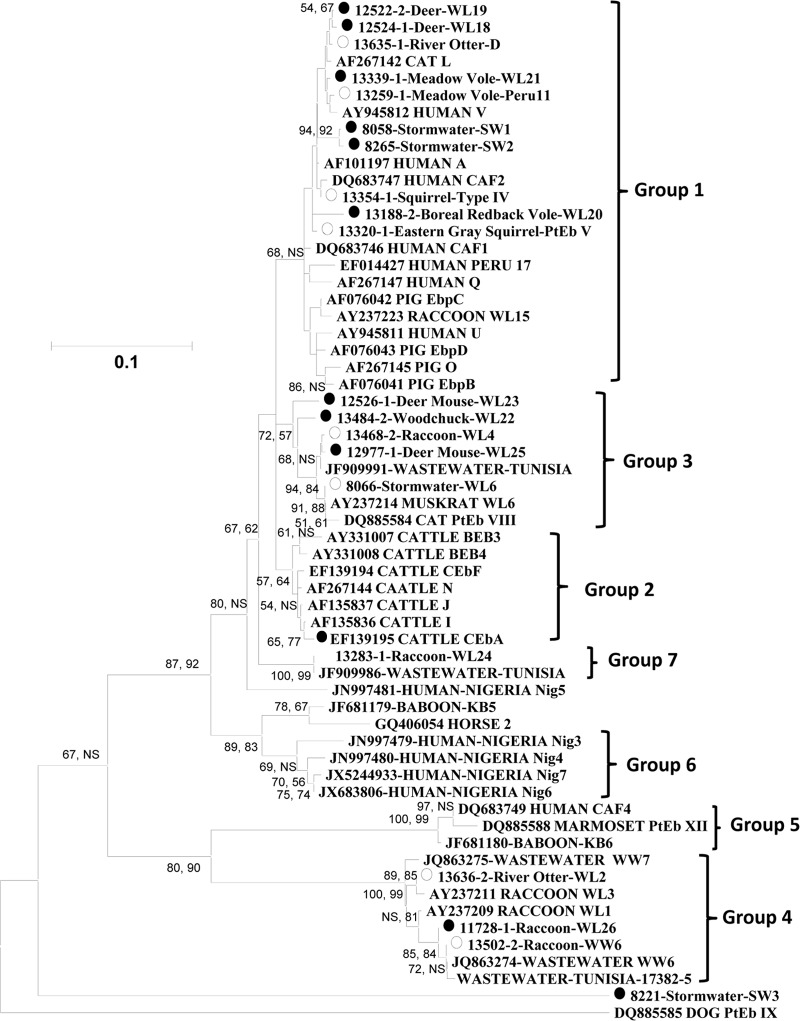
Altogether, 20 E. bieneusi genotypes were found in this study. Both the neighbor-joining and maximum parsimony analyses suggested that these genotypes belonged to five genotype groups or genogroups: groups 1, 3, 4, and 7 and an outlier (Fig. 2). Four of the five genogroups, including groups 1, 3, 4, and 7, were seen in 20, 40, 12, and 1 animal, respectively (Table 1). Groups 1 and 3 had a broad host range; each group was detected in 10 animal species belonging to the rodent, carnivore, lagomorph, and ruminant groups. In contrast, group 4 was detected in only four animal species belonging to the carnivore and rodent groups, with raccoons being the major host and members of the squirrel family being minor hosts. The only detection of group 7 was in a raccoon (Table 1). Three of the five genogroups were found in stormwater samples, including groups 1 and 3 and an outlier, with the majority (28/32) of the genotyped samples having group 3 genotypes (Table 2). An almost identical topology was produced by both phylogenetic approaches. The only exception was the placement of the outlier found in one stormwater sample. In the neighbor-joining analysis, the outlier was placed near the base of the tree but was separated from genotype PtEbIX in dogs (Fig. 2). In the maximum parsimony analysis, the outlier grouped together with genotype PtEbIX, with 94% bootstrap support.
FIG 2.
Phylogenetic relationship among various E. bieneusi genotypes in wildlife in the NYCDEP watershed and known E. bieneusi genotypes, as inferred by a neighbor-joining analysis of SSU rRNA sequences. Genotypes with open circles are known genotypes found in the NYCDEP watershed, and those with black circles are new genotypes. A similar tree topology was also produced by maximum parsimony analysis, with the exception that the SW3 genotype grouped together with genotype PtEbIX, with 94% bootstrap support. Numbers on branches are percent bootstrap values (>50) using 1,000 replicates, with the first number being generated by neighbor-joining analysis and the second number being generated by maximum parsimony analysis. NS, nonsignificant (bootstrap value of <50%).
Cryptosporidium genotypes in stormwater.
Forty-four (65.7%) of the 67 stormwater samples analyzed were PCR positive for Cryptosporidium spp. in at least one of the six replicate PCR analyses, including 3 of 4 (75.0%) samples from the E9 basin, 6 of 15 (40.0%) samples from Malcolm Brook, and 35 of 48 (72.9%) samples from the N5 basin (Table 2). Of the 44 Cryptosporidium-positive samples, 9, 15, 15, 24, 20, and 25 samples generated the expected SSU rRNA PCR products when 0.5, 1.0, 1.5, 2.0, 2.5, and 3.0 μl of the 100 μl of extracted DNA were used for PCR, respectively. In general, there were good agreements in the occurrences of Cryptosporidium and E. bieneusi, although the number of samples from one of the study sites, the E9 basin, was small. Thus, the overall prevalence of Cryptosporidium in these water samples was similar to that of E. bieneusi (72.9% and 68.8%, respectively); sites with higher E. bieneusi occurrences also had higher Cryptosporidium occurrences, and E. bieneusi-positive water samples were almost twice as likely to be positive for Cryptosporidium than were E. bieneusi-negative samples (32/39 [82.1%] versus 12/28 [42.8%]).
Altogether, 13 Cryptosporidium species/genotypes were found in stormwater, including deer mouse genotype III (W1), C. ubiquitum (W4), the shrew genotype (W5), muskrat genotype I (W7), opossum genotype II (W8), C. baileyi (W10), the snake genotype (W11), an unknown genotype (W12), the skunk genotype (W13), the vole genotype (W15), muskrat genotype II (W16), chipmunk genotype I (W17), and rat genotype IV (W19). The most common genotypes included C. ubiquitum, deer mouse genotype III, the skunk genotype, muskrat genotype I, and C. baileyi, being found in 33, 15, 14, 12, and 12 PCR replicates, respectively. The remaining genotypes were each found in only 1 to 5 PCR replicates. There were differences in the distributions of common Cryptosporidium species/genotypes among study sites. Thus, although C. baileyi and muskrat genotype I were found at all three sampling sites, deer mouse genotype III and the skunk genotype were found only in the N5 basin. Likewise, the most commonly detected Cryptosporidium species in the study, C. ubiquitum, was absent in the E9 basin (Table 2).
DISCUSSION
The results of this study suggest that the host range of group 3 E. bieneusi genotypes is much broader than previously believed. This group of genotypes was initially found in muskrats in the Chesapeake area. Subsequent ITS sequence analyses of numerous E. bieneusi isolates from domestic animals and humans have failed to detect them thus far. In this study, however, one known genotype and three new genotypes of this phylogenetically unique group were found in 10 species of mammals of four orders. This is about the same host range of eight group 1 genotypes in this study, which are well known to have a broad host range. In fact, one member of group 3, WL4, was by far the most commonly detected genotype in this study, being identified in nearly one-half (35/74) of the E. bieneusi-positive animals. Likewise, group 4 genotypes, which were previously considered raccoon specific, were also found in other carnivores in addition to raccoons as well as in several species of rodents in the squirrel family. Nevertheless, these genetically unique genotypes are rarely seen in domestic animals, implying that they do have some host preference. Indeed, in one of the most sampled animals in this study, deer mice, only group 3 genotypes were found in the 13 E. bieneusi-positive animals.
The dominant E. bieneusi genotype in animals, WL4, was also the dominant genotype in stormwater samples, suggesting that wildlife played a significant role in contamination of stormwater by E. bieneusi in the watershed. This was especially the case in the N5 basin, which is much larger and less commercially developed than the neighboring Malcolm Brook. The high usage of lands for commercial parks and high-density residential lots in the Malcolm Brook basin might have reduced the activity of wildlife, thus resulting in the disappearance of the WL4 genotype at this site. Interestingly, although the N5 basin consists mostly of large residential lots and Malcolm Brook has many high-density residential lots, no group 1 genotypes previously detected in humans were found in any stormwater samples, indicating that humans and companion animals were not major contributors to E. bieneusi contamination in stormwater in the watershed. This is in agreement with the genotyping results for Cryptosporidium oocysts isolated from different aliquots of these samples in a previous study, which showed that wildlife contributed exclusively to Cryptosporidium spp. in stormwater from the Malcolm Brook and N5 basins (32).
The lack of strict host specificity makes it difficult to track the source of contamination of E. bieneusi in stormwater to specific wildlife species or groups, especially when the dominant genotype, WL4, was found in 10 species of mammals of four orders in the watershed. Because WL4 was most often detected in members of the squirrel family, deer mice, and raccoons in this study and was shown to be a common genotype in muskrats, it is likely that these animals were the main source of E. bieneusi in stormwater. To facilitate the interpretation of contamination in stormwater, we analyzed the same DNA from stormwater samples in this study for Cryptosporidium spp. There was good agreement in the occurrences of Cryptosporidium and E. bieneusi in both the overall prevalence and sample-specific positivity. There was also good agreement between the nature of dominant E. bieneusi and Cryptosporidium genotypes at the Malcolm Brook and N5 basins (only four samples were collected from the E9 basin). Over one-half of the Cryptosporidium-positive stormwater samples (21/35) and 33 of 108 PCR-positive replicates had C. ubiquitum (W4), which was found mostly in members of the squirrel family in the watershed. This, together with the occurrence of another Cryptosporidium genotype of the squirrel family, chipmunk genotype I (W17), supports the role of these animals in E. bieneusi contamination in the watershed, especially in the N5 basin and Malcolm Brook. The role of raccoons in E. bieneusi contamination in the N5 basin was also supported by the common finding (in 11 samples and 14 PCR replicates) of the Cryptosporidium skunk genotype (commonly found in raccoons) in stormwater samples. Likewise, the relatively common finding of Cryptosporidium muskrat genotypes I (W7) and II (W16) and the vole genotype (W15) indicates that muskrats contributed to E. bieneusi contamination in water in the N5 and E9 basins. In contrast, Cryptosporidium genotyping results do not support a significant role of deer mice in E. bieneusi contamination at all these sampling sites. Although Cryptosporidium deer mouse genotype III (W1) was commonly seen in stormwater samples (6 samples and 15 PCR replicates), another common Cryptosporidium genotype in deer mice, deer mouse genotype IV (W3), was absent. As deer mouse genotype III also occurs in squirrels at a high frequency, members of the squirrel family could be responsible for the occurrence of deer mouse genotype III in stormwater. Taken together, data from E. bieneusi and Cryptosporidium genotyping indicate that muskrats were a major source of pathogen contamination in stormwater in the E9 basin, and squirrel family members were a major source in Malcolm Brook, while squirrel family members, raccoons, and muskrats were all major sources of pathogens in stormwater in the N5 basin (Table 2). These results are in agreement with the environmental ecology and land use at these sampling sites.
Most of the E. bieneusi isolates found in wildlife and stormwater samples are not from human-pathogenic genotypes. Among the 20 E. bieneusi genotypes found in this study, only 3 have ever been detected in humans: type IV, Peru11, and D, which were detected in 7, 5, and 2 animals, respectively. None of them have been found in stormwater. Seven other genotypes in the study also belong to the potentially human-pathogenic group 1, but altogether, group 1 genotypes were found in only 20 of 255 (7.8%) wildlife samples and 20 of 74 (27.0%) E. bieneusi-positive animals. Likewise, only 3 of 67 (4.5%) stormwater samples and 3 of 39 (7.7%) E. bieneusi-positive stormwater samples had group 1 genotypes. Some of the group 1 genotypes in wildlife and stormwater samples are newly discovered genotypes. Because host adaptation also exists within group 1 genotypes (36), it is likely that not all group 1 genotypes are human pathogenic. Thus, wildlife may contribute to E. bieneusi contamination in stormwater but most likely do not have public health significance because they are generally infected with non-human-pathogenic genotypes. Moreover, stormwater samples from this study were collected prior to entry into the reservoir and any treatment with chlorine or UV disinfection, which may further reduce the public health impact of E. bieneusi in source water.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grants 31229005 and 31110103901); Fundamental Research Funds for the Central Universities, China; the National Special Fund for the State Key Laboratory of Bioreactor Engineering (no. 2060204); and the Centers for Disease Control and Prevention.
We thank the NYCDEP Water Quality staff for the collection and shipping of wildlife and stormwater samples for this study.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print 18 October 2013
REFERENCES
- 1.Ghosh K, Weiss LM. 2009. Molecular diagnostic tests for microsporidia. Interdiscip. Perspect. Infect. Dis. 2009:926521. 10.1155/2009/926521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Didier ES, Weiss LM. 2006. Microsporidiosis: current status. Curr. Opin. Infect. Dis. 19:485–492. 10.1097/01.qco.0000244055.46382.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matos O, Lobo ML, Xiao L. 2012. Epidemiology of Enterocytozoon bieneusi infection in humans. J. Parasitol. Res. 2012:981424. 10.1155/2012/981424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santin M, Fayer R. 2011. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res. Vet. Sci. 90:363–371. 10.1016/j.rvsc.2010.07.014 [DOI] [PubMed] [Google Scholar]
- 5.Didier ES, Stovall ME, Green LC, Brindley PJ, Sestak K, Didier PJ. 2004. Epidemiology of microsporidiosis: sources and modes of transmission. Vet. Parasitol. 126:145–166. 10.1016/j.vetpar.2004.09.006 [DOI] [PubMed] [Google Scholar]
- 6.Wumba R, Longo-Mbenza B, Menotti J, Mandina M, Kintoki F, Situakibanza NH, Kakicha MK, Zanga J, Mbanzulu-Makola K, Nseka T, Mukendi JP, Kendjo E, Sala J, Thellier M. 2012. Epidemiology, clinical, immune, and molecular profiles of microsporidiosis and cryptosporidiosis among HIV/AIDS patients. Int. J. Gen. Med. 5:603–611. 10.2147/IJGM.S32344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotte L, Rabodonirina M, Chapuis F, Bailly F, Bissuel F, Raynal C, Gelas P, Persat F, Piens MA, Trepo C. 1999. Waterborne outbreak of intestinal microsporidiosis in persons with and without human immunodeficiency virus infection. J. Infect. Dis. 180:2003–2008. 10.1086/315112 [DOI] [PubMed] [Google Scholar]
- 8.Hunter PR. 2000. Waterborne outbreak of microsporidiosis. J. Infect. Dis. 182:380–381. 10.1086/315654 [DOI] [PubMed] [Google Scholar]
- 9.Graczyk TK, Sunderland D, Awantang GN, Mashinski Y, Lucy FE, Graczyk Z, Chomicz L, Breysse PN. 2010. Relationships among bather density, levels of human waterborne pathogens, and fecal coliform counts in marine recreational beach water. Parasitol. Res. 106:1103–1108. 10.1007/s00436-010-1769-2 [DOI] [PubMed] [Google Scholar]
- 10.Graczyk TK, Girouard AS, Tamang L, Nappier SP, Schwab KJ. 2006. Recovery, bioaccumulation, and inactivation of human waterborne pathogens by the Chesapeake Bay nonnative oyster, Crassostrea ariakensis. Appl. Environ. Microbiol. 72:3390–3395. 10.1128/AEM.72.5.3390-3395.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coupe S, Delabre K, Pouillot R, Houdart S, Santillana-Hayat M, Derouin F. 2006. Detection of Cryptosporidium, Giardia and Enterocytozoon bieneusi in surface water, including recreational areas: a one-year prospective study. FEMS Immunol. Med. Microbiol. 47:351–359. 10.1111/j.1574-695X.2006.00098.x [DOI] [PubMed] [Google Scholar]
- 12.Graczyk TK, Conn DB, Lucy F, Minchin D, Tamang L, Moura LN, DaSilva AJ. 2004. Human waterborne parasites in zebra mussels (Dreissena polymorpha) from the Shannon River drainage area, Ireland. Parasitol. Res. 93:385–391. 10.1007/s00436-004-1142-4 [DOI] [PubMed] [Google Scholar]
- 13.Ye J, Xiao L, Ma J, Guo M, Liu L, Feng Y. 2012. Anthroponotic enteric parasites in monkeys in public park, China. Emerg. Infect. Dis. 18:1640–1643. 10.3201/eid1810.120653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galvan AL, Magnet A, Izquierdo F, Fenoy S, Rueda C, Fernandez Vadillo C, Henriques-Gil N, del Aguila C. 2013. Molecular characterization of human-pathogenic microsporidia and Cyclospora cayetanensis isolated from various water sources in Spain: a year-long longitudinal study. Appl. Environ. Microbiol. 79:449–459. 10.1128/AEM.02737-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breton J, Bart-Delabesse E, Biligui S, Carbone A, Seiller X, Okome-Nkoumou M, Nzamba C, Kombila M, Accoceberry I, Thellier M. 2007. New highly divergent rRNA sequence among biodiverse genotypes of Enterocytozoon bieneusi strains isolated from humans in Gabon and Cameroon. J. Clin. Microbiol. 45:2580–2589. 10.1128/JCM.02554-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thellier M, Breton J. 2008. Enterocytozoon bieneusi in human and animals, focus on laboratory identification and molecular epidemiology. Parasite 15:349–358. 10.1051/parasite/2008153349 [DOI] [PubMed] [Google Scholar]
- 17.Henriques-Gil N, Haro M, Izquierdo F, Fenoy S, del Aguila C. 2010. Phylogenetic approach to the variability of the microsporidian Enterocytozoon bieneusi and its implications for inter- and intrahost transmission. Appl. Environ. Microbiol. 76:3333–3342. 10.1128/AEM.03026-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sulaiman IM, Fayer R, Lal AA, Trout JM, Schaefer FW, III, Xiao L. 2003. Molecular characterization of microsporidia indicates that wild mammals harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl. Environ. Microbiol. 69:4495–4501. 10.1128/AEM.69.8.4495-4501.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sulaiman IM, Fayer R, Yang C, Santin M, Matos O, Xiao L. 2004. Molecular characterization of Enterocytozoon bieneusi in cattle indicates that only some isolates have zoonotic potential. Parasitol. Res. 92:328–334. 10.1007/s00436-003-1049-5 [DOI] [PubMed] [Google Scholar]
- 20.Widmer G, Akiyoshi DE. 2010. Host-specific segregation of ribosomal nucleotide sequence diversity in the microsporidian Enterocytozoon bieneusi. Infect. Genet. Evol. 10:122–128. 10.1016/j.meegid.2009.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Y, Li N, Dearen T, Lobo ML, Matos O, Cama V, Xiao L. 2011. Development of a multilocus sequence typing tool for high-resolution genotyping of Enterocytozoon bieneusi. Appl. Environ. Microbiol. 77:4822–4828. 10.1128/AEM.02803-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li N, Xiao L, Wang L, Zhao S, Zhao X, Duan L, Guo M, Liu L, Feng Y. 2012. Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in wastewater. PLoS Negl. Trop. Dis. 6:e1809. 10.1371/journal.pntd.0001809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akinbo FO, Okaka CE, Omoregie R, Adamu H, Xiao L. 2013. Unusual Enterocytozoon bieneusi genotypes and Cryptosporidium hominis subtypes in HIV-infected patients on highly active antiretroviral therapy. Am. J. Trop. Med. Hyg. 89:157–161. 10.4269/ajtmh.12-0635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santin M, Cortes Vecino JA, Fayer R. 2008. Enterocytozoon bieneusi genotypes in dogs in Bogota, Colombia. Am. J. Trop. Med. Hyg. 79:215–217 http://www.ajtmh.org/content/79/2/215.long [PubMed] [Google Scholar]
- 25.Li W, Kiulia NM, Mwenda JM, Nyachieo A, Taylor MB, Zhang X, Xiao L. 2011. Cyclospora papionis, Cryptosporidium hominis, and human-pathogenic Enterocytozoon bieneusi in captive baboons in Kenya. J. Clin. Microbiol. 49:4326–4329. 10.1128/JCM.05051-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santin M, Fayer R. 2009. A longitudinal study of Enterocytozoon bieneusi in dairy cattle. Parasitol. Res. 105:141–144. 10.1007/s00436-009-1374-4 [DOI] [PubMed] [Google Scholar]
- 27.Fayer R, Santin M, Trout JM. 2007. Enterocytozoon bieneusi in mature dairy cattle on farms in the eastern United States. Parasitol. Res. 102:15–20. 10.1007/s00436-007-0746-x [DOI] [PubMed] [Google Scholar]
- 28.Santin M, Trout JM, Fayer R. 2005. Enterocytozoon bieneusi genotypes in dairy cattle in the eastern United States. Parasitol. Res. 97:535–538. 10.1007/s00436-005-1482-8 [DOI] [PubMed] [Google Scholar]
- 29.Lee JH. 2007. Prevalence and molecular characteristics of Enterocytozoon bieneusi in cattle in Korea. Parasitol. Res. 101:391–396. 10.1007/s00436-007-0468-0 [DOI] [PubMed] [Google Scholar]
- 30.Ben Ayed L, Yang W, Widmer G, Cama V, Ortega Y, Xiao L. 2012. Survey and genetic characterization of wastewater in Tunisia for Cryptosporidium spp., Giardia duodenalis, Enterocytozoon bieneusi, Cyclospora cayetanensis and Eimeria spp. J. Water Health 10:431–444. 10.2166/wh.2012.204 [DOI] [PubMed] [Google Scholar]
- 31.Feng Y, Alderisio KA, Yang W, Blancero LA, Kuhne WG, Nadareski CA, Reid M, Xiao L. 2007. Cryptosporidium genotypes in wildlife from a New York watershed. Appl. Environ. Microbiol. 73:6475–6483. 10.1128/AEM.01034-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang J, Alderisio KA, Xiao L. 2005. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl. Environ. Microbiol. 71:4446–4454. 10.1128/AEM.71.8.4446-4454.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.US EPA. 2001. Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FAEPA 821-R-01–025. Office of Water, US Environmental Protection Agency, Washington, DC [Google Scholar]
- 34.Jiang J, Alderisio KA, Singh A, Xiao L. 2005. Development of procedures for direct extraction of Cryptosporidium DNA from water concentrates and for relief of PCR inhibitors. Appl. Environ. Microbiol. 71:1135–1141. 10.1128/AEM.71.3.1135-1141.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santin M, Fayer R. 2009. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J. Eukaryot. Microbiol. 56:34–38. 10.1111/j.1550-7408.2008.00380.x [DOI] [PubMed] [Google Scholar]
- 36.Li W, Cama V, Akinbo FO, Ganguly S, Kiulia NM, Zhang X, Xiao L. 2013. Multilocus sequence typing of Enterocytozoon bieneusi: lack of geographic segregation and existence of genetically isolated sub-populations. Infect. Genet. Evol. 14:111–119. 10.1016/j.meegid.2012.11.021 [DOI] [PubMed] [Google Scholar]