Abstract
Few data are available on the prevalence and molecular typing of species belonging to the genus Anaplasma in Mediterranean ruminants. In this study, PCR analysis and sequencing of both 16S rRNA and groEL genes were combined to investigate the presence, prevalence, and molecular traits of Anaplasma spp. in ruminants sampled on the Island of Sardinia, chosen as a subtropical representative area. The results demonstrate a high prevalence of Anaplasma spp. in ruminants, with animals infected by at least four of six Anaplasma species (Anaplasma marginale, A. bovis, A. ovis, and A. phagocytophilum). Moreover, ruminants host a number of neutrophil-tropic strains genetically closely related to the canine pathogen A. platys. The high Anaplasma spp. prevalence and the identification of as-yet-unclassified neutrophil-tropic strains raise concerns about the specificity of serological tests routinely used in ruminants and provide additional background for reconstructing the evolutionary history of species genetically related to A. phagocytophilum.
INTRODUCTION
Tick-borne rickettsial diseases bring about considerable economic constraints to livestock management throughout the world (1, 2). Furthermore, over the last several decades, bacteria within the order Rickettsiales have emerged as zoonotic agents, particularly in Europe but also in Africa and in the Americas (3, 4). Consequently, surveillance on rickettsial species circulating among animals and humans has increased appreciably, resulting in a proliferation of studies seeking to detect and molecularly characterize strains in different regions of the world (5).
Among Rickettsiales, the genus Anaplasma has been especially studied for its pathogenicity in farm animals and also, to a lesser extent, in people. Anaplasmosis, caused by various species of Anaplasma, is an important issue for animal breeders. Indeed, infection by Anaplasma spp. generates additional costs to veterinary care by causing reduction in animal body weight, decreases in milk production, abortions, and frequently death (6–9).
The six species included in the genus Anaplasma show different preferential host and cell tropism (10–12). Three species infect the red blood cells of small ruminants (A. ovis), cattle (A. marginale and A. centrale), and wild ruminants (A. marginale and A. centrale). A. bovis causes anaplasmosis in ruminants and small mammals and infects monocytes. A. phagocytophilum is the agent of human and animal granulocytic anaplasmosis and preferentially infects neutrophil granulocytes of ruminants, dogs, horses, and humans. Finally, A. platys shows unique tropism for the platelets of dogs, being the etiological agent of the infectious canine cyclic thrombocytopenia. According to what was stated above, ruminants can be infected by five of six species belonging to the genus Anaplasma. Infections are commonly recorded in wild and domestic ruminants worldwide (11, 13).
In Europe, wild and domestic ruminants play an active role as Anaplasma carriers acting as infection reservoirs potentially able to indirectly transmit strains to other host species in which the same strains are pathogenic. For instance, A. phagocytophilum infections are commonly reported in asymptomatic wild ruminants in several Mediterranean countries, usually in the same areas as infected dogs, and humans develop serious symptoms (14–16). Other Anaplasma species are scarcely reported in Mediterranean Countries, and the molecular characterization of most of strains circulating in this area has yet to be uncovered.
Our group previously reported A. phagocytophilum infections in Sardinian symptomatic horses and dogs. In addition, we detected A. platys in a symptomatic dog living in the same area (14, 15, 17). To date, there is a lack of data regarding the presence of A. phagocytophilum and other Anaplasma species in Sardinian ruminants in particular and in Mediterranean countries in general.
We show in the present study that Sardinian domestic ruminants are commonly infected by distinct Anaplasma species, and we report the presence of novel Anaplasma sp. strains genetically closely related to the canine species A. platys. Preferential cellular tropism and phylogeny of these latter strains are also investigated by confocal laser scanning microscopy and network analyses, respectively.
MATERIALS AND METHODS
Samples and DNA extraction.
From 2010 to 2012, EDTA blood samples were obtained from 99 asymptomatic domestic ruminants (43 calves, 22 sheep, and 34 goats) farmed in different areas of the Island of Sardinia, Italy (Table 1 and see Fig. S1 in the supplemental material). In addition, a blood sample was collected from a Sardinian red deer ( Cervus elaphus corsicanum) kept in captivity in a wildlife recovery facility (Monastir, Sardegna). DNA was extracted from all of the samples by using a DNeasy blood and tissue kit (Qiagen, Italy) according to vendor's recommendations for blood extraction. A. phagocytophilum genomic DNA was extracted from FA substrate slides (Fuller Laboratories, Fullerton, CA) and used as a positive control in Anaplasma sp.-specific PCRs. A DNA preparation of an A. platys strain isolated in Southern Italy (17) was also used as a positive control (in A. platys-specific PCRs). Sampling was approved by the Ethical Committee of the University of Sassari.
TABLE 1.
Origin of ruminants investigated in this study and summary of PCR results
| No. of positive animals/no. examinedb |
|||||||
|---|---|---|---|---|---|---|---|
| Hosta | Geographic location | No. of sampled animals | Positive for 16S rRNA | Positive for groEL |
|||
| A. phagocytophilum | A. platys | A. marginale, A. ovis, and A. centrale | Total | ||||
| Sheep | Villacidro | 13 | 12/13 | 0/13 | 0/13 | 12/13 | 12/13 |
| Perfugas | 1 | 1/1 | 0/1 | 0/1 | 1/1 | 1/1 | |
| Bulzi | 4 | 1/1 | 0/4 | 1/4 | 4/4 | 4/4 | |
| S. M. Coghinas | 2 | 1/2 | 0/2 | 0/2 | 1/2 | 1/2 | |
| Ittiri | 2 | 2/2 | 0/2 | 0/2 | 0/2 | 0/2 | |
| Total | 22 | 17/19 | 0/22 | 1/22 | 18/22 | 18/22 | |
| Bovine | Sedini | 1 | 0/1 | 0/1 | 0/1 | 1/1 | 1/1 |
| Bortigiadas | 14 | 2/3 | 0/14 | 12/14 | 14/14 | 14/14 | |
| Badesi | 5 | 4/4 | 0/5 | 4/5 | 5/5 | 5/5 | |
| Tula | 1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | |
| Perfugas | 9 | 6/7 | 0/9 | 3/9 | 7/9 | 8/9 | |
| Erula | 4 | 0/1 | 0/4 | 3/4 | 4/4 | 4/4 | |
| Samugheo | 3 | 0/3 | 3/3 | 3/3 | 3/3 | ||
| Lanusei | 6 | 3/3 | 0/6 | 2/6 | 5/6 | 5/6 | |
| Total | 43 | 15/20 | 0/43 | 27/43 | 39/43 | 40/43 | |
| Goat | Perfugas | 5 | 4/4 | 0/5 | 5/5 | 4/5 | 5/5 |
| Tisiennari | 21 | 12/12 | 0/21 | 17/21 | 21/21 | 21/21 | |
| Sassari | 8 | 0/8 | 5/8 | 4/8 | 6/8 | ||
| Total | 34 | 16/16 | 0/34 | 27/34 | 29/34 | 32/34 | |
| Total | 99 | 48/55 | 0/99 | 55/99 | 86/99 | 90/99 | |
The red deer sample is not included in this table.
Data are presented as the number positive/the total number of animals examined.
Anaplasma PCR strategies and profiles.
In order to investigate the presence of Anaplasma spp. in Sardinian domestic ruminants, 53 of the 99 blood DNA extractions were initially tested with two primers (AnaplsppF, 5′-AGAAGAAGTCCCGGCAAACT-3′; AnaplR3, 5′-GAGACGACTTTTACGGATTAGCTC-3′) targeting ∼800 bp of the 16S rRNA gene of species belonging to the genus Anaplasma. Briefly, 100 to 150 ng of DNA extractions were used in a 50-μl PCR mixture containing 200 μM deoxynucleoside triphosphates (dNTPs), 0.3 μM concentrations each of the two primers, and 1.25 U of Taq DNA polymerase (Qiagen, Italy). PCR amplifications were performed with an initial denaturation at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C (30 s), annealing at 50°C (30 s), and extension at 72°C (1 min), followed by a final extension at 72°C for 10 min. Based on the 16S rRNA gene PCR results, in order to mine deeper into the presence of selected Anaplasma species in ruminants, we tested the 99 DNA extractions with a set of four primers combined in two heminested PCRs designed for the molecular identification and characterization of the A. phagocytophilum and A. platys groEL genes (14, 15, 17). In order to target the corresponding groEL region of A. centrale, A. marginale, and A. ovis, three new primers (AmaceovgroELF, 5′-ACGGTATGCAGTTTGACCGC-3′; AmaceovgroELR1, 5′-TCAACCCTATCCTTACGCTC-3; AmaceovgroELR2, 5′-GTCGTAGTCAGAAGAAGAAAC-3′) were designed and combined in a heminested PCR to detect ∼518 bp of this gene. In the first PCR round 100 to 150 ng of DNA extractions were used in a 50-μl PCR mixture containing 200 μM dNTPs, 0.3 μM concentrations of each of the AmaceovgroELF and AmaceovgroELR1 primers, and 1.25 U of Taq DNA polymerase. Then, 1 μl of the first PCR product was subjected to a second PCR round with the primers AmaceovgroELF and AmaceovgroELR2. Both amplifications were performed with an initial denaturation at 94°C for 3 min, followed by 30 cycles of denaturation (30 s) at 94°C, annealing (30 s) alternatively at 55°C (first PCR round) or 60°C (second PCR round), and extension (1 min) at 72°C, followed in turn by a final extension at 72°C for 10 min. Since the groEL gene sequence of A. bovis is still not available in the GenBank database, a groEL-specific PCR could not be designed for this bacterium, and A. bovis was excluded from this analysis.
Cloning, sequencing, and phylogenetic analyses.
An ABI Prism BigDye terminator cycle sequencing ready reaction kit (Life Technologies, Italy) was used for direct cycle sequencing of 14 PCR products obtained with the 16S rRNA gene primers and representative of host species and geographic location, according to the manufacturer's protocol. Ambiguous nucleotide positions were resolved by cloning amplicons into the vector pCR2.1-TOPO and by universal M13 primers sequencing. Similarly, 37 PCR products obtained with the groEL-specific primers were sequenced either directly or after cloning into pCR2.1-TOPO. The generated sequences were edited with Chromas 2.2 (Technelysium, Helensvale, Australia) and aligned with CLUSTAL W (18) in order to assign them to unique sequence types. Sequence types, named after the host species and sequentially ordered, were checked against the GenBank database with nucleotide blast BLASTN (19). The 16S rRNA gene sequence types obtained here were aligned among them and with a set of 14 sequences representative of the 16S rRNA gene variability of the six different species belonging to the genus Anaplasma. In particular, the set was composed of three A. marginale sequences (CP001079, JQ839012, and AF414873), two A. centrale sequences (EF520690 and JQ839010), three A. ovis sequences (JQ917905, JQ917880, and JQ917902), three A. bovis sequences (JN558825, JN558819, and AB196475), one A. platys sequence (EU439943), and two A. phagocytophilum sequences (AB196721 and JX173652). Also, the groEL sequence types generated here were aligned among them and with a set of 16 sequences representative of the groEL variability of the six different species belonging to the genus Anaplasma. The groEL reference sequences were as follows: A. marginale (JQ839015, CP001079, and JQ839003), A. centrale (AF414866, EF520695, and CP001059), A. ovis (FJ460441 and AF441131), A. platys (EF201806 and FJ460441), A. phagocytophilum (HM057225, CP000235, AY848749, and AY848752), and A. bovis (JX092099 and JX092095). Rickettsia rickettsii (CP003318) and Ehrlichia canis (EU439944) were included as outgroups in both of the 16S rRNA and groEL sequence alignments.
Pairwise/multiple sequence alignments and sequence similarities were calculated using the CLUSTAL W and the identity matrix options of Bioedit (20), respectively. Genetic distances among the operational taxonomic units were computed by the maximum composite likelihood method (21) or by the Kimura two-parameter method using MEGA version 2.1 (22) and were used to construct neighbor-joining trees (23). Statistical support for internal branches of the trees was evaluated by bootstrapping with 1,000 iterations (24). Maximum-parsimony trees and consensus values were generated using the same software. MEGA was also used to investigate the variability of the nucleotide positions in the groEL and 16S rRNA sequence alignments. In order to reduce systematic error, to investigate more deeply the taxon evolutionary relationships, and to test the robustness of the phylogenetic analyses, groEL sequence clusters were also detected by the analysis of phylogenetic networks inferred from uncorrected p-distances with the Neighbor-Net method in SplitsTree4 (25), not considering invariable sites.
Blood fractionation and thin-layer slide preparation.
One set of EDTA blood samples (5 ml) was collected from two goats (M2 and 324) that tested positive by three independent A. platys groEL heminested PCRs and negative by the analogue PCR specifically designed for detecting the A. marginale-A. centrale-A. ovis groEL gene. Next, 3-ml samples were layered onto 3 ml of Histopaque-1077 (Sigma-Aldrich, Italy) and centrifuged at 400 × g for 30 min at room temperature in order to isolate the buffy coat according to the manufacturer's instructions. Both the buffy coat and the erythrocytes were opportunely diluted in phosphate-buffered saline (PBS) and centrifuged for 6 min at 127 × g with a Thermo Shandon Cytospin 4 cytocentrifuge (Thermo Scientific, Italy) to obtain thin-layer cell slide preparations. In order to obtain slide preparations of platelets, sodium citrate blood samples (5 ml) were also collected from the same goats and centrifuged (827 × g for 30 min) at room temperature. Platelet-enriched plasma fractions were collected and centrifuged at 1,146 × g for 10 min. Pellets were resuspended in 500 μl of plasma and cytocentrifuged as described below. Blood smears were also prepared from the same goats according to standard procedures. Platelets, red blood cells, buffy coats, and blood smears were always layered on Superfrost positively charged glass slides (Fisher Scientific, Italy).
PCR synthesis of digoxigenin-, fluorescein-, and rhodamine-labeled DNA probes.
A PCR DIG probe synthesis kit (Roche, Italy) was used to generate a 107-bp digoxigenin-labeled DNA probe specific for A. platys by PCR. Fifty-microliter PCRs were set up by mixing 5 μl of PCR DIG probe synthesis mix (containing 200 μM concentrations of each dNTP and 70 μM DIG-dUTP) with 5 μl of 10× PCR buffer containing MgCl2, 0.75 μl of enzyme mix, 29.25 μl of water, 100 pg of an amplicon obtained by A. platys groEL heminested PCR of a bovine source (N194), and 5 μl each of the primers aplgroELdigprobF (5′-TAGAAGGGGAAGCTTTAAGCA-3′), and aplgroELdigprobR (5′-ACTGCGATATCACCAAGCATA-3′). An unlabeled probe was generated according to the same protocol, without incorporation of digoxigenin in the PCR mix. PCR profiles were designed according to the manufacturer's instructions for probe synthesis. Both digoxigenin-labeled and unlabeled probes were used (independently) for in situ hybridization experiments. Similarly, fluorescein- and rhodamine-labeled versions of the 107-bp A. platys groEL-specific probe were also originated by using a PCR fluorescein labeling mix (Roche) and a tetramethyl-rhodamine-5-dUTP (Roche), respectively. PCR protocols were adjusted to vendor recommendations. A rhodamine-labeled 98-bp DNA probe was designed targeting the groEL gene of A. marginale, A. centrale, and A. ovis. This probe was synthesized starting from an amplicon obtained by a A. marginale-, A. centrale-, and A, ovis-specific groEL heminested PCR and by using the primers Amacenov_probe_F (5′-GGTCAGGAAGGCCATTGCTG-3′) and Amacenov_probe_R (5′-TGCAACCTGCAAGCCTCCGC-3′). PCR was performed by according to the same protocols used for generating probes specific for the groEL region of A. platys.
Combined in situ hybridization and immunofluorescence.
Buffy coats, red blood cells, platelets, and blood smears layered on positively charged glass slides were fixed in 4% paraformaldehyde) for 10 min, digested with 10 μg of proteinase K/ml for 10 min, dehydrated by passage through 25, 50, 75, and 100% methanol in PBS (1 to 2 min in each solution), and air dried. Slides were stored at −20°C for later use.
Prior to hybridization, the slides were rehydrated in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 15 min and then prehybridized in prehybridization solution at 42°C for 1 h (50% molecular-grade Fluka hybridization solution II, 43% formamide, 7% Milli-Q water) in a programmable temperature-controlled slide processing system (StatSpin ThermoBrite, Italy).
After the prehybridization solution was removed, the slides were covered with a cover glass, treated with 200 μl of hybridization solution (obtained by adding 1.5 μg of a denatured molecular probe to 1 ml of hybridization solution), placed at 95°C for 8 min, and incubated overnight. The hybridization solution varied depending on the type of sample analyzed (buffy coat, red blood cells, platelets, or blood smear).
Slides obtained from blood smears were treated with hybridization solution containing the A. platys digoxigenated groEL-specific probe or, alternatively, an unlabeled version of the same probe. After hybridization, unhybridized probes were removed by washing the slides twice with Tris-buffered saline (5 min per wash). The slides were treated with decreasing concentrations of Sigma-Aldrich molecular-biology-grade SSPE (5-min washes at 25°C with 2×, 1×, and 0.5× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA; pH 7.7], a 30-min wash at 50°C with 0.1× SSPE and bovine serum albumin [BSA] 0.2%, and a 5-min wash at 25°C with 0.1× SSPE and BSA 0.2%). Preparations were then washed for 5 min with buffer I (0.1 M Tris, 0.15 M NaCl; pH 7.6). Nonspecific endogenous alkaline phosphatase was blocked with 200 μl of blocking solution (20 μl of normal rabbit serum, 3.1 μl of 0.3% Triton X-100, 980 μl of buffer I) at room temperature for 45 min. For the detection of the digoxigenin-labeled hybrids, slides were incubated for 2 h with 1:25-diluted anti-digoxigenin-AP Fab fragments (Sigma-Aldrich) at room temperature. Slides were rinsed twice and incubated for 50 min at 37°C in the dark with BCIP (5-bromo-4-chloro-3-indolylphosphate) and 4-nitroblue tetrazolium chloride. Color development was stopped with Tris-EDTA buffer (pH 8.0). The slides were mounted with Top-Water mount (W. Pabisch SpA, Italy) and photographed with a Nikon Eclipse 80i microscope equipped with a Nikon DS-L2 camera control unit.
Buffy coats and platelet slides were mounted and probed either with rhodamine- or fluorescein-labeled DNA probes synthesized by using a A. platys groEL heminested PCR. Similarly, the same samples were hybridized with probes synthesized by A. marginale-A. centrale-A. ovis groEL heminested PCR. Buffy coats and platelet slides were incubated in a programmable temperature-controlled slide processing system (StatSpin ThermoBrite) at 42°C for 12 to 14 h (overnight) in hybridization solution containing 5 ng of the opportune DNA probe (in 70 μl of prehybridization solution). On the following day, the coverslips were removed, and the slides were washed in decreasing concentrations of SSC at 48°C, with a final rinse in PBS.
In order to label neutrophils, buffy coat slides were incubated overnight at 4°C with 1:100 biotinylated monoclonal rat anti-mouse neutrophil antibodies (Caltag Laboratories). Before incubation with the primary antibody, nonspecific binding was blocked with 2% BSA in PBS. Buffy coat slides were then incubated for 1 h at room temperature with Histostain-Plus (Life Technologies) and subsequently treated with Alexa Fluor 555 streptavidin (Invitrogen) for 1 h. Slides were counterstained with Hoechst blue and then coverslipped in ProLong Gold antifade reagent (Invitrogen).
Platelet and erythrocyte slides were incubated overnight at 4°C with 1:100 mouse monoclonal anti platelet antibody labeled with fluorescein isothiocyanate (Antibodies-Online, Inc.) and 1:100 rabbit fluorescein isothiocyanate conjugate anti-goat RBC antibody (Fitzgerald, USA), respectively. The slides were then washed once with PBS, once with water, counterstained with Hoechst blue, and eventually coverslipped in ProLong Gold antifade reagent.
Confocal laser scanning microscopy.
Buffy coat, red blood cell, and platelet slides were analyzed by confocal laser scanning microscopy using a Leica DMI4000 B automated inverted research microscope. Images were acquired by using Leica LAS AF Lite software in combination with a 40× oil immersion objective lens and a numerical aperture of 1.25. Excitation wavelengths used were 488 nm (green) and 568 nm (red). All images were edited using Adobe Photoshop CS4. Manipulations did not change the data content. In each experiment, unlabeled probes were used as a negative control. Also, experiments were repeated by omitting the first antibody or the DNA probe. All experiments were run at least in triplicate.
Nucleotide sequence accession numbers.
The 16S rRNA and groEL nucleotide sequences determined in the present study have been submitted to GenBank under the accession numbers given in Tables 2 and 3.
TABLE 2.
Designation of the eight 16S rRNA sequence types identified in this study
| Sequence type | Host | Geographical location | GenBank accession no. | BLAST analysis |
|---|---|---|---|---|
| BovineCaprine1 | Bos taurus 204934 | Perfugas (northern Sardinia) | KC335221 | 99% A. platys (invariable) |
| Capra hircus 50 | Perfugas (northern Sardinia) | KC335222 | 99% A. platys (invariable) | |
| BovineCaprine2 | Bos taurus 197 | Badesi (northern Sardinia) | KC335223 | 100% A. marginale (worldwide) |
| Capra hircus 71 | Perfugas (northern Sardinia) | KC335224 | 100% A. marginale (worldwide) | |
| Bovine1 | Bos taurus 199 | Badesi (northern Sardinia) | KC335218 | 100% A. marginale (Asia, USA) |
| Bos taurus 204927 | Perfugas (northern Sardinia) | KC335219 | 100% A. marginale (Asia, USA) | |
| Bovine2 | Bos taurus 194 | Samugheo (central Sardinia) | KC335220 | 99% A. platys (invariable) |
| Ovine1 | Ovis aries 230 | Villacidro (southwestern Sardinia) | KC335227 | 99% A. phagocytophilum (worldwide) |
| Ovine2 | Ovis aries 725 | S.M. Coghinas (northern Sardinia) | KC335228 | 99 to 100% A. bovis (Asia, USA) |
| Ovis aries C3 | Ittiri (northwestern Sardinia) | KC335229 | 99 to 100% A. bovis (Asia, USA) | |
| Ovis aries A1 | Ittiri (northwestern Sardinia) | KC335230 | 99 to 100% A. bovis (Asia, USA) | |
| Ovine3 | Ovis aries 208 | Bulzi (northern Sardinia) | KC335231 | 99 to 100% A. ovis (Asia, Africa, USA) |
| OvineCaprine1 | Capra hircus 70 | Perfugas (northern Sardinia) | KC335225 | 99 to 100% A. ovis (Asia, Africa, Europe) |
| Ovis aries 235 | Villacidro (southwestern Sardinia) | KC335226 | 99 to 100% A. ovis (Asia, Africa, Europe) |
TABLE 3.
Designation of the 25 groEL sequence types identified in this study
| Sequence type | Host | Geographical location | GenBank accession no. | BLAST analysis |
|---|---|---|---|---|
| Ovine1 | Ovis aries 228 | Villacidro (southwestern Sardinia) | KC335242 | 99% A. marginale, A centrale (worldwide) |
| Bovine1 | Bos taurus 125554 | Lanusei (central eastern Sardinia) | KC335245 | 99 to 100% A. marginale (worldwide) |
| Bos taurus 199 | Badesi (northern Sardinia) | KC335244 | 99 to 100% A. marginale (worldwide) | |
| Bos taurus 195 | Badesi (northern Sardinia) | KC335243 | 99 to 100% A. marginale (worldwide) | |
| Bovine2 | Bos taurus 199 | Badesi (northern Sardinia) | KC335241 | 99% A. marginale, A centrale (worldwide) |
| Ovine2 | Ovis aries 232 | Villacidro (southwestern Sardinia) | KC335266 | 99% A. marginale (Philippines) |
| OvineBovine1 | Ovis aries 230 | Villacidro (southwestern Sardinia) | KC335238 | 99% A. marginale (Philippines) |
| Bos taurus 032 | Lanusei (central eastern Sardinia) | KC335237 | 99% A. marginale (Philippines) | |
| Bos taurus 204934 | Perfugas (northern Sardinia) | KC335236 | 99% A. marginale (Philippines) | |
| Ovine3 | Ovis aries 208 | Bulzi (northern Sardinia) | KC335240 | 98 to 99% A. marginale (worldwide) |
| Ovine4 | Ovis aries 208 | Bulzi (northern Sardinia) | KC335239 | 99% A. marginale (Philippines) |
| Bovine4 | Bos taurus 204927 | Perfugas (northern Sardinia) | KC335235 | 99% A. marginale (Philippines) |
| Bovine5 | Bos taurus 032 | Lanusei (central eastern Sardinia) | KC335232 | 99% A. marginale (Philippines) |
| Ovine5 | Ovis aries 231 | Villacidro (southwestern Sardinia) | KC335234 | 99% A. marginale (Philippines) |
| Ovine6 | Ovis aries 233 | Villacidro (southwestern Sardinia) | KC335233 | 99% A. marginale (Philippines) |
| Caprine1 | Capra hircus 322 | Sassari (northwestern Sardinia) | KC335267 | 99% A. ovis (South Africa) |
| Caprine2 | Capra hircus G | Bortigiadas (northern Sardinia) | KC335268 | 99% A. ovis (South Africa) |
| Caprine3 | Capra hircus 50 | Perfugas (northern Sardinia) | KC335269 | 99% A. ovis (South Africa) |
| Bovine6 | Bos taurus 306 | Samugheo (central Sardinia) | KC335256 | 93% A. platys (invariable) |
| CaprineCervus1 | Capra hircus 50 | Perfugas (northern Sardinia) | KC335249 | 93% A. platys (invariable) |
| Capra hircus 71 | Perfugas (northern Sardinia) | KC335248 | 93% A. platys (invariable) | |
| Cervus elaphus 1 | Monastir (southern Sardinia) | KC335247 | 93% A. platys (invariable) | |
| Bovine3 | Bos taurus 194 | Badesi (northern Sardinia) | KC335270 | 99% A. marginale, A centrale (worldwide) |
| Bovine7 | Bos taurus 194 | Badesi (northern Sardinia) | KC335265 | 93% A. platys (invariable) |
| Bos taurus 303 | Samugheo (central Sardinia) | KC335264 | 93% A. platys (invariable) | |
| Bos taurus 195 | Badesi (northern Sardinia) | KC335263 | 93% A. platys (invariable) | |
| Bos taurus 199 | Badesi (northern Sardinia) | KC335262 | 93% A. platys (invariable) | |
| Bos taurus 204 | Bulzi (northern Sardinia) | KC335261 | 93% A. platys (invariable) | |
| Caprine4 | Capra hircus 324 | Sassari (northwestern Sardinia) | KC335260 | 92% A. platys (invariable) |
| Caprine5 | Capra hircus 84 | Bortigiadas (northern Sardinia) | KC335259 | 93% A. platys (invariable) |
| Capra hircus C | Bortigiadas (northern Sardinia) | KC335258 | 93% A. platys (invariable) | |
| Capra hircus E | Bortigiadas (northern Sardinia) | KC335257 | 93% A. platys (invariable) | |
| Cervus1 | Cervus elaphus 1 | Monastir (southern Sardinia) | KC335246 | 93% A. platys (invariable) |
| Bovine8 | Bos taurus 304 | Samugheo (central Sardinia) | KC335255 | 93% A. platys (invariable) |
| Caprine6 | Capra hircus M2 | Sassari (northwestern Sardinia) | KC335254 | 93% A. platys (invariable) |
| Capra hircus 332 | Sassari (northwestern Sardinia) | KC335253 | 93% A. platys (invariable) | |
| Capra hircus 334 | Sassari (northwestern Sardinia) | KC335252 | 93% A. platys (invariable) | |
| Bovine9 | Bos taurus 204927 | Perfugas (northern Sardinia) | KC335251 | 93% A. platys (invariable) |
| Ovine7 | Ovis aries 208 | Bulzi (northern Sardinia) | KC335250 | 92% A. platys (invariable) |
RESULTS
Anaplasma sp. 16S rRNA PCR and phylogenetic analysis.
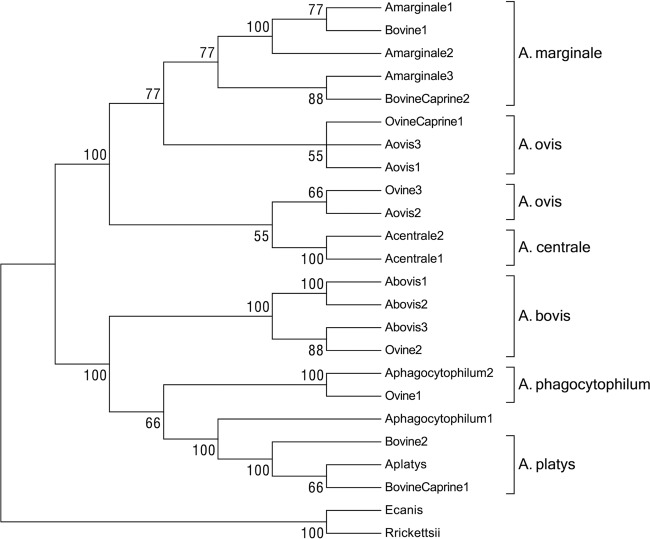
A total of 48 of 55 tested ruminants (87.3%) were determined to be positive by Anaplasma sp. 16S rRNA PCR. In particular, 17 of 19 sheep (89.5%), 15 of 20 bovines (75%), and 16 of 16 goats (100%), homogeneously distributed in the study area, showed the expected 800-bp band on agarose electrophoresis (Table 1 and see Fig. S1 in the supplemental material). Controls (Milli-Q water samples) were always negative. Upon sequencing and CLUSTAL W alignment, 14 PCR products chosen as representative of different hosts and geographic locations resulted in eight different 16S rRNA sequence types (Table 2). The eight sequence types were named after the host species in which they were isolated and were identified by a progressive number. Sequence types BovineCaprine2 and Bovine1, identified in three calves and one goat, shared 100% homology with A. marginale strains isolated worldwide. Two sequence types (Ovine3 and OvineCaprine1) derived from two sheep and one goat were 99 to 100% similar to A. ovis sequences isolated in Africa, Asia, Europe, and the United States. One sequence type (Ovine2) containing sequences exclusively derived from sheep shared 99 to 100% homology with Asiatic and U.S. A. bovis sequences. Sequence type Ovine1, derived from a sheep, was 99% similar to A. phagocytophilum sequences isolated worldwide. Notably, two sequence types (BovineCaprine1 and Bovine2) shared 99% identity with the invariable 16S rRNA gene sequence of A. platys. Table S1 in the supplemental material reports the variability among the CLUSTAL W alignment of the eight identified sequence types with the most similar sequences representative of the six species belonging to the genus Anaplasma. A total of 48 phylogenetic informative sites out of 55 variable sites are present, with 47 substitutions and a single deletion of one nucleotide. Transitions (40/48) are much more prevalent than transversions (5/48). Three positions are hypervariable, with three or four nucleotides alternatively present. These observations are congruent with the evolutionary model of the bacterial 16S rRNA locus and indicate that this genetic variability is suitable for phylogenetic analyses. Coinciding character- and distance-based trees (Fig. 1) obtained from the same alignment indicates that the 16S rRNA sequence types generated in our study group into two major clades strongly supported by bootstrap statistics. A first clade defined by the reference sequences of A. marginale, A. centrale, and A. ovis contains four sequence types derived from ovine, caprine, and bovine hosts. In particular, the sequence types Bovine1 and BovineCaprine2 group into separated subclades with A. marginale, which appears polyphyletic in the trees. Sequence types Ovine3 and OvineCaprine1 also group in two subclades with A. ovis, which similarly appears as polyphyletic. The second major clade is composed by two subclades. One subclade contains the 16S sequences of A. bovis, and both in accordance with BLAST analyses and with the alignment shown in Table S1 in the supplemental material, the sequence type Ovine2. The second subclade contains the sequence types Bovine2 and BovineCaprine1 and the 16S sequences of A. platys. The sequences of A. phagocytophilum group in the two different subclades and with the sequence type Ovine1.
FIG 1.
16S rRNA-based phylogenetic analyses of the strains identified in the present study and of eight sequences representative of the different species of the genus Anaplasma. Even though both character-based and distance-based evolutionary analyses generated coinciding trees, only the evolutionary history inferred using the neighbor-joining method is shown, with sum of branch length of 0.32723954. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. All positions containing gaps and missing data were eliminated. There were a total of 760 positions in the final data set. E. canis and R. rickettsii were used as outgroups.
groEL PCR and phylogenetic analysis.
In order to further investigate the genetic variability of the Anaplasma spp. in Sardinian domestic ruminants, 99 animals (Table 1) were tested by three heminested PCRs specifically designed to amplify the same groEL gene of A. phagocytophilum, of A. platys, and of A. marginale, A. centrale, and/or A. ovis, respectively (see Materials and Methods).
Only 9 of 99 (9.1%) animals were Anaplasma-free since they were simultaneously negative to the 3 groEL PCRs. Ninety ruminants (90.9%) were positive in at least one test, and this finding is consistent with observations based on 16S. More specifically (Table 1), 1 of 22 sheep (4.5%), 27 of 43 calves (62.8%), and 27 of 34 goats (79.4%) were positive when tested with the PCR test specific for the groEL gene of A. platys. When the same 99 samples were tested by PCR for the presence of A. marginale, A. centrale, and/or A. ovis, 18 of 22 sheep (81.8%), 39 of 43 calves (90.7%), and 29 of 34 goats (85.3%) generated the expected band, as revealed by agarose electrophoresis and gel imaging. Coinfections with at least two different Anaplasma species were observed in 51 (56.7%) of 90 infected domestic ruminants, since they tested simultaneously positive to the two PCRs designed to amplify the groEL gene of A. platys and of A. marginale, A. centrale, and/or A. ovis. Four animals were exclusively positive to A. platys (4.4%), while 35 of them (38.9%) were infected with A. marginale, A. centrale, and/or A. ovis only. All of the 99 bovine, caprine, and ovine samples were negative when tested for the presence of A. phagocytophilum. Also, a blood sample obtained from a red deer was found to be positive to the A. platys groEL-specific PCR.
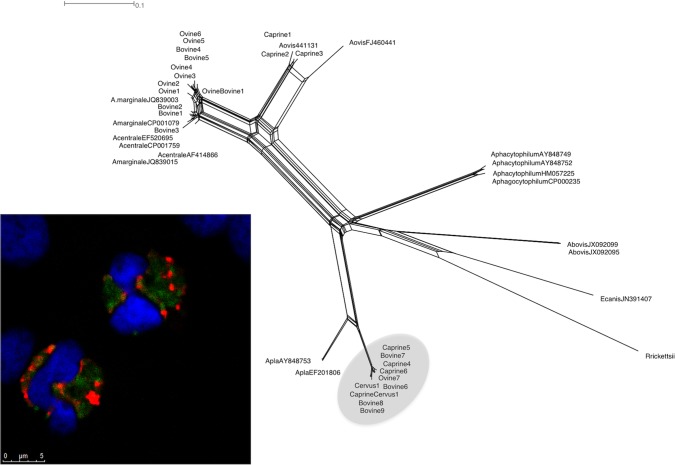
Upon cloning and sequencing of 35 groEL amplicons (19 obtained by A. platys-specific PCR and 16 by A. marginale, A. centrale, and/or A. ovis PCR) representative of different hosts and geographic locations, 39 sequences were obtained. Alignment with CLUSTAL W allowed grouping the 39 sequences in 25 different sequence types (Table 3). On BLAST analyses (Table 3), nine sequence types of both ovine and bovine origin shared the highest level of similarity with strains of A. marginale isolated worldwide (98 to 100%). Three ovine and bovine sequences showed the same levels of similarity (99%) with a number of A. marginale and A. centrale strains associated to a large geographical distribution. Three caprine sequence types were 99% similar to an A. ovis strain isolated in South Africa. Ten sequence types obtained from ovine, caprine, and bovine samples shared 92 to 93% similarity with the groEL gene of A. platys (e value of 0.0). Sequence alignment of these last 10 sequences with the groEL sequence of A. platys allowed scoring 44 variable nucleotide positions (see Table S4 in the supplemental material). Remarkably, 32 positions contained nucleotide substitutions shared by all of the sequences obtained here and therefore represent nucleotide signatures of the groEL region of the corresponding Anaplasma strains infecting ruminants. A total of 30 of 44 variable positions were transitions, and 14 were transversions. No variation other than nucleotide substitution was observed along the alignment. Genetic variability was not related to the host species from which the sequence types were derived. Neighbor-joining and maximum-parsimony analyses generated coinciding trees in which 22 of 25 sequence types group with reference sequences in statistically supported clusters (Fig. 2). In particular, 10 sequence types derived from sheep and calves group with A. marginale. Three sequence types derived from goats group with A. ovis, whereas two sequence types cluster with A. marginale, A. centrale, and A. ovis but cannot be unambiguously assigned to one of these three species. Similar to what observed with 16S phylogeny, 10 sequence types derived from sheep, goats, calves, and a red deer generate a distinct clade with A. platys and group in a separate subclade. None of the groEL sequence types appear to be related to A. phagocytophilum.
FIG 2.
groEL-based phylogenetic analyses of the strains identified in the present study and of 16 sequences representative of the different species of the genus Anaplasma. As in the case of 16S rRNA-based analyses, both character-based and distance-based evolutionary analyses generated coinciding trees, but only the evolutionary history inferred using the neighbor-joining method is shown, with the sum of the branch length being 0.82216554. The evolutionary distances were computed using the Kimura two-parameter method and are in the units of the number of transitional substitution per site. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. All positions containing gaps and missing data were eliminated. There were a total of 470 positions in the final data set. E. canis and R. rickettsii were used as outgroups.
Network analysis of the same groEL sequences (see the Discussion and Fig. 4) confirms what was observed with traditional trees, and allows assigning the sequences Bovine2 and Bovine3, unresolved in the groEL trees, to A. marginale. Notably, ten sequences group with A. platys in a separate subclade, according to results graphically represented by phylogenetic trees based on the16S rRNA and groEL genes (Fig. 1 and 2).
FIG 4.
Identification of A. platys-like strains by network analyses (right) and cellular distribution in neutrophil granulocytes (lower left). Neutrophil granulocytes were contemporarily probed with a FITC-labeled A. platys DNA-probe and with TRITC-labeled antineutrophil antibodies.
In situ hybridization and confocal laser scanning microscopy.
To investigate the cellular tropism of the Anaplasma sp. closely related but distinct from A. platys, an A. platys-specific digoxigenin-labeled DNA probe was successfully generated by PCR. When blood smears obtained from two goats positive to A. platys-specific PCR were treated with the A. platys-specific DNA probe, no reaction of the probe could be observed with platelets (see Fig. S2, lower panels, in the supplemental material). Interestingly, the digoxigenin-labeled probe reacted with polymorphonucleated cells morphologically resembling neutrophils (see Fig. S2, upper panels, in the supplemental material). To confirm a neutrophil tropism for this Anaplasma strains, fluorescein- and rhodamine-labeled versions of the A. platys-specific probe were produced and tested against buffy coat, red blood cells, and platelet-enriched blood fractions obtained from the same animals. The reactivity of the probes with these blood components, analyzed by confocal laser scanning microscopy, confirmed what has already been observed with the digoxigenin-labeled probe. No reactivity of the fluorescent probes could be observed either with platelet preparations or red blood cells (see Fig. S3, lower and middle panels, in the supplemental material), whereas neutrophil-shaped cells strongly reacted with the same probes (see Fig. S3, upper panels, in the supplemental material). Unlabeled probes used as controls never reacted with buffy coat, red blood cells, and platelet-enriched blood preparations. Similarly, no signal was detected when probes specific for A. marginale, A. centrale, and/or A. ovis were tested on the same samples. Experiments were repeated by probing neutrophils with antineutrophil antibodies (Fig. 3, upper panels). Confocal laser scanner microscopy confirmed that only neutrophils showing a positive reaction to the specific antibodies reacted at the same time with the A. platys-specific probe. Again, preparations of platelets obtained from the same animals only reacted with specific anti-platelet antibodies, confirming the absence of a specific signal generated by the A. platys-specific DNA probe. Notably, the DNA probe signal in neutrophils did not colocalize with the antineutrophil-specific signal but was homogeneously spread in the neutrophil cytoplasm (Fig. 4).
FIG 3.
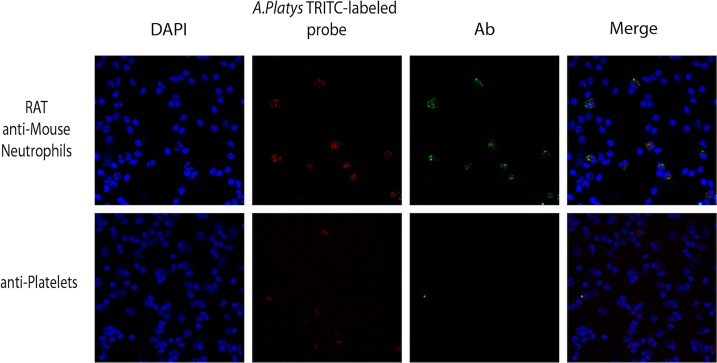
Detection of A. platys-like strains in goat neutrophil granulocytes. Buffy coats obtained from PCR-positive goats were layered in slides by centrifugation and probed with TRITC (tetramethyl rhodamine isothiocyanate)-labeled A. platys DNA probes alternatively combined with fluorescein isothiocyanate (FITC)-labeled anti neutrophil antibodies (upper row) or FITC-labeled antiplatelet antibodies (lower row). DAPI was used to counterstain peripheral blood mononuclear cell nuclei.
DISCUSSION
With the exception of A. platys, all of the bacterial species belonging to the genus Anaplasma infect domestic and wild ruminants with worldwide geographical distributions mirroring that of tick vectors. Ruminants represent therefore key species in the epidemiology of the genus Anaplasma by either acting as definitive hosts or as infection reservoirs. In the present study, the presence and molecular traits of the six species belonging to the genus Anaplasma were investigated in ruminants of a typical Mediterranean environment. Several factors make the island of Sardinia a natural laboratory for the study of Anaplasma incidence and prevalence: (i) its geographical location in the middle of the Mediterranean Sea, (ii) its being a hot spot for tick-transmitted diseases, and (iii) its climate favoring severe tick seasonal infestations caused by at least six tick genera (Rhipicephalus, Haemaphysalis, Hyalomma, Boophilus, Dermacentor, and Ixodes) present in the Island at different relative abundances (15, 26, 27). In the present study, we report a comprehensive study focused on the genetic characterization of the Anaplasma strains circulating in Mediterranean domestic ruminants. Indeed, although several studies evaluating the incidence and economic impact of ruminant anaplasmosis in Africa and in the Americas are available (28, 29, 30), similar studies are still lacking for the Mediterranean area, in which the investigation of the genus Anaplasma is still lacking, and such studies are mainly limited to the detection of species impacting humans and pets, such as the zoonotic A. phagocytophilum and the canine species A. platys. Here, we demonstrated that Sardinian ruminants are commonly infected by different Anaplasma species (Table 1). Indeed, the levels of Anaplasma prevalence, i.e., the percentage of animals positive to at least one Anaplasma species, were high and comparable in Sardinian ruminants when independently calculated by groEL and 16S rRNA PCRs (ca. 87 and 91%, respectively). A high Anaplasma prevalence could also be individually observed in sheep (82% 16S versus 89% groEL), bovines (75% 16S versus 93% groEL), and goats (100% 16S versus 94% groEL), with ca. 50% of ruminants coinfected by at least two Anaplasma species (data based on the groEL gene [not shown]).
These data confirm the relevance of ruminants as important hosts and reservoirs of different Anaplasma species in the subtropical Mediterranean areas. Furthermore, the contemporary presence of more than one Anaplasma species in the same animal suggests carefully reinterpreting and reconsidering previous studies in which prevalence was investigated by serology and brings about new concerns regarding the development of diagnostic serological and molecular tools. This is of particular importance if one considers that several studies reported high degrees of cross-reactivity among species of the genus Anaplasma in serological tests, for instance, between A. phagocytophilum and A. marginale (31) and between A. marginale and A. centrale, as well as cross-reactivity among A. phagocytophilum, Ehrlichia spp., and A. platys (15, 17, 31, 32).
BLASTN analysis (Table 2), alignment (see Table S1 in the supplemental material), and phylogenies (Fig. 1) of the 16S rRNA sequence types obtained here confirmed that Sardinian ruminants are infected by different Anaplasma species. Some of these species were relatively frequent, such as A. ovis in sheep and goats, A. marginale in calves and goats, and A. bovis in sheep. On the contrary, A. phagocytophilum was only detected in a single sheep. This last result was unexpected, since this species was previously detected in Sardinian symptomatic horses, dogs, ticks, and humans (14, 27). Therefore, it can be postulated that in this geographic area ruminants are not relevant reservoirs for granulocytic anaplasmosis, but alternative vertebrate species can act as maintenance hosts. This could be also related to the scarce presence of Ixodes ticks in the island, representing only 0.3% of the total tick population. Two 16S rRNA sequence types detected in calves and in a goat shared 99% homology with the 16S rRNA sequence of the canine A. platys. Upon phylogenetic analyses, the two A. platys-like sequence types grouped with A. platys in a distinct subcluster closely related to a second subcluster composed by A. phagocytophilum and A. bovis (Fig. 1). Similar strains have already been reported in central and southern China (33), but they were never included in phylogenetic analyses. BLASTN analysis (Table 3) and phylogenies (Fig. 2) of the 25 unique groEL sequence types obtained in the present study confirmed the presence of distinct strains of A. marginale in sheep and calves and of A. ovis in goats. None of the 99 ruminants tested positive for A. phagocytophilum. These results confirm what was observed with the 16S rRNA gene and allow pointing out that, unlike previous observations made in other European countries, the implications of ruminants in the epidemiology of A. phagocytophilum are weak in this geographic area. Notably, a great proportion (10/25) of the groEL gene sequence types (Table 3; see also Table S2 in the supplemental material) identified in ruminants clustered with A. platys in phylogenetic trees, as already observed using 16S rRNA phylogenetic analyses. These results were also confirmed by network analyses (Fig. 4) and indicate that ruminants harbor a number of Anaplasma strains closely related to the canine, platelet-infecting A. platys species.
In situ hybridization experiments conducted on peripheral blood fractions (platelets, buffy coat, and erythrocytes) of goats PCR positive to A. platys with DNA probes labeled either with DIG or fluorochromes unexpectedly resulted in the absence of a specific signal in the platelet fraction but in a clear signal in neutrophil granulocytes (Fig. 3; see also Fig. S2 and S3 in the supplemental material). Confocal microscopy experiments pointed out that the signal was homogeneously distributed into the neutrophil granulocyte cytoplasm (Fig. 4). The genetic similarity of these new strains identified in ruminants with A. platys (92 to 93% groEL identity, 99% 16S rRNA identity), and their weaker relation to A. phagocytophilum (79 to 80% groEL identity) would indicate that ruminants host a number of strains belonging to the species A. platys. However, their peculiar host and cell tropism, which are reminiscent of A. phagocytophilum, suggest the assignment of the A. platys-like strains to a separate taxon. The presence of A. platys-like strains opens new concerns about the specificity of the direct and indirect diagnostic tests routinely used to detect different Anaplasma species in ruminants, which should be reconsidered on the basis of potential cross-reactivity, especially when coinfection is present. Also, the identification of A. platys-like strains provides additional background helpful for reconstructing the evolutionary history of the A. phagocytophilum cluster. Considering that the groEL sequence of A. platys is invariable in strains isolated in different regions of the world, one could postulate that this Anaplasma species originated recently. Most probably, the ancestor of A. platys had host and cellular tropism more similar to the one of A. phagocytophilum, which is able to infect both ruminant and carnivore neutrophil granulocytes. A. platys could have originated form A. phagocytophilum-like strains through host range specialization (from mixed host tropism to exclusively canine tropism) and through a shift in cellular tropism (from neutrophil granulocytes to platelets). Under this evolutionary scenario the A. platys-like strains found in ruminants could represent ancestral A. platys strains in which the original host and cellular tropism has been maintained. An open question that cannot be answered based on our data is the identification of factors that acted (and still act) as barriers generating and maintaining a genetic separation between A. phagocytophilum and the A. platys-like strains (79/80% groEL identity) in ruminants, since they share the same host and cell type. Possible causes could be related to the different tick vectors transmitting the two species (Ixodes ticks for A. phagocytophilum and Rhipicephalus for A. platys). In conclusion, considering the high prevalence of these tick vectors, more investigations are needed to assess the economic impact of the different Anaplasma species in Mediterranean ruminants and to develop more specific direct and indirect tools for Anaplasma sp. detection in wild and domestic species.
Supplementary Material
ACKNOWLEDGMENTS
Rosanna Zobba was supported by the Regione Autonoma della Sardegna through the program Promozione della Ricerca Scientifica e dell'Innovazione Tecnologica in Sardegna (LR 7/2007; PO Sardegna FSE 2007-2013).
Footnotes
Published ahead of print 25 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03129-13.
REFERENCES
- 1.Uilenberg G. 1997. General review of tick-borne diseases of sheep and goats worldwide. Parasitologia 39:161–165 [PubMed] [Google Scholar]
- 2.Bekker CP, de Vos S, Taoufik A, Sparagano OA, Jongejan F. 2002. Simultaneous detection of Anaplasma and Ehrlichia species in ruminants and detection of Ehrlichia ruminantium in Amblyomma variegatum ticks by reverse line blot hybridization. Vet. Microbiol. 89:223–238. 10.1016/S0378-1135(02)00179-7 [DOI] [PubMed] [Google Scholar]
- 3.Parola P, Davoust B, Raoult D. 2005. Tick- and flea-borne rickettsial emerging zoonoses. Vet. Res. 36:469–492. 10.1051/vetres:2005004 [DOI] [PubMed] [Google Scholar]
- 4.Parola P, Paddock CD, Raoult D. 2005. Tick-borne rickettsioses around the world: emerging diseases challenging old concepts. Clin. Microbiol. Rev. 18:719–756. 10.1128/CMR.18.4.719-756.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicholson WL, Allen KE, McQuiston JH, Breitschwerdt EB, Little SE. 2010. The increasing recognition of rickettsial pathogens in dogs and people. Trends Parasitol. 26:205–212. 10.1016/j.pt.2010.01.007 [DOI] [PubMed] [Google Scholar]
- 6.Sainz A, Amusategui I, Tesouro MA. 1999. Ehrlichia platys infection and disease in dogs in Spain. J. Vet. Diagn. Invest. 11:382–384. 10.1177/104063879901100419 [DOI] [PubMed] [Google Scholar]
- 7.Melendez RD. 2000. Future perspective on veterinary hemoparasite research in the tropic at the start of this century. Ann. N. Y. Acad. Sci. 916:253–258. 10.1111/j.1749-6632.2000.tb05297.x. [DOI] [PubMed] [Google Scholar]
- 8.Stuen S, Bergstrom K, Palmer E. 2002. Reduced weight gain due to subclinical Anaplasma phagocytophilum (formerly Ehrlichia phagocytophila) infection. Exp. Appl. Acarol. 28:209–215. 10.1023/A:1025350517733 [DOI] [PubMed] [Google Scholar]
- 9.Stuen S, Nevland S, Moum T. 2003. Fatal cases of tick-borne fever (TBF) in sheep caused by several 16S rRNA gene variants of Anaplasma phagocytophilum. Ann. N. Y. Acad. Sci. 990:433–434. 10.1111/j.1749-6632.2003.tb07407.x [DOI] [PubMed] [Google Scholar]
- 10.Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, description of six new species combinations and designation of Ehrlichia equi and HGE agent as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145–2165. 10.1099/00207713-51-6-2145 [DOI] [PubMed] [Google Scholar]
- 11.Inokuma H. 2007. Vectors and reservoir hosts of Anaplasmataceae, p 199–212 In Raoult D, Parola P. (ed), Rickettsial diseases. Taylor & Francis Group, LLC, New York, NY [Google Scholar]
- 12.Rar V, Golovljova I. 2011. Anaplasma, Ehrlichia, and “Candidatus Neoehrlichia” bacteria: pathogenicity, biodiversity, and molecular genetic characteristics, a review. Infect. Genet. Evol. 11:1842–1861. 10.1016/j.meegid.2011.09.019 [DOI] [PubMed] [Google Scholar]
- 13.Kawahara M, Rikihisa Y, Lin Q, Isogai E, Tahara K, Itagaki A, Hiramitsu Y, Tajima T. 2006. Novel genetic variants of Anaplasma phagocytophilum, Anaplasma bovis, Anaplasma centrale, and a novel Ehrlichia sp. in wild deer and ticks on two major islands in Japan. Appl. Environ. Microbiol. 72:1102–1109. 10.1128/AEM.72.2.1102-1109.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberti A, Zobba R, Chessa B, Addis MF, Sparagano O, Pinna Parpaglia ML, Cubeddu T, Pintori G, Pittau M. 2005. Equine and canine Anaplasma phagocytophilum strains isolated on the island of Sardinia (Italy) are phylogenetically related to pathogenic strains from the United States. Appl. Environ. Microbiol. 71:6418–6422. 10.1128/AEM.71.10.6418-6422.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alberti A, Addis MF, Sparagano O, Zobba R, Chessa B, Cubeddu T, Parpaglia ML, Ardu M, Pittau M. 2005. Anaplasma phagocytophilum, Sardinia, Italy. Emerg. Infect. Dis. 11:1322–1324. 10.3201/eid1108.050085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woldehiwet Z. 2006. Anaplasma phagocytophilum in ruminants in Europe. Ann. N. Y. Acad. Sci. 1078:446–460. 10.1196/annals.1374.084 [DOI] [PubMed] [Google Scholar]
- 17.Alberti A, Sparagano OA. 2006. Molecular diagnosis of granulocytic anaplasmosis and infectious cyclic thrombocytopenia by PCR-RFLP. Ann. N. Y. Acad. Sci. 1081:371–378. 10.1196/annals.1373.055 [DOI] [PubMed] [Google Scholar]
- 18.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 20.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. (Oxford) 41:95–98 [Google Scholar]
- 21.Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030–11035. 10.1073/pnas.0404206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S, Tamura K, Jakobsen IB, Nei M. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244–1245. 10.1093/bioinformatics/17.12.1244 [DOI] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- 25.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 26.Di Todaro N, Piazza C, Otranto D, Giangaspero A. 1999. Ticks infesting domestic animals in Italy: current acarological studies carried out in Sardinia and Basilicata regions. Parasitologia 41(Suppl):39–40 [PubMed] [Google Scholar]
- 27.Satta G, Chisu V, Cabras P, Fois F, Masala G. 2011. Pathogens and symbionts in ticks: a survey on tick species distribution and presence of tick-transmitted micro-organisms in Sardinia, Italy. J. Med. Microbiol. 60:63–68. 10.1099/jmm.0.021543-0 [DOI] [PubMed] [Google Scholar]
- 28.Kocan KM. 2003. Antigens and alternatives for control of Anaplasma marginale infection in cattle: U.S.A. and South America. Clin. Microbiol. Rev. 16:698–712. 10.1128/CMR.16.4.698-712.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kivaria FM. 2006. Estimated direct economic costs associated with tick-borne diseases on cattle in Tanzania. Trop. Anim. Health Prod. 38:291–299. 10.1007/s11250-006-4181-2 [DOI] [PubMed] [Google Scholar]
- 30.Minjauw B, McLeod A. 2003. Tick-borne diseases and poverty: impact of ticks and tick-borne diseases on the livelihood of small-scale and marginal livestock owners in India and eastern and southern Africa. DFID Animal Health Programme, Centre for Tropical Veterinary Medicine, University of Edinburgh, Edinburgh, United Kingdom: http://www.dfid.gov.uk/r4d/PDF/Outputs/RLAHTickBorn_Book.pdf [Google Scholar]
- 31.Dreher UM, de la Fuente J, Hofmann-Lehmann R, Meli ML, Pusterla N, Kocan KM, Woldehiwet Z, Braun U, Regula G, Staerk KDC, Lutz H. 2005. Serologic cross-reactivity between Anaplasma marginale and Anaplasma phagocytophilum. Clin. Diagn. Lab. Immunol. 12:1177–1183. 10.1128/CDLI.12.10.1177-1183.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Adhami B, Scandrett WB, Lobanov VA, Gajadhar AA. 2011. Serological cross-reactivity between Anaplasma marginale and an Ehrlichia species in naturally and experimentally infected cattle. J. Vet. Diagn. Invest. 23:1181–1188. 10.1177/1040638711425593 [DOI] [PubMed] [Google Scholar]
- 33.Liu Z, Ma M, Wang Z, Wang J, Peng Y, Li Y, Guan G, Luo J, Yin H. 2012. Molecular survey and genetic identification of Anaplasma species in goats from central and southern China. Appl. Environ. Microbiol. 78:464–470. 10.1128/AEM.06848-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.