Abstract
To understand phage infection and host cell lysis mechanisms in pathogenic Salmonella, a novel Salmonella enterica serovar Typhimurium-targeting bacteriophage, SPN9CC, belonging to the Podoviridae family was isolated and characterized. The phage infects S. Typhimurium via the O antigen of lipopolysaccharide (LPS) and forms clear plaques with cloudy centers due to lysogen formation. Phylogenetic analysis of phage major capsid proteins revealed that this phage is a member of the lysogen-forming P22-like phage group. However, comparative genomic analysis of SPN9CC with P22-like phages indicated that their lysogeny control regions and host cell lysis gene clusters show very low levels of identity, suggesting that lysogen formation and host cell lysis mechanisms may be diverse among phages in this group. Analysis of the expression of SPN9CC host cell lysis genes encoding holin, endolysin, and Rz/Rz1-like proteins individually or in combinations in S. Typhimurium and Escherichia coli hosts revealed that collaboration of these lysis proteins is important for the lysis of both hosts and that holin is a key protein. To further investigate the role of the lysogeny control region in phage SPN9CC, a ΔcI mutant (SPN9CCM) of phage SPN9CC was constructed. The mutant does not produce a cloudy center in the plaques, suggesting that this mutant phage is virulent and no longer temperate. Subsequent comparative one-step growth analysis and challenge assays revealed that SPN9CCM has shorter eclipse/latency periods and a larger burst size, as well as higher host cell lysis activity, than SPN9CC. The present work indicates the possibility of engineering temperate phages as promising biocontrol agents similar to virulent phages.
INTRODUCTION
Food poisoning is generally caused by the intake of a food or drink contaminated with food-borne pathogenic bacteria, such as Salmonella, Escherichia coli, Listeria, and Campylobacter (1). Salmonella causes salmonellosis with various symptoms, such as diarrhea, vomiting, high fever, and even death (2, 3). In the United States, more than 1.4 million cases of salmonellosis have been reported every year, and the number has increased by more than 10% annually in recent years (1, 3, 4). Although antibiotics have been widely used to control the pathogen responsible for salmonellosis, multidrug-resistant Salmonella strains, such as Salmonella enterica serovar Typhimurium DT104, have appeared (5, 6).
Because of the emergence of antibiotic-resistant Salmonella strains, an approach using bacteriophage has been proposed to control them (7, 8). To take advantage of phage treatment against salmonellosis, it is necessary to characterize Salmonella phages phenotypically and genotypically. Moreover, understanding of mechanisms of Salmonella host cell infection by Salmonella-targeting phages is important for this purpose. The major processes of host infection by phages include phage attachment via a host receptor, control of the host lytic-lysogenic cycle, and the host cell lysis mechanism.
Several Salmonella host receptors for phage infection have been experimentally determined and characterized, such as flagella (9, 10), Vi capsular antigen (11), lipopolysaccharide (LPS) (12), and host outer membrane proteins (OmpC [13], BtuB [14, 15], TolC [16], and FhuA [17]). These receptors play a role in the determination of phage host specificity, suggesting that host receptor study would be able to provide novel insights into the mechanisms of phage infection of Salmonella host cells. Lambdoid lysogenic phages generally contain a lysogeny control region consisting of cro, cI, cII, cIII, N, and Q (18, 19). Constitutive bacteriophage promoters, PL and PR, express N and Cro proteins. N protein binds to all terminators for antitermination. During this early gene expression, CII, CIII, and Q proteins are produced. Among these proteins, the CII-CIII complex activates PRE and PI promoters, resulting in the lysogenic cycle by the production of integrase and CI protein, which are related to phage genome integration and blocking of all phage gene expression. At this point, if the host HflA proteolytic enzyme is activated in the presence of a low concentration of cyclic AMP because of a sufficient supply of glucose to the host, it digests CII protein such that the CII-CIII complex cannot produce CI protein, resulting in prevention of the lysogenic cycle. Furthermore, Q protein activates gene expression related to phage structure and host cell lysis. Therefore, the study of the lysogeny control region is important to understand the phage lytic/lysogenic cycles in the host. Holin and endolysin are known to be important for host cell lysis (20). Holin creates holes in the cytoplasmic membrane. These holes are used as transport channels for endolysin, which digests the peptidoglycan layer. In addition, Rz/Rz1-like proteins often enhance endolysin activity as endolysin accessory proteins (21).
Salmonella-targeting phage P22 belongs to the family of Podoviridae morphologically and has been well characterized to develop genetic transfer tools via lysogenization (18, 19). Host receptor studies have revealed that the phage tailspike protein plays a role in the interaction with the host by interacting with the O antigen of LPS in S. Typhimurium (22, 23). Complete analysis of the phage P22 genome sequence also revealed the presence of functional genes related to lysogenization and host specificity determination (18, 24). In addition, comparative genomic analysis of P22 and closely related phages revealed the presence of the P22-like phage group (25). This group includes ε34, ST104, ST64T, SE1, c341, and HK620. They share phage morphogenesis and assembly genes for similar morphology and generally infect Salmonella, E. coli, and Shigella in the Enterobacteriaceae family. However, while ant moron regions in phage P22 have been known to be involved in the regulation of gene expression, these regions are completely or partially missing from other P22-like phages (26). Although the role of this region is not clearly understood, it may be related to lysogeny conversion (27). Further studies of the genomes of these P22-like phages indicate that morphogenesis-related genes are highly conserved, but other genes are variable, suggesting that even though they have similar phage morphologies, the host specificity of these P22-like phages may differ among them. Therefore, further study of these P22-like phages would provide new information about host infection by phages in this group.
To understand the infection mechanisms of the bacteriophage at the genomic level, the complete genome of SPN9CC was analyzed and compared with P22-like phage genomes. In addition, a ΔcI mutant of the lysogen-forming P22-like phage SPN9CC was constructed and characterized. This study will be useful for increasing our knowledge of the host infection and lysis mechanisms of P22-like phages, including SPN9CC.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study and gene knockout mutant strains for the host receptor study are listed in Table 1. Prophage-free Salmonella enterica serovar Typhimurium LT2C was used for the isolation and propagation of S. Typhimurium-targeting phages (28) (Cancer Research Center, Columbia, MO). All of the bacteria listed in Table 1 were cultivated at 37°C for 12 h in Luria-Bertani (LB) broth medium (Difco, Detroit, MI), and the agar medium was prepared by 1.5% agar supplementation (Difco) of the broth medium.
TABLE 1.
Host range of SPN9CC bacteriophage
| Bacterial host | SPN9CCa | Sourceb or reference |
|---|---|---|
| Salmonella enterica serovar Typhimurium | ||
| LT2 | +++ | 73 |
| LT2C | +++ | 28 |
| SL1344 | +++ | NCTC |
| UK1 | +++ | 74 |
| ATCC 14028s | +++ | ATCC |
| DT104 | + | 6 |
| ATCC 43174 | ++ | ATCC |
| Salmonella enterica serovar Enteritidis ATCC 13076 | + | ATCC |
| Salmonella enterica serovar Paratyphi | ||
| A IB 211 | ++ | IVI |
| B IB 231 | − | IVI |
| C IB 216 | − | IVI |
| Salmonella enterica Dublin IB 2973 | + | IVI |
| Escherichia coli | ||
| K-12 MG1655 | − | 75 |
| DH5α | − | ATCC |
| O157:H7 ATCC 35150 | − | ATCC |
| O157:H7 ATCC 43890 | − | ATCC |
| Gram-negative bacteria | ||
| Shigella flexneri 2a strain 2457T | − | IVI |
| Shigella boydii IB 2474 | − | IVI |
| Vibrio fischeri ATCC 700601 | − | ATCC |
| Pseudomonas aeruginosa ATCC 27853 | − | ATCC |
| Cronobacter sakazakii ATCC 29544 | − | ATCC |
| Gram-positive bacteria | ||
| Enterococcus faecalis ATCC 29212 | − | ATCC |
| Staphylococcus aureus ATCC 29213 | − | ATCC |
| Bacillus cereus ATCC 14579 | − | ATCC |
| Listeria monocytogenes ATCC 19114 | − | ATCC |
| Salmonella Typhimurium SL1344 mutants | ||
| ΔflgK mutant | +++ | 29 |
| ΔbtuB mutant | +++ | 15 |
| ΔrfaL mutant | − | 31 |
| ΔrfaL (pUHE21-lacIq::rfaL) mutant | +++ | 31 |
+++, EOP of 1 to 0.01; ++, EOP of 0.01 to 0.0001; +, EOP of, <0.0001; −, not susceptible to SPN9CC.
NCTC, National Collection of Type Cultures; ATCC, American Type Culture Collection; KCTC, Korean Collection for Type Cultures; IVI, International Vaccine Institute.
Bacteriophage isolation and propagation.
Commercially processed broiler chicken skin samples were collected from the Moran traditional market, Seongnam, South Korea, and used for isolation of S. Typhimurium-targeting bacteriophage SPN9CC with S. Typhimurium strain LT2C. The basic procedures for the isolation and propagation of bacteriophage SPN9CC were previously described by Shin et al. (29).
Lysogen induction.
Selected SPN9CC lysogens of S. Typhimurium LT2C were cultivated at 37°C until the optical density at 600 nm (OD600) reached 1.0, and 0.5 μg/ml of mitomycin C (Sigma, St. Louis, MO) was added to the cultures. Then, these cultures were additionally incubated at 37°C for 2 h. After incubation, the cells were removed by centrifugation and filtration, and the supernatant was collected. The spotting assay of this supernatant with S. Typhimurium LT2C was conducted to confirm the presence of induced phage SPN9CC.
Electron microscopy.
A transmission electron microscope (TEM) was used for morphological analysis of purified phage SPN9CC. This TEM analysis was performed as described by Shin et al. (29). The morphological classification of phage SPN9CC was conducted according to the guidelines of the International Committee on Taxonomy of Viruses (30).
Host range determination by spotting assay.
The host range and comparative efficiency of plating (EOP) of phage SPN9CC were determined with a spotting assay using S. Typhimurium, S. Paratyphi, E. coli, and other Gram-negative and Gram-positive bacterial strains by the procedure previously described by Park et al. (31).
Genome sequencing and bioinformatic analysis.
Genomic DNA of phage SPN9CC was isolated and purified as described by Sambrook and Russell (32). The construction of a genomic DNA library and pyrosequencing with Genome Sequencer FLX (GS-FLX) Titanium (Roche, Mannheim, Germany) were conducted by Macrogen, Seoul, South Korea. The prediction of open reading frames (ORFs) was conducted with Glimmer 3.02 (33), GeneMarkS (34), and FgenesV (Softberry, Inc., Mount Kisco, NY). The prediction of ribosomal binding sites of ORFs was performed with RBSfinder (J. Craig Venter Institute, Rockville, MD). The annotation of predicted ORFs was conducted with BLASTP (35) and InterProScan (36) by using conserved protein domain databases. The GenBank data file was generated with the GAMOLA (37) and Sequin programs (National Center for Biotechnology Information, Bethesda, MD). The phylogenetic analysis of major capsid proteins (MCPs) from bacteriophages, including SPN9CC, was performed with MEGA5 by the neighbor-joining method by using p distance values (38). The program Mobyle was used for comparative codon usage analysis of the S. Typhimurium SL1344 host and phage SPN9CC (39). Comparative genomic analysis of SPN9CC with other P22-like phages and visualization were conducted with BLASTN (35) and ACT12 (40).
Expression of the host cell lysis gene cluster.
The SPN9CC_0042, SPN9CC_0043, SPN9CC_0044, and SPN9CC_0044_1 genes, encoding holin, endolysin, and Rz/Rz1 endopeptidases, respectively, in the host cell lysis gene cluster were amplified by PCR with the primers listed in Table 2. These PCR products were doubly digested with EcoRI and SalI and cloned into the multiple cloning site of pBAD18 (41) individually or in combination with more than two genes. These cloned plasmids are listed in Table 3. S. Typhimurium SL1344 and E. coli MG1655 were used as gene expression hosts of the cloned pBAD18 plasmids after transformation. The expression of the cloned genes was induced by the addition of 0.2% (final concentration) arabinose after 2 h of incubation of the subinoculated cultures. To test the lysis activity of host cell lysis proteins during incubation and the induction of the cultures, the OD600 was measured every hour.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′)a | Reference |
|---|---|---|
| SPN9CC_0042_F | TAAAAGAATTCAAATCCCCTCAATAAAGGGGGTAGAG | This study |
| SPN9CC_0042_R | TTTTTGTCGACTTATCGCCGCTATTACGCTATTTC | This study |
| SPN9CC_0043_F | AAAAAGAATTCAAACGCAAAGAGCGTGAGGACAG | This study |
| SPN9CC_0043_R | TTTTTGTCGACATAATCGCGGTTACTCTGCTCATTG | This study |
| SPN9CC_0044_F | AAATTGAATTCTTGAGCGTGAAGTCTGTTTGTGGG | This study |
| SPN9CC_0044_R | AAAAAGTCGACTATGTGATGGAAATTATTTCAGGCATTG | This study |
| 9CC-BRED_C | TCTTAAAAGTGAACTCATCACCACATAACCTTGCAATGCAAAAAGCTTCGCTATGTCATACCAGTTCATTTTCATCCTTAAATTATACA | This study |
| 9CC-BRED_CF | TTGTAGGAATACTTGTCCGCTGTCTTTGATGAGCTTCTTAAAAGTGAACTCATCATGTAGGCTGGAGCTGCTTCG | This study |
| 9CC-BRED_CR | TTTACGATTTGTGACTGTTCTTGTTTGATACAAATTGTATAATTTAAGGATGAAAATTCCGGGGATCCGTCGACC | This study |
| 9CC-BRED_conf_F | TATCTCATCAGGCCATTGGCTGGCTACAAC | This study |
| 9CC-BRED_conf_R | TAATGACAAACTGCACCACGCGTACAACCG | This study |
Specific restriction enzymes used for cloning are underlined. Forward and reverse primers contain EcoRI and SalI sites, respectively.
TABLE 3.
Plasmids used in this study
| Plasmid | Description | Reference |
|---|---|---|
| pBAD18 | ParaC ColE1 ori Ampr | 41 |
| pBAD18-42 | pBAD18 expressing SPN9CC_0042 | This study |
| pBAD18-43 | pBAD18 expressing SPN9CC_0043 | This study |
| pBAD18-44 | pBAD18 expressing SPN9CC_0044 | This study |
| pBAD18-42/43 | pBAD18 expressing SPN9CC_0042 and SPN9CC_0043 | This study |
| pBAD18-43/44 | pBAD18 expressing SPN9CC_0043 and SPN9CC_0044 | This study |
| pBAD18-42/43/44 | pBAD18 expressing SPN9CC_0042, SPN9CC_0043, and SPN9CC_0044 | This study |
| pUHE21-2 lacIq | pMB1 ori Ampr lacIq | 76 |
| pUHE21-2 lacIq::rfaL | pUHE21-2 lacIq expressing rfaL | 31 |
| pUHE21-2 lacIq::flgK | pUHE21-2 lacIq expressing flgK | 29 |
| pACYC184 | p15A ori Cmr Tetr | 77 |
| pMS100 | pACYC184 expressing btuB | 15 |
Deletion of the cI gene from the SPN9CC genome by BRED.
The cI gene of SPN9CC was specifically deleted by the bacteriophage recombineering of electroporated DNA (BRED) method previously described by Marinelli et al. (42). To delete the cI gene by the BRED method, a 200-bp double-stranded DNA substrate containing a 100-bp region upstream and the other 100-bp region downstream of the cI target gene was PCR amplified with primers 9CC-BRED_C, 9CC-BRED_CF, and 9CC-BRED_CR (Table 2). An S. Typhimurium SL1344 electroporation host with pKD46 encoding recombinase was induced with arabinose and used for electrocompetent cell preparation (43). The phage genomic DNA and 200-bp DNA substrate were coelectroporated into the arabinose-induced electrocompetent cells for homologous recombination. After a 1-h shaking incubation of the transformants at 37°C, 6 ml of 0.4% molten LB top agar containing 200 μl of the transformant culture was overlaid on the 1.5% LB base agar and incubated overnight. Plaques were randomly picked, and plaque PCR was performed with specific primers 9CC-BRED_conf_F and 9CC-BRED_conf_R (Table 2). The plaque PCR products were partially sequenced with the same primers to confirm the deletion of the cI gene. Phage SPN9CCM with the cI gene deleted was purified by the single-picking method and propagated as described above.
One-step growth curve and bacterial challenge test.
S. Typhimurium SL1344 was used as the host strain for one-step growth curve determination and a bacterial challenge test. The overall procedures used for the one-step growth curve assay and the challenge test were previously described by Park et al. (31).
Nucleotide sequence accession number.
The GenBank accession number of the complete genome sequence and annotation information of bacteriophage SPN9CC is JF900176.1.
RESULTS
Isolation and morphology of phage SPN9CC.
For the development of new biocontrol agents, Salmonella-targeting bacteriophages were isolated from a commercially processed broiler chicken skin sample with the host strain S. Typhimurium LT2C. Of these phages, SPN9CC produced distinct clear plaques with cloudy centers (see Fig. S1A in the supplemental material), suggesting the possibility of lysogen formation in the cloudy center. Mitomycin C treatment of the colonies isolated from the cloudy centers of the clear plaques revealed the induction of phage SPN9CC, confirming lysogen formation (data not shown). TEM morphological observation revealed that this phage has the short tail typical of members of the Podoviridae family (see Fig. S1B).
Host range and host receptor study.
The host range test of phage SPN9CC demonstrated specific inhibition of S. Typhimurium, S. Paratyphi, and S. Dublin. However, various Gram-positive and Gram-negative bacteria, including other Salmonella strains, were not inhibited by this phage, suggesting that it specifically infects certain Salmonella strains (Table 1). To determine the host receptor for phage SPN9CC, previously constructed mutants of S. Typhimurium SL1344 were used, including a ΔflgK mutant (flgK encodes a flagellar hook-associated protein), a ΔbtuB mutant (btuB encodes a vitamin B12 uptake protein), and a ΔrfaL mutant (rfaL encodes O-antigen ligase) (15, 29, 31). Only the ΔrfaL mutant displayed resistance to phage SPN9CC, suggesting that the O antigen of LPS is a host receptor for phage infection. Subsequent complementation of this mutant with the pUHE21-lacIq::rfaL expression vector (31) confirmed O antigen as a receptor of SPN9CC (Table 1).
Bacteriophage genome analysis.
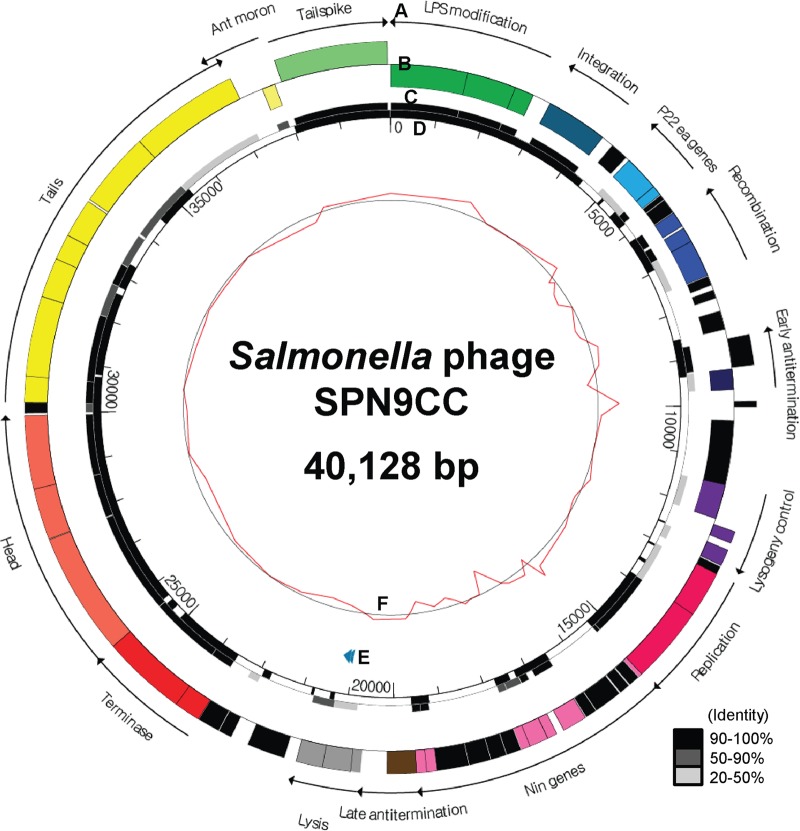
Sequencing of the complete genome of SPN9CC was performed with approximately 90 times coverage by next-generation sequencing (NGS) technology with a 454 pyrosequencer, revealing 40,128 bp with a GC content of 47.33%, 63 putative ORFs, and two tRNAs (tRNA_Thr and tRNA_Asn) (Fig. 1). Comparative analysis of the codon usage preferences of tRNA_Thr of the S. Typhimurium SL1344 host and phage SPN9CC indicated a different preference in threonine, suggesting that this tRNA may play a role in the translation of phage mRNA and not of host mRNA. In addition, the gene density was observed to be 1.545 genes/kb and the coding percentage was 90.9%. The average length of each ORF was determined to be 588 bp. A comparative phylogenetic analysis using MCPs from various phages revealed that SPN9CC is closely related to Salmonella-targeting P22-like phages such as P22, ST64T, ST104, and ε34 (Fig. 2).
FIG 1.
Genome map of phage SPN9CC. (A) Functions of gene clusters. (B) Predicted ORFs by strand. The colors indicate the functions of the gene clusters. Black ORFs encode hypothetical proteins. (C) Comparative analysis of phage SPN9CC and P22 ORFs at the amino acid sequence level. Different degrees of homology between phage SPN9CC and P22 ORFs are indicated by different levels of darkness, as shown in the lower right corner. (D) Comparative genomic analysis of phages SPN9CC and P22 at the DNA sequence level. (E) tRNA prediction is indicated by the blue arrowheads. (F) GC content of phage SPN9CC. The scale values are in base pairs.
FIG 2.
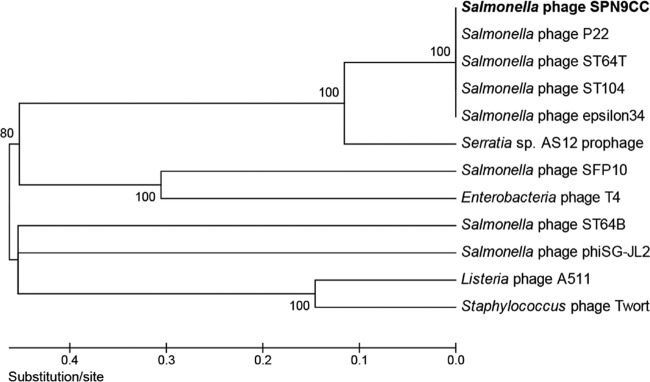
Comparative phylogenetic analysis of MCPs from various bacteriophages. The MCPs were compared with the ClustalW multiple-alignment algorithm, and the phylogenetic tree was generated with MEGA5 by the neighbor-joining method by using p distance values.
Annotation and functional analysis of the 63 ORFs in this genome revealed that 44 of them have putative functions. Functional categorization of these genes revealed 14 groups, such as LPS modification and superinfection exclusion (O-antigen conversion proteins GtrABC and superinfection exclusion protein B), integration (phage integrase), P22 ea genes (Eaa and Eai), recombination (Erf recombination protein, Abc1, and Abc2 anti-RecBCD proteins), antitermination (antitermination proteins N and Q), lysogeny control (Cro, CI, and CII), replication (DNA replication protein and helicase), nin genes (NinABEFHXZ), host cell lysis (holin, endolysin, and Rz/Rz1 endopeptidases), DNA packaging (terminase large and small subunits), head (portal protein, scaffolding protein, and MCP), tail (DNA stabilization proteins/tail accessory proteins [Gp4, Gp10, and Gp26], head assembly protein, and DNA transfer proteins/ejection proteins), Ant moron (Mnt regulatory protein), and host specificity (tailspike protein).
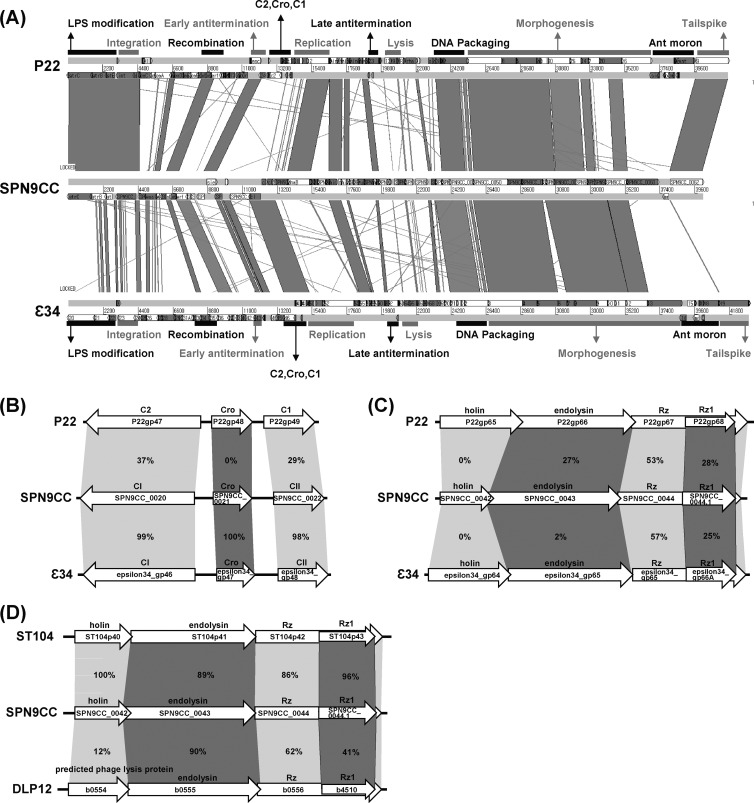
Comparative genomic analysis of SPN9CC with P22-like phages.
Comparative genomic analysis of phage SPN9CC with P22-like phages such as P22 and ε34 revealed that DNA packaging and morphogenesis (heads and tails) gene clusters are highly conserved, indicating that P22-like phages commonly share phage structure genes and belong to the Podoviridae family (Fig. 3A). A recent comparative genomic study of P22-like phages supports our analysis result (25). However, the tailspike protein of ε34 differs enough from those of phages P22 and SPN9CC that it most likely has a different host specificity (Fig. 3A). While host range analyses of phages P22 and SPN9CC displayed the same inhibition spectrum (data not shown), the specific infection of S. Anatum by phage ε34 substantiates this (44). The lysogeny control region (Cro, CI, and CII) of phage SPN9CC differs from that of phage P22 but is similar to that of phage ε34, suggesting that SPN9CC and ε34 may share lytic/lysogenic decision and lysogen formation mechanisms (Fig. 3B). Comparative analysis of the host cell lysis gene clusters of phages SPN9CC, P22, and ε34s revealed that they are not conserved among them, suggesting that they most likely lyse their host strains in different manners (Fig. 3C). Although the functions of the genes in this gene cluster of phage P22 were experimentally confirmed (45–47), the function of each gene in the host cell lysis gene cluster of phage SPN9CC cannot be deduced from those in the gene cluster of phage P22 because of the low levels of identity between the amino acid sequences encoded by these genes of P22 and phage SPN9CC. To understand the host cell lysis mechanism of phage SPN9CC, the function of each gene in the host cell lysis gene cluster of phage SPN9CC should be confirmed experimentally. Interestingly, the genes in this gene cluster of phage SPN9CC are similar to those of ST104 and even E. coli K-12 prophage DLP12, suggesting that they may use the same mechanism for host cell lysis (Fig. 3D). Successful S. Typhi cell lysis results obtained with endolysin from E. coli phage DLP12 support this (48). However, whereas the amino acid sequence identity levels of host cell lysis proteins, such as holin, endolysin, and Rz/Rz1-like proteins, between two host cell lysis gene clusters in phages SPN9CC and ST104 are extremely high, the functions of the genes in the gene cluster of ST104 have not been experimentally confirmed. Therefore, the expression of these genes in S. Typhimurium and E. coli host strains needs to be examined to elucidate the functions of all of the genes in the host cell lysis gene cluster of phage SPN9CC and their cooperation effect on host cell lysis.
FIG 3.
Comparative genomic analysis of P22-like phages (SPN9CC, P22, ST104, and ε34) and E. coli K-12 prophage DLP12. (A) Comparative analysis of the complete genome sequences of SPN9CC, P22, and ε34 with BLASTN and ACT12. Black and gray bars indicate the functions of gene clusters in the genomes. (B and C) Comparative analyses of lysogeny control regions (B) and host cell lysis gene clusters (C) in SPN9CC, P22, and ε34. (D) Comparative analysis of host cell lysis gene clusters in SPN9CC, ST104, and E. coli K-12 prophage DLP12. The percentages of amino acid identity between homologous genes are indicated.
Function of host cell lysis gene cluster.
Interestingly, the high level of amino acid sequence identity of the host cell lysis proteins (except for holin) encoded by the host cell lysis gene clusters of S. Typhimurium-targeting phage SPN9CC and E. coli K-12 prophage DLP12 suggests that host cell lysis proteins encoded by the genes in this cluster of phage SPN9CC should function in both Salmonella and E. coli. To elucidate the host cell lysis mechanism of this phage, each gene in this cluster was cloned into pBAD18 and transformed into S. Typhimurium and E. coli host cells, respectively.
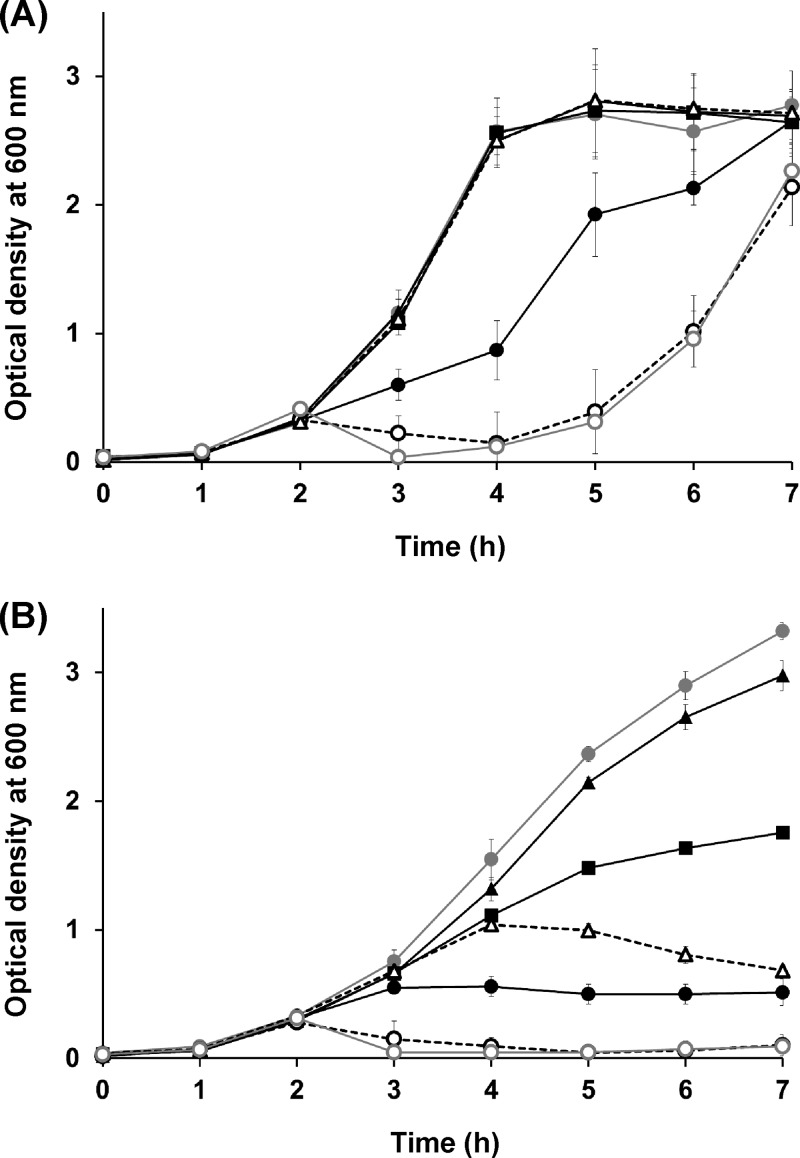
The expression of a single gene encoding holin (SPN9CC_0042) in S. Typhimurium resulted in host cell lysis (Fig. 4A). However, the expression of a single gene encoding endolysin (SPN9CC_0043) or Rz/Rz1-like proteins (SPN9CC_0044) in S. Typhimurium did not, suggesting that the endolysin needs holin to cross the cytoplasmic membrane. To elucidate their cooperation effects on S. Typhimurium host cell lysis, various combinations for the expression of more than two genes were prepared and those genes were coexpressed in S. Typhimurium. The expression of combinations of the genes for holin and endolysin or all four cell lysis proteins (holin plus endolysin or holin plus endolysin plus Rz/Rz1-like proteins) in S. Typhimurium resulted in much higher host cell lysis efficiency than expression of the holin gene alone (Fig. 4A). However, gene expression combinations without holin (endolysin plus Rz/Rz1-like proteins) did not lyse the host cells, suggesting that holin is a key protein for the lysis of S. Typhimurium (Fig. 4A).
FIG 4.
Confirmation of the host cell lysis system of phage SPN9CC via the expression of host cell lysis genes encoding holin, endolysin, and Rz/Rz1 endopeptidases in S. Typhimurium SL1344 (A) and E. coli MG1655 (B). Closed gray circles represent the negative control without gene expression. Closed black circles, triangles, and squares indicate the expression levels of the SPN9CC_0042 (holin), SPN9CC_0043 (endolysin), and SPN9CC_0044/0044.1 (Rz/Rz1) genes, respectively. Open black circles and triangles with dotted lines indicate the coexpression of SPN9CC_0042/0043 and SPN9CC_0043/0044/0044.1, respectively. Open gray circles indicate the coexpression of all of the genes, SPN9CC_0042/0043/0044/0044.1.
However, the expression of these genes in E. coli host cells displayed different host cell lysis patterns (Fig. 4B). As for the S. Typhimurium host cells, endolysin alone did not contribute to the lysis of E. coli host cells but the coexpression of endolysin and other proteins (endolysin plus holin or endolysin plus Rz/Rz1-like proteins) in E. coli host cells did result in host cell lysis, suggesting that endolysin may need support to cross the E. coli cytoplasmic membrane and that either holin or Rz/Rz1-like proteins could help endolysin to cross the membrane (Fig. 4B). It is intriguing that the main difference between the patterns of E. coli and Salmonella host cell growth inhibition by the SPN9CC lysis gene cluster is the role of Rz/Rz1-like proteins, which inhibit only E. coli host cell growth bacteriostatically (Fig. 4B).
Conversion of phenotypes in phage SPN9CC by deletion of the cI gene.
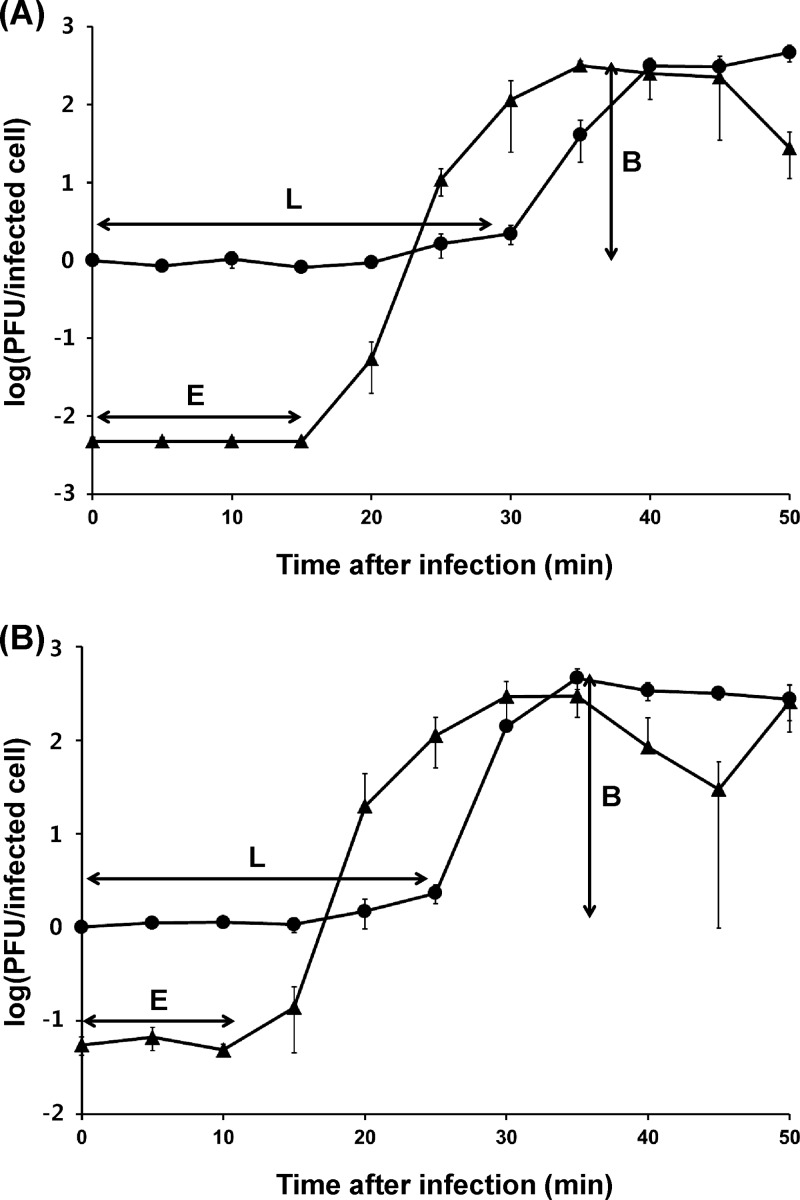
CI, CII, and Cro are key proteins in the lysogeny control region (49–51). Among them, CI is a repressor causing termination of gene expression in the phage genome. Therefore, mutation of the cI gene can inhibit lysogen formation. The effects of cI gene deletion on the life cycle of phage SPN9CC was studied by constructing the ΔcI mutant phage by the BRED method (42). Interestingly, whereas phage SPN9CC generates distinct clear plaques with cloudy centers as lysogens, the ΔcI mutant phage SPN9CCM does not produce cloudy centers in the plaques, suggesting that the phenotype of ΔcI mutant phage may be converted from temperate to virulent (see Fig. S1C in the supplemental material). To further understand the plaque morphology change caused by cI deletion, one-step growth analyses and bacterial challenge assays of phages SPN9CC and SPN9CCM were compared. The one-step growth analyses revealed that while phage SPN9CC has relatively long eclipse and latency periods and a small burst size, phage SPN9CCM has much shorter eclipse and latency periods and a larger burst size (Fig. 5). The eclipse and latency periods of SPN9CC and SPN9CCM were 15 and 30 min and 10 and 20 min, respectively. The average burst sizes of phages SPN9CC and SPN9CCM were 220 and 280 PFU per cell, respectively, suggesting that the efficiency of phage multiplication was increased for SPN9CCM most likely because of an inability to form lysogens. Furthermore, bacterial challenge assays of phages SPN9CC and SPN9CCM with S. Typhimurium SL1344 demonstrated that the inhibition activity of phage SPN9CCM is much higher than that of phage SPN9CC (Fig. 6).
FIG 5.
One-step growth curve analysis of phages SPN9CC (A) and SPN9CCM (B). Circles represent non-chloroform-treated samples, and triangles represent chloroform-treated samples. The error bars indicate the standard deviations of triplicate experiments. E, eclipse period; L, latency period; B, burst size.
FIG 6.
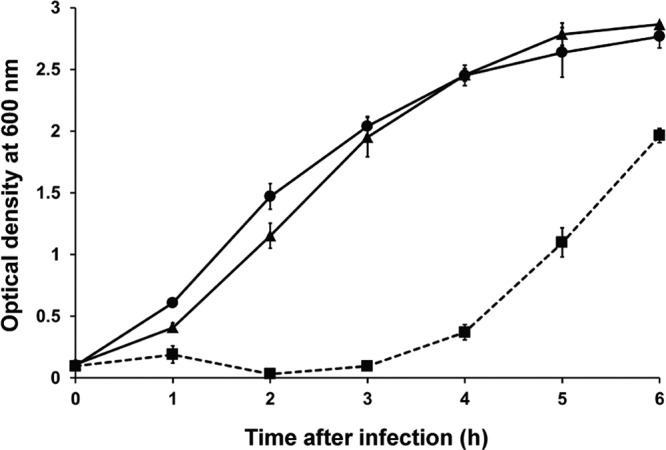
Assay of S. Typhimurium LT2C challenge with phages SPN9CC and SPN9CCM. Circles represent non-phage-treated samples, triangles represent SPN9CC-treated samples, and squares with a broken line represent SPN9CCM-treated samples. The SPN9CC- and SPN9CCM-treated samples were tested at a multiplicity of infection of 10.
DISCUSSION
Salmonellosis is one of the most common types of food poisoning caused by food-borne pathogens all over the world. To reduce this food poisoning, the bacteriophage approach has recently been appearing more attractive than antibiotic treatment because of the emergence of multidrug-resistant Salmonella strains (5, 6). To maximize the efficiency of this phage approach, further understanding of phage infection and host cell lysis mechanisms is required (15, 52). Phage P22 has been studied in the context of the development of a molecular transduction tool (18, 19), the identification of host cell specificity and a receptor (22, 23), the tail structure for host cell interaction (53, 54), the lysogeny control region (55, 56), superinfection exclusion (57, 58), and other areas. The P22-like phage group was previously proposed on the basis of the homology of virion assembly genes, which include those for ST104, ε34, ST64T, L, Sf6, c341, and HK620, among others (25). Recent improvement of genome sequencing and analysis technologies, such as NGS and bioinformatic tools, enabled us to analyze the full genome sequences of these P22-like phages and to study their characteristics at the genomic level. Recent comparative genomic analysis revealed that while their genomic characteristics are diverse, most likely because of horizontal gene transfer/exchange in the group, morphogenesis and DNA packaging are highly conserved (25, 26). However, the diversity of other genomic features may determine the specific characteristics of each phage in the group, such as host cell specificity, the lysogeny control region, and the host cell lysis system, involved in the mechanisms of host cell infection and lysis.
Salmonella-targeting temperate phage SPN9CC was isolated from a commercially processed broiler chicken skin sample, and its complete genome analysis suggests that phage SPN9CC is in the P22-like phage group. One-step growth analysis of phage SPN9CC revealed a longer latency period and a smaller burst size than those of other lytic phages, such as T7-like Podoviridae family phages (Fig. 5A) (59–61), suggesting that lysogen formation during phage infection may affect the host cell lysis activity of phage SPN9CC. A high frequency of observed mutants insensitive to this phage during a bacterial challenge test also supports this (see Fig. S2 in the supplemental material). Generally, superinfection of a lysogen by other phages is prevented by repression of the expression of superinfecting phage genes by CI repressor proteins by lysogen or host cell receptor modification (62). It is well known that the host cell receptor is modified once the host cell is lysogenized by phage (18, 26, 63). SPN9CC has LPS modification proteins homologous to GtrABC (SPN9CC_003, SPN9CC_002, and SPN9CC_001, respectively), which modify LPS to prevent superinfection of the SPN9CC lysogen. Furthermore, the resistance activity of the host cell lysogen caused by LPS modification during lysogenization may contribute to the formation of cloudy centers in SPN9CC plaques (see Fig. S1A). In the center of the plaques, a high phage concentration may promote lysogenization of phage SPN9CC, similar to phage P22 (64, 65) or the first lysogens formed may expand from the middle outward (see Fig. S1A).
The role of LPS modification proteins GtrABC and superinfection exclusion protein B is the prevention of other phage infections after lysogen formation via modification of the host cell O antigen of LPS (18, 19). Among the recombination proteins, the Abc1 and Abc2 anti-RecBCD proteins are involved in phage recombination and protect both ends of the linear phage genome from host cell RecBCD exonuclease and Erf recombination protein circularizes this linear genome by the ligation of both ends of the phage genome (66). Lysogeny control and antitermination determine the phage lytic/lysogenic cycles, depending on the host cell status. Replication proteins are produced during early gene expression, and they are responsible for phage genome replication. However, the functions of the ea and nin genes are unknown (67). Host cell lysis proteins such as holin, endolysin, and Rz/Rz1-like proteins have been suggested to cooperate in bursting the host cell after replication and reconstruction of the phage (21). Holin creates pinholes in the host cell inner membrane, and the subsequent secretion of endolysin via these pinholes results in host cell lysis. Although ant moron regions have been found in P22-like phages, these regions are highly variable among them (26) and the function of the ant moron is not clearly understood. Phage SPN9CC also has only one gene in this region, a mnt gene encoding a repressor protein, which is very similar to phages ST104 and ST64T. This Mnt repressor has been suggested to control the expression of ant gene encoding an antirepressor (68). As with other P22-like phages in the Podoviridae family, phage SPN9CC has only a tailspike protein without a tail fiber protein. This tailspike protein is homologous to other tailspike proteins observed in certain P22-like phages that target S. Typhimurium.
The complete genome sequence of phage SPN9CC and comparative genomic analyses with other P22-like phages revealed a diversity of phage infection and host cell lysis mechanisms in the group (Fig. 1 and 3). P22-like phages are in the family Podoviridae and have short tails, indicating that the tailspike protein is a major determinant of host specificity in P22-like phages (23, 69, 70). However, the tailspike protein is variable in the group, suggesting that the host specificity and host range of P22-like phages could be variable. Whereas phages P22, ST104, and ST64T with homologous tailspike proteins infect S. Typhimurium, phages ε34 and Sf6 with different types of tailspike proteins infect S. Anatum and even Shigella, respectively, supporting the notion of variable host range and specificity (25, 44, 71). Comparative analysis of the lysogeny control regions of P22-like phages indicated that the region of SPN9CC is nearly identical to that of phage ε34 but quite different from that of phage P22, suggesting that P22-like phages may have diverse lytic/lysogenic decision mechanisms (Fig. 3B).
Characterization of the host cell lysis gene cluster of SPN9CC is important to understanding the host cell lysis mechanism of SPN9CC. The host cell lysis gene cluster encodes putative holin, endolysin, and Rz/Rz1-like proteins. This gene cluster of phage SPN9CC is quite different from those of P22 and phage ε34s but very similar to those of ST104 and even the E. coli K-12 DLP12 prophage, suggesting the possibility of E. coli cell lysis via the activity of lysis proteins that are encoded in the gene cluster of phage SPN9CC. The expression of these genes individually or in combinations in S. Typhimurium or E. coli host cells revealed that holin is a key protein for the lysis of the cells of both hosts, but endolysin could not achieve lysis by itself (Fig. 4A and B). These results indicate that endolysin of SPN9CC requires holin to cross the cytoplasmic membrane to act on the peptidoglycan in the periplasm. Rz/Rz1-like proteins are known accessory proteins of endolysin (21), and Rz/Rz1-like proteins alone or in combination with endolysin in S. Typhimurium did not exhibit cell lysis activity. Interestingly, Rz/Rz1-like proteins alone and even in combination with endolysin resulted in growth inhibition of E. coli even though the degree of inhibition was relatively low. However, the lysis activity of Rz/Rz1-like proteins in E. coli host cells is not fully understood. Comparative functional analysis of the S. Typhimurium and E. coli host cell lysis gene clusters revealed that this lysis protein combination works better in E. coli than in S. Typhimurium (Fig. 4A and B).
Comparison of host cell lysis activities by the bacterial challenge assay revealed that phage SPN9CCM had higher host cell lysis activity than phage SPN9CC. However, an SPN9CCM-resistant strain emerged 4 h after infection (Fig. 6). The phage adsorption assay conducted with the SPN9CCM-resistant strain indicated that more than 98% of phage SPN9CCM adsorbed to the wild-type S. Typhimurium host in 10 min but less than 5% of phage SPN9CCM adsorbed to the SPN9CCM-resistant strain under the same conditions (data not shown), suggesting that the host receptor for phage infection may be modified. Various LPS modification mechanisms (72) are known, and the exact growth recovery mechanism of SPN9CCM-infected host strains needs to be elucidated in the future.
In this study, comparative analysis of phage SPN9CC and P22-like phages provided novel insights into phage infection and S. Typhimurium host strain lysis mechanisms. We therefore believe that this study contributes to a better understanding of the new approach to bacteriophage treatment to inhibit food-borne pathogens, as well as to the development of newly optimized phages for therapy.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Ministry of Education, Science and Technology (20090078983) and the R&D Convergence Center Support Program, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea.
Staphylococcus aureus was kindly provided by Water Borne Virus Bank in the Republic of Korea.
Footnotes
Published ahead of print 1 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02279-13.
REFERENCES
- 1.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. 1999. Food-related illness and death in the United States. Centers for Disease Control and Prevention, Atlanta, GA: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbara G, Stanghellini V, Berti-Ceroni C, De Giorgio R, Salvioli B, Corradi F, Cremon C, Corinaldesi R. 2000. Role of antibiotic therapy on long-term germ excretion in faeces and digestive symptoms after Salmonella infection. Aliment. Pharmacol. Ther. 14:1127–1131. 10.1046/j.1365-2036.2000.00818.x [DOI] [PubMed] [Google Scholar]
- 3.CDC 2007. Bacterial foodborne and diarrheal disease national case surveillance, annual report, 2005. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/nationalsurveillance/pdfs/fbsurvsumm2005.pdf [Google Scholar]
- 4.Gilliss D, Cronquist A, Cartter M, Tobin-D'Angelo M, Blythe D, Smith K, Lathrop S, Birkhead G, Cieslak P, Dunn J, Holt KG, Guzewich JJ, Henao OL, Mahon B, Griffin P, Tauxe RV, Crim SM. 2011. Vital signs: incidence and trends of infection with pathogens transmitted commonly through food—foodborne diseases active surveillance network, 10 U.S. sites, 1996-2010. Morb. Mortal. Wkly. Rep. 60:749–755 http://www.cdc.gov/mmwr/preview/mmwrhtml/mm6022a5.htm [PubMed] [Google Scholar]
- 5.CDC 2008. Salmonella surveillance, annual summary, 2006. Centers for Disease Control and Prevention, Atlanta, GA: http://198.246.124.22/ncidod/dbmd/phlisdata/salmtab/2006/SalmonellaIntroduction2006.pdf [Google Scholar]
- 6.Poppe C, Smart N, Khakhria R, Johnson W, Spika J, Prescott J. 1998. Salmonella typhimurium DT104: a virulent and drug-resistant pathogen. Can. Vet. J. 39:559–565 [PMC free article] [PubMed] [Google Scholar]
- 7.García P, Martínez B, Obeso JM, Rodríguez A. 2008. Bacteriophages and their application in food safety. Lett. Appl. Microbiol. 47:479–485. 10.1111/j.1472-765X.2008.02458.x [DOI] [PubMed] [Google Scholar]
- 8.O'Flaherty S, Ross RP, Coffey A. 2009. Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol. Rev. 33:801–819. 10.1111/j.1574-6976.2009.00176.x [DOI] [PubMed] [Google Scholar]
- 9.Kagawa H, Ono N, Enomoto M, Komeda Y. 1984. Bacteriophage chi sensitivity and motility of Escherichia coli K-12 and Salmonella typhimurium Fla− mutants possessing the hook structure. J. Bacteriol. 157:649–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samuel ADT, Pitta TP, Ryu WS, Danese PN, Leung ECW, Berg HC. 1999. Flagellar determinants of bacterial sensitivity to x-phage. Proc. Natl. Acad. Sci. U. S. A. 96:9863–9866. 10.1073/pnas.96.17.9863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickard D, Toribio AL, Petty NK, de Tonder A, Yu L, Goulding D, Barrell B, Rance R, Harris D, Wetter M, Wain J, Choudhary J, Thomson N, Dougan G. 2010. A conserved acetyl esterase domain targets diverse bacteriophage to the Vi capsular receptor of Salmonella enterica serovar Typhi. J. Bacteriol. 192:5746–5754. 10.1128/JB.00659-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salgado CJ, Zayas M, Villafane R. 2004. Homology between two different Salmonella phages: Salmonella enterica serovar Typhimurium phage P22 and Salmonella enterica serovar Anatum var. 15 + phage ε34. Virus Genes 29:87–98. 10.1023/B:VIRU.0000032792.86188.fb [DOI] [PubMed] [Google Scholar]
- 13.Ho TD, Slauch JM. 2001. OmpC is the receptor for gifsy-1 and gifsy-2 bacteriophages of Salmonella. J. Bacteriol. 183:1495–1498. 10.1128/JB.183.4.1495-1498.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong J, Kim K-P, Heu S, Lee SJ, Adhya S, Ryu S. 2008. Identification of host receptor and receptor-binding module of a newly sequenced T5-like phage EPS7. FEMS Microbiol. Lett. 289:202–209. 10.1111/j.1574-6968.2008.01397.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim M, Ryu S. 2011. Characterization of a T5-like coliphage SPC35 and differential development of resistance to SPC35 in Salmonella Typhimurium and Escherichia coli. Appl. Environ. Microbiol. 77:2042–2050. 10.1128/AEM.02504-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ricci V, Piddock LJV. 2010. Exploiting the role of TolC in pathogenicity: identification of a bacteriophage for eradication of Salmonella serovars from poultry. Appl. Environ. Microbiol. 76:1704–1706. 10.1128/AEM.02681-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casjens SR, Gilcrease EB, Winn-Stapley DA, Schicklmaier P, Schmieger H, Pedulla ML, Ford ME, Houtz JM, Hatfull GF, Hendrix RW. 2005. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J. Bacteriol. 187:1091–1104. 10.1128/JB.187.3.1091-1104.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vander Byl C, Kropinski AM. 2000. Sequence of the genome of Salmonella bacteriophage P22. J. Bacteriol. 182:6472–6481. 10.1128/JB.182.22.6472-6481.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Susskind MM, Botstein D. 1978. Molecular genetics of bacteriophage P22. Microbiol. Rev. 42:385–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang I-N, Smith DL, Young R. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799–825. 10.1146/annurev.micro.54.1.799 [DOI] [PubMed] [Google Scholar]
- 21.Krupovic M, Cvirkaite-Krupovic V, Bamford DH. 2008. Identification and functional analysis of the Rz/Rz1-like accessory lysis genes in the membrane-containing bacteriophage PRD1. Mol. Microbiol. 68:492–503. 10.1111/j.1365-2958.2008.06165.x [DOI] [PubMed] [Google Scholar]
- 22.Baxa U, Steinbacher S, Miller S, Weintraub A, Huber R, Seckler R. 1996. Interactions of phage P22 tails with their cellular receptor, Salmonella O-antigen polysaccharide. Biophys. J. 71:2040–2048. 10.1016/S0006-3495(96)79402-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venza Colon CJ, Vasquez Leon AY, Villafane RJ. 2004. Initial interaction of the P22 phage with the Salmonella typhimurium surface. P. R. Health Sci. J. 23:95–101 [PubMed] [Google Scholar]
- 24.Pedulla ML, Ford ME, Karthikeyan T, Houtz JM, Hendrix RW, Hatfull GF, Poteete AR, Gilcrease EB, Winn-Stapley DA, Casjens SR. 2003. Corrected sequence of the bacteriophage P22 genome. J. Bacteriol. 185:1475–1477. 10.1128/JB.185.4.1475-1477.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casjens SR, Thuman-Commike PA. 2011. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology 411:393–415. 10.1016/j.virol.2010.12.046 [DOI] [PubMed] [Google Scholar]
- 26.Villafane R, Zayas M, Gilcrease EB, Kropinski AM, Casjens SR. 2008. Genomic analysis of bacteriophage ε34 of Salmonella enterica serovar Anatum (15+). BMC Microbiol. 8:227. 10.1186/1471-2180-8-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Los M, Kuzio J, McConnell M, Kropinski A, Wegrzyn G, Christie G. 2010. Bacteriophages in the control of food- and waterborne pathogens. ASM Press, Washington, DC [Google Scholar]
- 28.Erickson M, Newman D, Helm RA, Dino A, Calcutt M, French W, Eisenstark A. 2009. Competition among isolates of Salmonella enterica subsp. enterica serovar Typhimurium: role of prophage/phage in archived cultures. FEMS Microbiol. Lett. 294:37–44. 10.1111/j.1574-6968.2009.01554.x [DOI] [PubMed] [Google Scholar]
- 29.Shin H, Lee J-H, Kim H, Choi Y, Heu S, Ryu S. 2012. Receptor diversity and host interaction of bacteriophages infecting Salmonella enterica serovar Typhimurium. PLoS One 7:e43392. 10.1371/journal.pone.0043392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fauquet C. 2005. Virus taxonomy: classification and nomenclature of viruses. Eighth report of the International Committee on the Taxonomy of Viruses Elsevier Academic Press, San Diego, CA [Google Scholar]
- 31.Park M, Lee J-H, Shin H, Kim M, Choi J, Kang D-H, Heu S, Ryu S. 2012. Characterization and comparative genomic analysis of a novel bacteriophage SFP10 simultaneously inhibiting both Salmonella and Escherichia coli O157:H7. Appl. Environ. Microbiol. 78:58–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33.Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. 10.1093/bioinformatics/btm009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29:2607–2618 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC55746/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 36.Zdobnov EM, Apweiler R. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848. 10.1093/bioinformatics/17.9.847 [DOI] [PubMed] [Google Scholar]
- 37.Altermann E, Klaenhammer TR. 2003. GAMOLA: a new local solution for sequence annotation and analyzing draft and finished prokaryotic genomes. Omics 7:161–169. 10.1089/153623103322246557 [DOI] [PubMed] [Google Scholar]
- 38.Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9:299–306. 10.1093/bib/bbn017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Néron B, Ménager H, Maufrais C, Joly N, Maupetit J, Letort S, Carrere S, Tuffery P, Letondal C. 2009. Mobyle: a new full web bioinformatics framework. Bioinformatics 25:3005–3011. 10.1093/bioinformatics/btp493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carver T, Berriman M, Tivey A, Patel C, Bohme U, Barrell BG, Parkhill J, Rajandream MA. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676. 10.1093/bioinformatics/btn529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marinelli LJ, Piuri M, Swigoňová Z, Balachandran A, Oldfield LM, van Kessel JC, Hatfull GF. 2008. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS One 3:e3957. 10.1371/journal.pone.0003957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uetake H, Luria SE, Burrous JW. 1958. Conversion of somatic antigens in Salmonella by phage infection leading to lysis or lysogeny. Virology 5:68–91. 10.1016/0042-6822(58)90006-0 [DOI] [PubMed] [Google Scholar]
- 45.Casjens S, Eppler K, Parr R, Poteete AR. 1989. Nucleotide sequence of the bacteriophage P22 gene 19 to 3 region: identification of a new gene required for lysis. Virology 171:588–598. 10.1016/0042-6822(89)90628-4 [DOI] [PubMed] [Google Scholar]
- 46.Rao GR, Burma DP. 1971. Purification and properties of phage P22-induced lysozyme. J. Biol. Chem. 246:6474–6479 [PubMed] [Google Scholar]
- 47.Nam K, Bläsi U, Zagotta MT, Young R. 1990. Conservation of a dual-start motif in P22 lysis gene regulation. J. Bacteriol. 172:204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srividhya KV, Krishnaswamy S. 2007. Subclassification and targeted characterization of prophage-encoded two-component cell lysis cassette. J. Biosci. 32:979–990. 10.1007/s12038-007-0097-x [DOI] [PubMed] [Google Scholar]
- 49.Ho YS, Pfarr D, Strickler J, Rosenberg M. 1992. Characterization of the transcription activator protein C1 of bacteriophage P22. J. Biol. Chem. 267:14388–14397 [PubMed] [Google Scholar]
- 50.Poteete AR, Hehir K, Sauer RT. 1986. Bacteriophage P22 Cro protein: sequence, purification, and properties. Biochemistry 25:251–256. 10.1021/bi00349a035 [DOI] [PubMed] [Google Scholar]
- 51.Watkins D, Hsiao C, Woods KK, Koudelka GB, Williams LD. 2008. P22 c2 repressor-operator complex: mechanisms of direct and indirect readout. Biochemistry 47:2325–2338. 10.1021/bi701826f [DOI] [PubMed] [Google Scholar]
- 52.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8:317–327. 10.1038/nrmicro2315 [DOI] [PubMed] [Google Scholar]
- 53.Chang J, Weigele P, King J, Chiu W, Jiang W. 2006. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure 14:1073–1082. 10.1016/j.str.2006.05.007 [DOI] [PubMed] [Google Scholar]
- 54.Landström J, Nordmark EL, Eklund R, Weintraub A, Seckler R, Widmalm G. 2008. Interaction of a Salmonella enteritidis O-antigen octasaccharide with the phage P22 tailspike protein by NMR spectroscopy and docking studies. Glycoconj. J. 25:137–143. 10.1007/s10719-007-9065-9 [DOI] [PubMed] [Google Scholar]
- 55.Levine M, Truesdell S, Ramakrishnan T, Bronson MJ. 1975. Dual control of lysogeny by bacteriophage P22: an antirepressor locus and its controlling elements. J. Mol. Biol. 91:421–438. 10.1016/0022-2836(75)90270-3 [DOI] [PubMed] [Google Scholar]
- 56.Poteete AR, Ptashne M. 1982. Control of transcription by the bacteriophage P22 repressor. J. Mol. Biol. 157:21–48. 10.1016/0022-2836(82)90511-3 [DOI] [PubMed] [Google Scholar]
- 57.Hofer B, Ruge M, Dreiseikelmann B. 1995. The superinfection exclusion gene (sieA) of bacteriophage P22: identification and overexpression of the gene and localization of the gene product. J. Bacteriol. 177:3080–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ranade K, Poteete AR. 1993. Superinfection exclusion (sieB) genes of bacteriophages P22 and lambda. J. Bacteriol. 175:4712–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kwon HJ, Cho SH, Kim TE, Won YJ, Jeong J, Park SC, Kim JH, Yoo HS, Park YH, Kim SJ. 2008. Characterization of a T7-like lytic bacteriophage (phiSG-JL2) of Salmonella enterica serovar Gallinarum biovar Gallinarum. Appl. Environ. Microbiol. 74:6970–6979. 10.1128/AEM.01088-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pajunen M, Kiljunen S, Skurnik M. 2000. Bacteriophage phiYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 182:5114–5120. 10.1128/JB.182.18.5114-5120.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verma V, Harjai K, Chhibber S. 2009. Characterization of a T7-like lytic bacteriophage of Klebsiella pneumoniae B5055: a potential therapeutic agent. Curr. Microbiol. 59:274–281. 10.1007/s00284-009-9430-y [DOI] [PubMed] [Google Scholar]
- 62.Ptashne M. 2004. A genetic switch—phage lambda revisited, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 63.Rundell K, Shuster CW. 1975. Membrane-associated nucleotide sugar reactions: influence of mutations affecting lipopolysaccharide on the first enzyme of O-antigen synthesis. J. Bacteriol. 123:928–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kourilsky P, Knapp A. 1974. Lysogenization by bacteriophage lambda. III. Multiplicity dependent phenomena occuring upon infection by lambda. Biochimie 56:1517–1523. 10.1016/S0300-9084(75)80275-6 [DOI] [PubMed] [Google Scholar]
- 65.Levine M. 1957. Mutations in the temperate phage P22 and lysogeny in Salmonella. Virology 3:22–41. 10.1016/0042-6822(57)90021-1 [DOI] [PubMed] [Google Scholar]
- 66.Poteete AR, Fenton AC, Murphy KC. 1988. Modulation of Escherichia coli RecBCD activity by the bacteriophage lambda Gam and P22 Abc functions. J. Bacteriol. 170:2012–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng S, Court DL, Friedman DI. 1995. Transcription termination signals in the nin region of bacteriophage lambda: identification of rho-dependent termination regions. Genetics 140:875–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vershon AK, Liao S-M, McClure WR, Sauer RT. 1987. Bacteriophage P22 Mnt repressor: DNA binding and effects on transcription in vitro. J. Mol. Biol. 195:311–322. 10.1016/0022-2836(87)90652-8 [DOI] [PubMed] [Google Scholar]
- 69.Greenberg M, Dunlap J, Villafane R. 1995. Identification of the tailspike protein from the Salmonella newington phage ε34 and partial characterization of its phage-associated properties. J. Struct. Biol. 115:283–289. 10.1006/jsbi.1995.1053 [DOI] [PubMed] [Google Scholar]
- 70.Steinbacher S, Miller S, Baxa U, Weintraub A, Seckler R. 1997. Interaction of Salmonella phage P22 with its O-antigen receptor studied by X-ray crystallography. Biol. Chem. 378:337–343 [DOI] [PubMed] [Google Scholar]
- 71.Lindberg AA, Wollin R, Gemski P, Wohlhieter JA. 1978. Interaction between bacteriophage Sf6 and Shigella flexneri. J. Virol. 27:38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim M, Ryu S. 2012. Spontaneous and transient defence against bacteriophage by phase-variable glucosylation of O-antigen in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 86:411–425. 10.1111/j.1365-2958.2012.08202.x [DOI] [PubMed] [Google Scholar]
- 73.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856. 10.1038/35101614 [DOI] [PubMed] [Google Scholar]
- 74.Curtiss R, Porter SB, III, Munson M, Tinge SA, Hassan JO, Gentry-Weeks C, Kelly SM. 1991. Colonization control of human bacterial enteropathogens in poultry. Academic Press, San Diego, CA [Google Scholar]
- 75.Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 2:2006.0007. 10.1038/msb4100049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Soncini FC, Vescovi EG, Groisman EA. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 177:4364–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chang AC, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.