Abstract
Diversity, abundance, and activity of ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA) were investigated using the ammonia monooxygenase α subunit (amoA) in the intertidal sediments of the Yangtze Estuary. Generally, AOB had a lower diversity of amoA genes than did AOA in this study. Clone library analysis revealed great spatial variations in both AOB and AOA communities along the estuary. The UniFrac distance matrix showed that all the AOB communities and 6 out of 7 AOA communities in the Yangtze Estuary were statistically indistinguishable between summer and winter. The studied AOB and AOA community structures were observed to correlate with environmental parameters, of which salinity, pH, ammonium, total phosphorus, and organic carbon had significant correlations with the composition and distribution of both communities. Also, the AOA communities were significantly correlated with sediment clay content. Quantitative PCR (qPCR) results indicated that the abundance of AOB amoA genes was greater than that of AOA amoA genes in 10 of the 14 samples analyzed in this study. Potential nitrification rates were significantly greater in summer than in winter and had a significant negative correlation with salinity. In addition, potential nitrification rates were correlated strongly only with archaeal amoA gene abundance and not with bacterial amoA gene abundance. However, no significant differences were observed between rates measured with and without ampicillin (AOB inhibitor). These results implied that archaea might play a more important role in mediating the oxidation of ammonia to nitrite in the Yangtze estuarine sediments.
INTRODUCTION
Over the past several decades, anthropogenic production of reactive nitrogen has increased by 120% (1), and global nitrogen overload has been identified as a main emerging environmental issue in this century (2, 3). Much of anthropogenic nitrogen is transported into estuarine and coastal regions via rivers, groundwater, and atmosphere (3, 4), which has already exerted a serious threat to the environmental quality of estuarine and coastal ecosystems. Nitrification, the sequential oxidation of ammonia to nitrite and then to nitrate, is an important bioremediation process in these nitrogen-enriched environments. Excessive nitrogen load in these ecosystems can be greatly reduced via a tight coupling between nitrification and denitrification or anaerobic ammonium oxidation (5, 6, 7), thereby diminishing the risk of nitrogen pollution. However, the removal of nitrogen from aquatic environments often depends directly on the supply of products (nitrite and nitrate) from nitrification (8). Thus, nitrification plays a critical biogeochemical role in the estuarine and coastal ecosystems.
As the first and rate-limiting step in nitrification, ammonia oxidation has been widely studied because of its ecological significance in the global nitrogen cycle and environmental implications (9). The process of ammonia oxidation was long recognized to be restricted to two groups of ammonia-oxidizing bacteria (AOB), the beta- and gammaproteobacteria (10). However, this view was challenged by the discovery of genes encoding proteins with homology to ammonia monooxygenases (amoA) in genome fragments of archaea which are affiliated with the phylum Thaumarchaeota (11, 12, 13). Since ammonia-oxidizing archaea (AOA) were discovered, they have been detected in various environments, including soils and sediments (12, 14, 15, 16, 17), estuaries (11, 18, 19, 20), subterranean environments (21), oxic and suboxic marine layers (22, 23, 24), sponges (25), corals (26), and wastewater sludge (27). So far, most studies have shown that a large proportion of the Thaumarchaeota are autotrophic and capable of performing the oxidation of ammonia to nitrite (15, 22), although several studies reported that some of these archaea may also be able to assimilate organic compounds, like amino acids (13, 28). In marine environments, AOA have been assumed to be more important contributors to ammonia oxidation than bacteria, based on molecular and biogeochemical data (29, 30). In contrast, in estuarine ecosystems, their relative importance in ammonia oxidation remains more ambiguous and uncertain (19, 20, 31, 32), which may imply a more complex interaction between dynamics of ammonia oxidizers and associated activity in these environments. Due to effects of land-sea interaction, estuaries and adjacent areas often experience tidal changes, salinity intrusions, and nutrient pulses, which may have important impacts on shaping dynamics of ammonia oxidizers. However, environmental variables that control diversity, distribution, and activity of ammonia oxidizers in these complex estuarine systems remain elusive.
The Yangtze River is the third-largest river in the world, and it delivers more than 7.5 × 1010 moles per year of N nutrients to the East China Sea through the estuarine areas (33). Especially in recent decades, the Yangtze Estuary has been receiving an increasing load of anthropogenic nitrogen from agricultural activities, fish farming, and domestic and industrial wastewater discharge, which has resulted in severely eutrophic status in the estuarine and coastal area (34, 35). Hence, the microbial nitrogen transformations are of major concern in the Yangtze Estuary. Although the microbial oxidation of ammonia to nitrite is known to play a central role in the nitrogen transformations, few studies have examined the dynamics of ammonia oxidizers and associated activity in the eutrophic estuarine environment. Thus, the relationships among physical/chemical variables, population structure, and activity of ammonia oxidizers still need to be further explored.
The objectives of the present study were (i) to elucidate the diversity and abundance of AOB (β-AOB) and AOA in the estuarine ecosystem based on the functional amoA gene marker, (ii) to investigate potential links of estuarine environmental variables with the dynamics of ammonia oxidizers, and (iii) to compare the relative contributions of AOB and AOA to nitrification at the study area.
MATERIALS AND METHODS
Field sampling.
Surface sediment samples were collected from 7 sites along the intertidal flats of the Yangtze Estuary (see Fig. S1 in the supplemental material), including Xupu (XP), Liuhekou (LHK), Wusongkou (WSK), Bailonggang (BLG), Daxingang (DXG), Yinyang (YY), and Luchao (LC). Fieldwork was conducted in January (winter) and August (summer) 2012. At each site, surface sediments (0 to 5 cm) were collected with stainless steel tubes from six to eight plots (50 cm by 50 cm). After collection, these sediment samples were stored in sterile plastic bags, sealed, and transported to the laboratory on ice within 4 h. Upon return to the laboratory, sediments from each site were immediately homogenized under a nitrogen atmosphere as one composite sample. Subsequently, one part of the composite sample from each site was stored at 4°C for nitrification rate measurements and sediment physiochemical analyses. The other part was preserved at −80°C for DNA extraction and subsequent molecular analysis.
Determination of environmental parameters.
At each sampling site, sediment temperature was determined in situ with a portable electronic thermometer. Sediment salinity and pH were measured using a YSI Model 30 salinity meter and a Mettler-Toledo pH meter, respectively, after sediments were mixed with deionized water free of CO2 at a ratio (sediment/water) of 1:2.5. Sediment water content was measured from weight loss of a known amount of wet sediment dried at 80°C to a constant value. A Beckman Coulter LS13320 laser granulometer (USA) was used for sediment particle analysis. Exchangeable ammonium (NH4+-N), nitrite (NO2−-N), and nitrate (NO3−-N) were extracted from fresh sediments with 2 M KCl and measured spectrophotometrically on a continuous-flow analyzer (SAN Plus, Skalar Analytical B.V., the Netherlands) with detection limits of 0.5 μM for NH4+-N and 0.1 μM for NO2−-N and NO3−-N (36). Organic carbon (OC) in sediments was measured using the K2Cr2O7 oxidation-reduction titration method. Total phosphorus (TP) in sediments was measured colorimetrically by the ascorbic acid-molybdate blue method (37) after 2 h of combustion (500°C) and 16 h of extraction with 1 M HCl.
DNA extraction and gene amplification.
Total DNA was extracted from sediment samples using Powersoil DNA isolation kits (MoBio, USA) according to the manufacturer's directions. AOB (β-AOB only) amoA gene fragments (491 bp) were amplified from the extracted DNA using the primer set composed of amoA-1F (5′-GGGGTTTCTACTGGTGGT-3′) and amoA-2R (5′-CCCCTCKGSAAAGCCTTCTTC-3′) (38). AOA amoA gene fragments (635 bp) were amplified using primers Arch-amoAF (5′-STAATGGTCTGGCTTAGACG-3′) and Arch-amoAR (5′-GCGGCCATCCATCTGTATGT-3′) (12). PCR was performed in a total volume of 50 μl containing 5 μl 10× PCR buffer (without MgCl2; Sangon, China), 4 μl MgCl2 (25 mM; Sangon), 1 μl deoxynucleoside triphosphate (dNTP) (each 10 mM; Sangon), 1 μl each primer (10 μM; Sangon), 1 μl Taq DNA polymerase (5 U μl−1; Sangon), and 1 μl template DNA. PCRs were carried out with 5 min at 95°C; 30 cycles of 94°C for 30 s, 55°C (for AOB) or 56°C (for AOA) for 45 s, and 72°C for 1 min; and a final 5-min extension cycle at 72°C. Appropriately sized fragments were separated by electrophoresis in 1% agarose gels and purified using the Gel Advance gel extraction system (Viogene, China). The purified fragments were cloned using the TOPO-TA cloning kit (Invitrogen, USA) in accordance with the manufacturer's instructions. Clones were randomly selected for further analysis.
Sequencing and phylogenetic analysis.
Screened clones were sequenced using an ABI Prism genetic analyzer (Applied Biosystems, Canada) in combination with a Big Dye Terminator kit (Applied Biosystems, Canada). The amoA gene sequences were edited using the DNAstar software package (DNAstar, USA). Possible chimeras were checked using the CHECK CHIMERA program of the Ribosomal Database Project (39). amoA gene sequences were analyzed initially using the BLASTn tool (http://www.ncbi.nlm.nih.gov/BLAST/) to aid the selection of the closest reference sequences. All of the sequences and their closest relative matches obtained from the NCBI BLAST were aligned using the ClustalX program (version 2.1) (40). The sequences displaying more than 95% identity with each other were grouped into one operational taxonomic unit (OTU) using Mothur software (http://www.mothur.org/wiki/Main_Page) by the furthest neighbor approach (41). Neighbor-joining phylogenetic trees were created using MEGA software (version 5.03) (42). The relative confidence of the tree topologies was evaluated by performing 1,000 bootstrap replicates (43).
Real-time qPCR.
Plasmids constructed in this study carrying a bacterial or archaeal amoA gene fragment were extracted from Escherichia coli hosts using a plasmid minipreparation kit (Tiangen, China) for use in standard curves. Concentrations of plasmid DNA were measured using a Nanodrop-2000 spectrophotometer (Thermo, USA). The standard curves spanned a range from 5.12 × 101 to 5.12 × 106 copies per μl for the AOB assay and from 2.64 × 101 to 2.64 × 106 copies per μl for the AOA assay. All samples were used and standard reactions were performed in triplicate with an ABI 7500 sequence detection system (Applied Biosystems, Canada) using the SYBR green quantitative PCR (qPCR) method. The primer set composed of amoA-1F and amoA-2R was used for the amplification of the AOB amoA gene. Arch-amoAF and Arch-amoAR were used to amplify the AOA amoA gene. The 25-μl qPCR mixture contained 12.5 μl of Maxima SYBR green/Rox qPCR master mix (Fermentas, Lithuania), 1 μl of each primer (10 μM), and 1 μl template DNA. All reactions were performed in 8-strip thin-well PCR tubes with ultraclean cap strips (ABgene, United Kingdom). The specificity of the qPCR amplification was determined by the melting curve and by gel electrophoresis. The PCR protocols were performed as follows: 50°C for 2 min and 95°C for 10 min, followed by 45 cycles of 30 s at 95°C, 40 s at 58°C (for AOB) and 56°C (for AOA), and 1 min at 72°C. In all experiments, negative controls containing no-template DNA were subjected to the same qPCR procedure to detect and exclude any possible contamination.
The consistency of the real-time quantitative PCR (qPCR) assay was confirmed by the strong linear inverse relationship between the threshold cycle (CT) and the log value of gene copy number for both primer sets (r2 = 0.996 for AOB and 0.989 for AOA). The amplification efficiencies were 91.1% and 99.1% for AOB and AOA, respectively. In addition, melting curve analyses showed only one observable peak at a melting temperature (Tm = 82.86 for AOB and 81.43 for AOA), and no detectable peaks associated with primer-dimer artifacts or other nonspecific PCR amplification products were observed. The gene abundances of each reaction were calculated based on the constructed standard curves and then converted to copies per gram of sediment, assuming 100% DNA extraction efficiency.
Potential nitrification rates.
Potential nitrification rates were estimated in triplicate based on the method of Bernhard et al. (44). Briefly, 1.0 g (wet weight) of sediment was added to 100-ml Erlenmeyer flasks containing 30 ml of artificial seawater with in situ salinity. NH4Cl (for ammonium) and KH2PO4 (for phosphate) were amended to the flasks with final concentrations of 300 μM and 60 μM, respectively. The suspension was incubated in the dark at near-in situ temperature with continuous shaking at 120 rpm. Within the incubations, subsamples were harvested at 0 h, 24 h, 48 h, and 72 h, respectively. Subsequently, they were centrifuged, filtered, and immediately frozen for the analysis of nitrate (plus nitrite) by the spectrophotometric method (36). Although nitrite concentrations over the nitrification incubation were negligible because nitrite can be quickly oxidized to nitrate by nitrite-oxidizing bacteria, the potential nitrification rates were calculated based on the changes in nitrate (plus nitrite) concentrations with time (44).
In a parallel experiment, an additional 1 g liter−1 ampicillin (final concentrations) was added to the flasks to separate the rates of bacterial and archaeal ammonia oxidation (45, 46). Ampicillin is a beta-lactam antibiotic and inhibits only bacteria (not archaea), as it targets cell wall production during cell growth in both Gram-positive and Gram-negative bacteria by acting as a competitive inhibitor of transpeptidase (46).
Statistical analysis.
The Mothur program was used to generate rarefaction curves for the observed unique OTUs, to form the heat maps of amoA genes based on the representative OTUs of each gene library, and to determine the species richness Chao1 estimator and diversity indices (Shannon-Weiner and Simpson) (41). The coverage of clone libraries was estimated by the percentage of the observed number of OTUs divided by Chao1 estimate (25). Correlations between community structures and environmental variables were explored with canonical correspondence analysis (CCA; the maximum gradient length was greater than 4 standard deviations [SD] for both AOB and AOA based on the detrended correspondence analysis, showing that the responses of OTUs to environmental variables were unimodal) using the software Canoco (version 4.5) (47). Community classifications of the sediment AOB and AOA were performed using the principal coordinate analysis (PCoA) (http://bmf.colorado.edu/unifrac/index.psp) (48, 49). The distances between each pair of gene libraries were determined by the UniFrac matrix, and all P values have been corrected for multiple comparisons by multiplying the calculated P value by the number of comparisons made (Bonferroni correction) (48, 49).
Nucleotide sequence accession numbers.
The bacterial and archaeal amoA sequences reported in this study (only the unique ones) have been deposited in GenBank under accession numbers KC735601 to KC736063 and KC735178 to KC735600, respectively (see Table S1 in the supplemental material).
RESULTS
Site characteristics.
In the Yangtze Estuary, sediment temperature in winter was about 2.4 to 3.7°C, whereas it increased up to 29.1 to 33.5°C in summer (Table 1). Salinity was generally low (0.7 to 1.5 practical salinity units [PSU]) in the upper estuary while it was relatively high (6.2 to 14.8 PSU) in the lower estuary. Sediment pH varied from 7.08 to 8.44 at the study area, with higher pH values at the high-salinity sites (8.16 to 8.44) than at the low-salinity sites (7.08 to 7.91). Sediment water contents were in the range of 21 to 31% with little spatial and seasonal variation. Sediments at most of the sampling sites were mainly composed of clay and silt with smaller amounts of sands. They were generally characterized by a high content of fine fractions with a grain size of <63 μm. Higher concentrations of organic carbon were detected at the low-salinity habitats (0.81 to 1.89%) than at the high-salinity sites (0.12 to 0.76%) (dry weight, similarly hereinafter). The concentrations of total phosphorus in sediments ranged from 639.2 to 1,095.0 μg g−1 with high spatial and temporal heterogeneity. The concentration of ammonium in sediments ranged from 17.5 to 117.2 μg g−1 with higher concentrations in summer than in winter. The concentrations of nitrate were in the range of 12.3 to 29.1 μg g−1, which positively correlated with ammonium concentrations (r = 0.539, P = 0.047, n = 14). Compared with ammonium and nitrate, nitrite had relatively low concentrations at the sampling sites, with values of 0.04 to 0.66 μg g−1.
TABLE 1.
Physicochemical variables of the sediment samples
| Season and sample | Mean ± SDb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Temp (°C) | Salinity (PSU) | pH | Moisture content (%) | Clay content (%) | Silt content (%) | Sand content (%) | OC (%)a | TP (μg g−1)a | NH4 (μg g−1)a | NO2 (μg g−1)a | NO3 (μg g−1)a | |
| Summer | ||||||||||||
| XP | 29.1 ± 0.3 | 0.9 ± 0.1 | 7.08 ± 0.00 | 21 ± 2 | 24.9 ± 2.1 | 67.2 ± 3.6 | 7.9 ± 0.5 | 1.58 ± 0.09 | 685.8 ± 17.2 | 79.2 ± 4.2 | 0.07 ± 0.01 | 16.4 ± 1.0 |
| LHK | 32.3 ± 0.2 | 1.0 ± 0.1 | 7.42 ± 0.03 | 28 ± 4 | 19.5 ± 2.3 | 66.8 ± 4.8 | 13.7 ± 1.3 | 1.58 ± 0.08 | 987.8 ± 37.5 | 85.4 ± 1.9 | 0.13 ± 0.08 | 20.9 ± 2.6 |
| WSK | 31.2 ± 0.2 | 0.7 ± 0.1 | 7.47 ± 0.04 | 25 ± 3 | 12.3 ± 1.9 | 67.7 ± 5.1 | 20.0 ± 1.9 | 1.89 ± 0.11 | 865.3 ± 33.6 | 117.2 ± 3.5 | 0.09 ± 0.00 | 17.9 ± 4.5 |
| BLG | 32.7 ± 0.5 | 1.5 ± 0.1 | 7.08 ± 0.03 | 26 ± 4 | 24.5 ± 2.2 | 74.6 ± 6.6 | 0.9 ± 0.2 | 1.14 ± 0.04 | 830.2 ± 0.8 | 46.8 ± 5.8 | 0.07 ± 0.01 | 18.9 ± 6.6 |
| DXG | 33.2 ± 0.6 | 0.8 ± 0.1 | 7.91 ± 0.04 | 34 ± 6 | 17.5 ± 1.3 | 69.1 ± 6.0 | 13.4 ± 1.5 | 1.08 ± 0.04 | 815.1 ± 47.5 | 31.7 ± 2.0 | 0.07 ± 0.04 | 14.9 ± 0.4 |
| YY | 32.4 ± 0.4 | 13.2 ± 0.1 | 8.17 ± 0.04 | 30 ± 5 | 10.0 ± 0.8 | 18.7 ± 2.7 | 71.3 ± 4.9 | 0.42 ± 0.01 | 731.1 ± 71.6 | 47.7 ± 6.0 | 0.07 ± 0.00 | 17.0 ± 0.6 |
| LC | 33.5 ± 0.3 | 6.2 ± 0.6 | 8.34 ± 0.01 | 23 ± 3 | 9.2 ± 0.8 | 77.5 ± 5.8 | 13.3 ± 1.3 | 0.12 ± 0.01 | 699.4 ± 17.7 | 24.7 ± 0.3 | 0.66 ± 0.04 | 14.1 ± 2.5 |
| Winter | ||||||||||||
| XP | 3.1 ± 0.2 | 1.0 ± 0.1 | 7.10 ± 0.01 | 25 ± 3 | 27.2 ± 1.9 | 69.3 ± 5.7 | 3.5 ± 0.2 | 0.81 ± 0.04 | 1095.0 ± 8.5 | 77.7 ± 5.6 | 0.08 ± 0.04 | 21.5 ± 3.3 |
| LHK | 3.6 ± 0.3 | 0.7 ± 0.1 | 7.36 ± 0.01 | 27 ± 4 | 18.4 ± 1.6 | 70.2 ± 6.8 | 11.4 ± 0.1 | 1.11 ± 0.03 | 943.6 ± 68.4 | 34.5 ± 1.8 | 0.08 ± 0.01 | 17.7 ± 3.2 |
| WSK | 3.7 ± 0.3 | 1.3 ± 0.1 | 7.46 ± 0.01 | 26 ± 3 | 21.9 ± 0.9 | 74.6 ± 7.1 | 3.5 ± 0.3 | 1.51 ± 0.09 | 921.9 ± 33.1 | 90.0 ± 2.0 | 0.09 ± 0.01 | 29.1 ± 4.5 |
| BLG | 2.9 ± 0.1 | 1.3 ± 0.2 | 7.62 ± 0.04 | 28 ± 2 | 25.4 ± 1.7 | 73.2 ± 6.9 | 1.4 ± 0.1 | 1.0 ± 0.06 | 781.5 ± 70.8 | 93.8 ± 0.4 | 0.10 ± 0.01 | 26.1 ± 4.3 |
| DXG | 2.5 ± 0.4 | 8.7 ± 0.2 | 8.36 ± 0.02 | 32 ± 5 | 13.6 ± 1.1 | 59.5 ± 4.5 | 26.9 ± 1.3 | 0.51 ± 0.01 | 678.7 ± 55.3 | 22.0 ± 2.1 | 0.11 ± 0.00 | 24.6 ± 2.3 |
| YY | 3.2 ± 0.3 | 14.8 ± 0.2 | 8.44 ± 0.02 | 28 ± 4 | 7.8 ± 0.6 | 16.2 ± 2.1 | 76.0 ± 5.6 | 0.59 ± 0.01 | 1063.5 ± 38.5 | 17.5 ± 2.5 | 0.04 ± 0.01 | 12.8 ± 3.6 |
| LC | 2.4 ± 0.1 | 13.5 ± 0.2 | 8.16 ± 0.01 | 22 ± 1 | 25.4 ± 1.3 | 73.5 ± 7.1 | 1.1 ± 0.2 | 0.76 ± 0.01 | 639.2 ± 10.6 | 23.2 ± 1.5 | 0.26 ± 0.01 | 12.3 ± 3.5 |
Dry weight.
Triplicate samples were analyzed to get means and standard deviations.
Phylogenetic diversity of bacterial and archaeal amoA gene sequences.
Both bacterial and archaeal amoA gene segments were amplified successfully in intertidal sediments of the Yangtze Estuary (see Fig. S2 in the supplemental material). A total of 1,200 bacterial and 1,329 archaeal amoA clones were sequenced from the 28 constructed clone libraries (Table 2). In each individual clone library, 6 to 18 AOB and 6 to 20 AOA OTUs occurred, as defined by <5% divergence in nucleotides (see Fig. S3). The diversity of the entire set of bacterial amoA genes was lower than that of archaeal amoA genes based on the values of the Shannon-Wiener index and the reciprocal of Simpson index (see Table S2), and this was consistent with the result of the rarefaction analysis (see Fig. S4). Generally, AOB had higher diversity in summer than in winter whereas the diversity of AOA was greater in winter as indicated by the measured indices (see Table S2). The estimated coverage values were between 90.9% and 100%, indicating that more than 90% of the amoA gene diversity was captured in all the libraries, and this was further confirmed by the gradually flattening rarefaction curves (see Fig. S4).
TABLE 2.
Diversity characteristics of clone libraries of AOB and AOA
| amoA gene and sample | Summer/winter value |
|||||
|---|---|---|---|---|---|---|
| No. of clones | OTUsa | Chao1b | Shannonc | 1/Simpsond | Coverage (%)e | |
| AOB | ||||||
| XP | 80/73 | 12/13 | 12.3/13.4 | 1.93/1.90 | 4.95/4.57 | 98.0/96.8 |
| LHK | 92/81 | 16/17 | 16.2/17.3 | 2.31/2.23 | 7.06/6.34 | 98.8/98.3 |
| WSK | 81/66 | 8/11 | 8.0/11.2 | 1.42/1.78 | 3.07/4.03 | 100.0/98.2 |
| BLG | 70/99 | 11/13 | 11.2/13.4 | 1.72/1.39 | 3.67/2.20 | 98.2/96.8 |
| DXG | 93/88 | 16/11 | 16.6/11.3 | 2.11/1.96 | 4.83/6.07 | 96.4/97.8 |
| YY | 95/86 | 18/12 | 18.4/12.3 | 2.49/2.06 | 10.19/6.19 | 98.0/97.3 |
| LC | 96/100 | 6/14 | 6.0/14.3 | 1.62/2.25 | 4.69/7.53 | 100.0/97.7 |
| AOA | ||||||
| XP | 104/83 | 7/16 | 7.5/16.4 | 1.42/2.16 | 3.67/5.79 | 93.3/97.7 |
| LHK | 101/98 | 6/13 | 6.0/13.5 | 1.37/2.20 | 3.31/7.38 | 100.0/96.3 |
| WSK | 101/80 | 15/13 | 15.6/13.3 | 2.17/2.32 | 6.53/10.03 | 96.2/98.1 |
| BLG | 94/79 | 10/15 | 11.0/15.5 | 1.89/2.29 | 5.42/8.15 | 90.9/96.8 |
| DXG | 101/101 | 11/20 | 11.8/20.5 | 1.74/2.77 | 4.64/15.93 | 93.6/97.6 |
| YY | 102/100 | 10/12 | 10.5/12.2 | 1.91/1.84 | 6.09/4.43 | 95.2/98.4 |
| LC | 88/97 | 13/15 | 14.2/15.5 | 1.83/1.99 | 4.08/4.94 | 91.5/96.8 |
OTUs are defined at 5% nucleotide acid divergence.
Nonparametric statistical predictions of total richness of OTUs based on distribution of singletons and doubles.
Shannon diversity index. A higher number represents more diversity.
Reciprocal of Simpson's diversity index. A higher number represents more diversity.
Percentage of coverage: percentage of observed number of OTUs divided by Chao1 estimate.
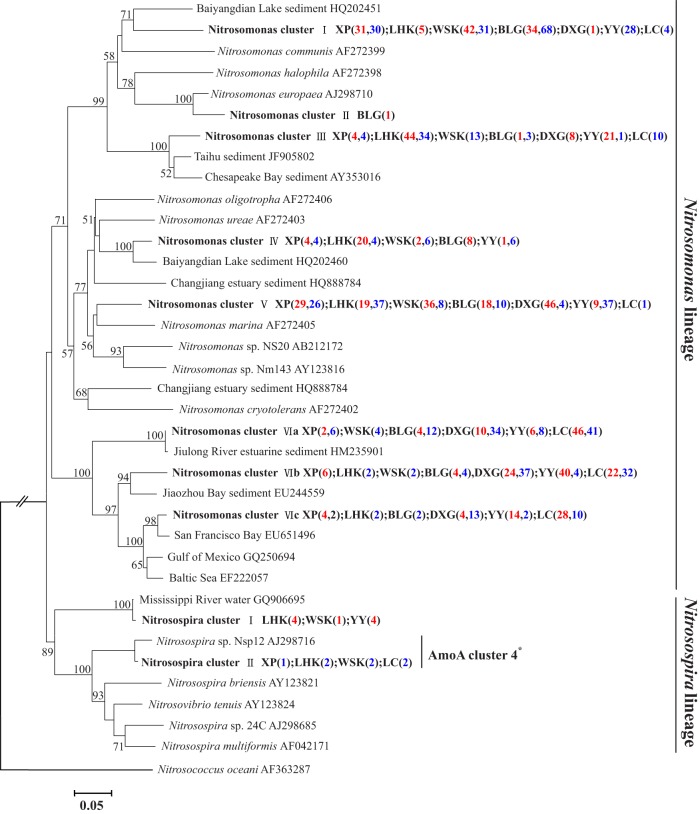
Phylogenetic analysis indicated that the bacterial amoA sequences acquired in this study were grouped with known AOB sequences in the Nitrosomonas and Nitrosospira genera (Fig. 1). The 6 Nitrosomonas-related clusters included 98.7% of the total AOB sequences and grouped with sequences in the lineages Nitrosomonas communis (AF272399), N. halophila (AF272398), N. europaea (AJ298710), N. oligotropha (AF272406), N. ureae (AF272403), N. marina (AF272405), and N. cryotolerans (AF272402) (10, 50). The first 5 Nitrosomonas clusters (Nitrosomonas clusters I to V) were the dominant group at the lower-salinity (0.7 to 1.5 psu) habitats of the Yangtze Estuary, accounting for 59.1% to 98.8% of the AOB communities. In the higher-salinity (6.2- to 14.8-PSU) habitats, however, the AOB communities were dominated by the phylogenetically distinct Nitrosomonas cluster VI (accounting for 63.2% to 100%, except winter YY), which was most closely related to Nitrosomonas sp. strain Nm143 (AY123816)/Nitrosomonas sp. strain NS20 (AB212172) (51, 52) and N. cryotolerans (AF272402) (with 73.1 to 77.6% sequence identity) (10). Only 1.3% of the total AOB sequences were clustered with Nitrosospira, which occupied 0% to 4.3% in all the samples analyzed in this study. The Nitrosospira cluster I was not grouped with any cultured AOB sequences but an environmental amoA sequence from Mississippi River water (GQ906695), while cluster II was grouped with Nitrosospira sp. strain Nsp12 (AJ298716) (50), and it was also named “Nitrosospira cluster 4” based on the nomenclature of Avrahami and Conrad (53).
FIG 1.
Neighbor-joining phylogenetic tree of bacterial amoA genes. The phylogeny is based on nucleotide sequences. Bootstrap values greater than 50% of 1,000 resamplings are shown near nodes. The scale indicates the number of nucleotide substitutions per site. GenBank accession numbers are shown for sequences from other studies. Numbers in parentheses following each cluster indicate the number of sequences recovered from each sampling site in summer (red) and winter (blue). The asterisk indicates the Nitrosospira clusters based on the nomenclature of Avrahami and Conrad (53). Representative accession numbers for each cluster and the relative abundance of clones from each site in each of the clusters are shown in Table S3 in the supplemental material.
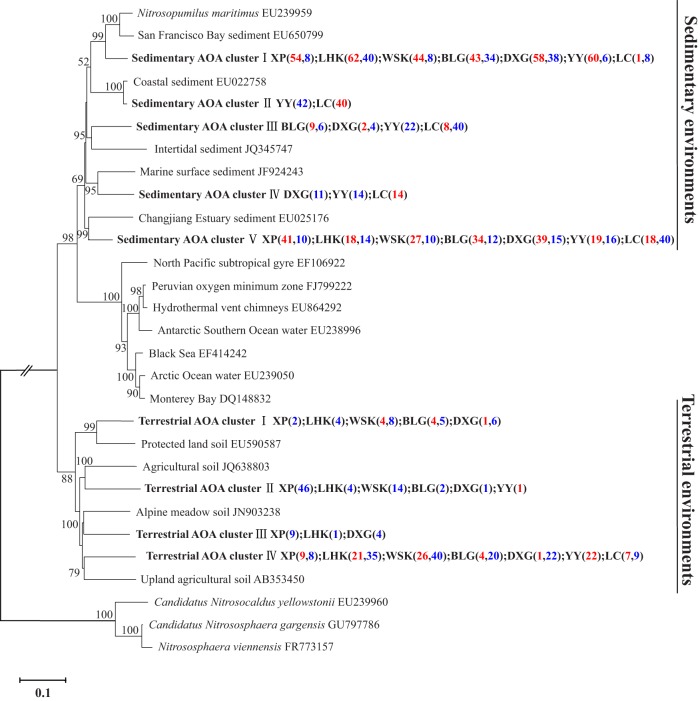
Phylogenetic analysis showed that the archaeal amoA sequences obtained in this study were clustered into two major evolutionary branches: a sedimentary cluster (marine group 1.1a) and a terrestrial cluster (soil group 1.1b) (Fig. 2). The two groups were distinct from each other, with any two sequences in these genera sharing only 69.0 to 77.8% identity at the nucleotide level. Of the total archaeal amoA sequences, 74.4% were affiliated with the marine group 1.1a AOA lineage and clustered with sequences recovered from estuary, intertidal, coastal, and marine sediments (EU025176, JQ345747, EU022758, and JF924243) (18, 21, 54). Sequences associated with terrestrial environments were affiliated with sequences mainly from protected land soil (EU590587), agricultural soil (JQ638803 and AB353450), and alpine meadow soil (JN903238) (55). The percentage of sequences in the soil group 1.1b decreased from the upper to the lower reaches of the Yangtze Estuary.
FIG 2.
Neighbor-joining phylogenetic tree of archaeal amoA genes. The phylogeny is based on nucleotide sequences. Bootstrap values greater than 50% of 1,000 resamplings are shown near nodes. The scale indicates the number of nucleotide substitutions per site. GenBank accession numbers are shown for sequences from other studies. Numbers in parentheses following each cluster indicate the number of sequences recovered from each sampling site in summer (red) and winter (blue). Representative accession numbers for each cluster and the relative abundance of clones from each site in each of the clusters are shown in Table S3 in the supplemental material.
Spatial and seasonal variations of AOB and AOA community structures.
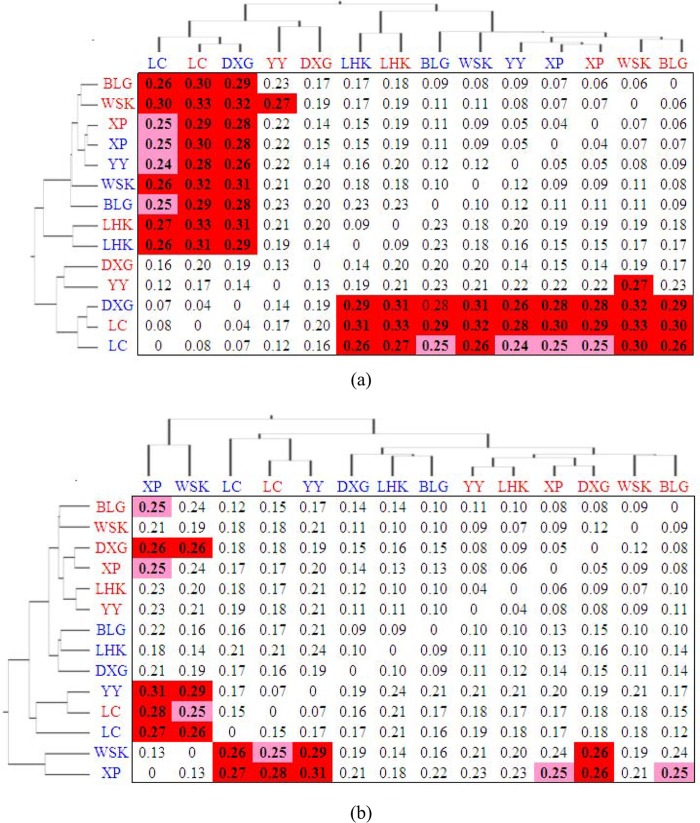
The observed variations in community composition were statistically compared using the UniFrac distance matrix (Fig. 3). Results showed that bacterial amoA clone libraries acquired from the lower-salinity sites (XP, LHK, WSK, and BLG) were significantly different from communities obtained from higher-salinity sites (LC and winter DXG) (P < 0.05) (Fig. 3a), and this was further confirmed by the PCoA (see Fig. S5a in the supplemental material). Also, the predominant groups between the lower- and higher-salinity sites were different based on the bacterial phylogenetic analysis (Fig. 1). Spatial variations of AOA communities were also observed in the sediments of the Yangtze Estuary. Archaeal amoA clone libraries acquired from the higher-salinity habitats (LC and winter YY) were significantly different from communities obtained from the lower-salinity sites (winter XP and winter WSK) (P < 0.05) (Fig. 3b). In addition, phylogenetic analysis revealed that the soil group 1.1b AOA lineage changed greatly along the salinity gradient of the Yangtze Estuary (Fig. 2).
FIG 3.
The UniFrac distance matrix of bacterial (a) and archaeal (b) amoA gene clone libraries. The summer samples are in red font, while the winter samples are in blue. The values show the UniFrac distances between each pair of clone libraries. Boldface values indicate that the libraries are drawn from significantly different communities (red background, P < 0.01; pink background, P < 0.05). All P values have been corrected for multiple comparisons by multiplying the calculated P value by the number of comparisons made (Bonferroni correction). The dendrograms represent clustering of the 14 clone libraries based on the distance matrix.
Based on the distance matrix, bacterial amoA gene libraries at all 7 sampling sites were statistically indistinguishable between summer and winter (P > 0.05) (Fig. 3a). This lack of statistically significant differences between summer and winter libraries suggests that AOB community compositions were stable in the sediments of the Yangtze Estuary. Only at site XP were the summer and winter archaeal amoA gene libraries significantly different from each other (P < 0.05) (Fig. 3b; see also Fig. S5b in the supplemental material).
Bacterial and archaeal amoA gene abundance.
The qPCR results indicated that the average abundance of bacterial amoA genes (8.17 × 105 copies g−1 dry sediment) was greater than that of archaeal amoA genes (1.72 × 105 copies g−1 dry sediment) in the sediments of the Yangtze Estuary (see Fig. S6a in the supplemental material). Specifically, 10 out of 14 samples analyzed in this study (except LHK, summer BLG, and winter WSK) showed higher AOB amoA gene abundance (7.36 × 104 to 5.20 × 106 copies g−1 dry sediment) than AOA amoA gene abundance (5.70 × 104 to 6.84 × 105 copies g−1 dry sediment) (Fig. 4; see also Table S4). The qPCR results also showed great heterogeneous distribution of the sediment bacterial amoA gene abundance among the sampling sites (Fig. 4). The highest number of copies (5.20 × 106 copies g−1 dry sediment) of bacterial amoA genes was detected at site XP in summer, whereas the lowest gene copy number (7.36 × 104 copies g−1 dry sediment) was recorded at site WSK in winter. However, no significant seasonal variation was observed at the study area (Student's t test, P > 0.05), with average bacterial amoA gene abundances of 1.04 × 106 and 5.91 × 105 copies per gram of dry sediment in summer and winter, respectively (see Fig. S6b). Sediment archaeal amoA gene copy numbers also showed great spatial heterogeneity in the Yangtze Estuary. Generally, the lower-salinity sites had a higher archaeal amoA gene abundance (8.11 × 104 to 6.84 × 105 copies g−1 dry sediment) than did the higher-salinity areas (5.70 × 104 to 6.78 × 104 copies g−1 dry sediment) (Fig. 4). Similarly, no significant difference in the abundance of archaeal amoA gene occurred between summer (2.01 × 105 copies g−1 dry sediment) and winter (1.43 × 105 copies g−1 dry sediment) (Student's t test, P > 0.05) (see Fig. S6c).
FIG 4.
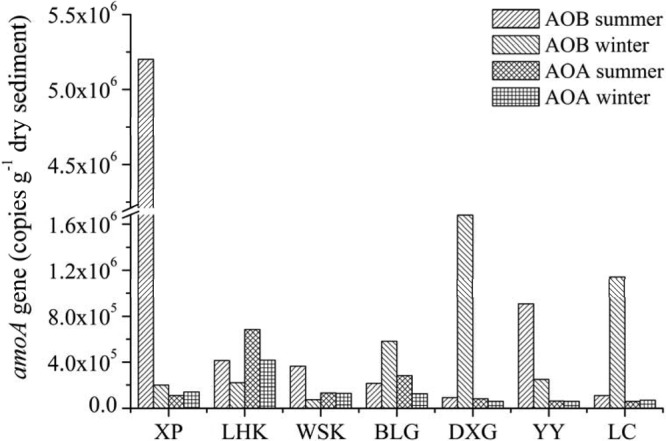
Bacterial and archaeal amoA gene copy numbers. The mean values of amoA copy numbers were obtained from triplicate real-time quantitative PCR assays.
Relationships of community structure and abundance with environmental variables.
Correlations of the AOB community structure with environmental parameters were analyzed via canonical correspondence analysis (CCA) (Fig. 5a). The environmental variables in the first two CCA dimensions explained 31.0% of the total variance in the AOB composition and 42.0% of the cumulative variance of the genotype-environment relationship. The results indicated that AOB community structures in the sediments of the Yangtze Estuary correlated with environmental parameters, including pH (P = 0.008, F = 1.72, 499 Monte Carlo permutations), total phosphorus (P = 0.012, F = 1.72, 499 Monte Carlo permutations), organic carbon (P = 0.016, F = 1.8, 499 Monte Carlo permutations), ammonium-N (P = 0.024, F = 1.61, 499 Monte Carlo permutations), and salinity (P = 0.046, F = 1.36, 499 Monte Carlo permutations). These factors had significant correlation with the composition and distribution of the communities, and they provided 41.5% of the total CCA explanatory power. Although the contribution of all other measured environmental factors (temperature, grain size, nitrite-N, and nitrate-N) was not statistically significant (P > 0.082, 499 Monte Carlo permutations), the combination of these variables provided additionally 32.5% of the total CCA explanatory power.
FIG 5.
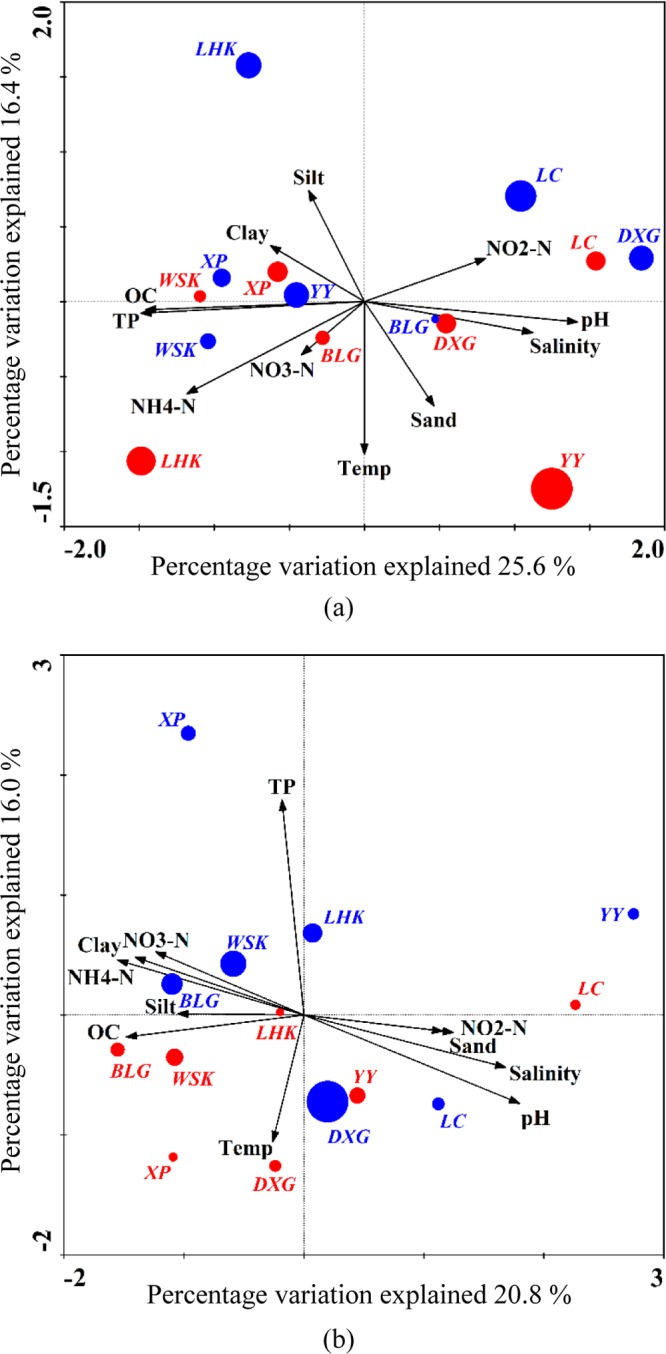
CCA ordination plots for the first two principal dimensions of the relationship between the AOB (a) and AOA (b) community compositions with the environmental parameters. The red circles represent the sampling stations in summer, and the blue circles represent the sampling stations in winter. The size of the circles corresponds to the Shannon diversity index in individual samples. TP, Temp, OC, NH4-N, NO3-N, and NO2-N represent total phosphorus, temperature, organic carbon, ammonium, nitrate, and nitrite, respectively.
Relationship studies between AOA communities and environmental parameters were also performed (Fig. 5b). The first two axes explained 29.5% of the total variance in the archaeal amoA genotype composition and 36.8% of the cumulative variance of the archaeal amoA-environment relationship. Results showed that the AOA communities correlated significantly with pH (P = 0.002, F = 1.96, 499 Monte Carlo permutations), clay content (P = 0.016, F = 1.58, 499 Monte Carlo permutations), ammonium-N (P = 0.018, F = 1.62, 499 Monte Carlo permutations), salinity (P = 0.026, F = 1.74, 499 Monte Carlo permutations), organic carbon (P = 0.026, F = 1.57, 499 Monte Carlo permutations), and total phosphorus (P = 0.046, F = 1.47, 499 Monte Carlo permutations), and these factors provided 51.5% of the total CCA explanatory power. Although the contributions of all other measured environmental factors (temperature, nitrite-N, nitrate-N, silt content, and sand content) were not statistically significant (P > 0.07, 499 Monte Carlo permutations), the combination of these variables provided additionally 28.7% of the total CCA explanatory power.
We also investigated the correlations of amoA gene abundance with environmental variables. In the present study, no significant correlations were observed between the bacterial amoA gene abundance and the environmental factors (P > 0.05, n = 14). However, the abundance of the Nitrosospira-related amoA gene was positively correlated with salinity (r = 0.487, P = 0.039, n = 14), while the amoA gene abundance of Nitrosomonas cluster VI was negatively correlated with total phosphorus (r = −0.673, P = 0.008, n = 14). Although relationships between archaeal amoA gene abundance and the environmental variables were not significant, the abundance of the soil group 1.1b was significantly correlated with salinity (r = −0.509, P = 0.032, n = 14), total phosphorus (r = 0.593, P = 0.025, n = 14), and pH (r = −0.534, P = 0.049, n = 14). Additionally, in the intertidal sediments of the Yangtze Estuary, the ratio of AOB to AOA amoA copy numbers was found to correlate with total phosphorus (r = −0.596, P = 0.025, n = 14).
Potential nitrification rates.
Potential nitrification rates in the intertidal sediments of the Yangtze Estuary were significantly higher in summer (0.10 to 4.48 μmol N g−1 day−1) than in winter (0.07 to 0.74 μmol N g−1 day−1) (Student's t test, t = 3.1, df = 12, P = 0.009) (Fig. 6; see also Fig. S6d in the supplemental material). No significant differences were observed between incubations with and without the ampicillin treatments (Student's t test, P > 0.05, decrease by only 13.1 to 26.6%) (Fig. 6). Strong negative correlations were found between the potential nitrification rates and salinity (r = −0.550, P = 0.042, n = 14). Furthermore, significant linear relationships were observed between potential nitrification rates and archaeal amoA gene abundance (r = 0.700, P = 0.005, n = 14), and the relationships were also significant in summer (r = 0.843, P = 0.017, n = 7) and winter (r = 0.909, P = 0.005, n = 7), respectively (see Table S5). Interestingly, potential nitrification rates were not related to the abundance of soil group 1.1b (r = 0.199, P = 0.494, n = 14) but were significantly correlated with the abundance of marine group 1.1a (r = 0.810, P = 0.000, n = 14). Meanwhile, no significant correlations were found between the bacterial amoA gene abundance and potential nitrification rates in this study (P > 0.05, n = 14) (see Table S5).
FIG 6.
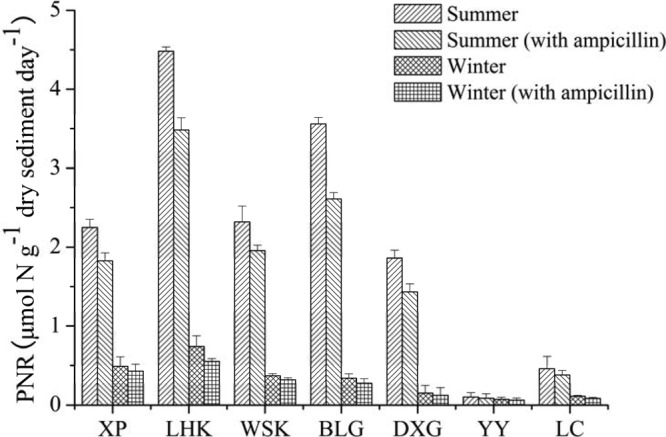
Potential nitrification rates (PNR) in the intertidal sediments of the Yangtze Estuary incubated with and without ampicillin. The error bars are standard deviations of triplicate incubations. Ampicillin at 1 g liter−1 was used as an antibiotic to inhibit ammonia-oxidizing bacteria.
DISCUSSION
In the present work, diversity, abundance, and activity of aerobic ammonia oxidizers were measured in the intertidal sediments of the Yangtze Estuary to provide insights into the microbial mechanisms driving nitrification in estuarine environments. Diversity estimators of the clone libraries in this study were within the same range previously reported at other environmental ecosystems (16, 19, 20, 21, 56, 57). Also, AOB generally had a lower diversity of amoA genes than did AOA in the intertidal sediments of the Yangtze Estuary, which was consistent with the results from other estuarine and coastal environments (16, 19, 20, 21).
The bacterial amoA sequences recovered from the intertidal sediments of the Yangtze Estuary showed substantial similarity to those retrieved in Baiyangdian Lake, Taihu Lake, Jiulong River, Jiaozhou Bay, San Francisco Bay, and the Mississippi River (17, 20, 56). Previous studies have suggested that Nitrosomonas species represent a widespread bacterial amoA sequence type, as Nitrosomonas is frequently recovered from estuarine and marine sediments and has a broad geographic distribution (57). Our results further confirmed this hypothesis, as 98.7% of the total AOB amoA sequences obtained from the Yangtze Estuary fell into this group. Interestingly, a phylogenetically distinct Nitrosomonas group, in which sequence identity with other known Nitrosomonas amoA sequences was <77.6%, was identified. This distinct Nitrosomonas group dominated the AOB communities at most of the high-salinity sampling sites in the Yangtze Estuary, and thus, we assumed that it might be a new group of Nitrosomonas species which can better adapt to the higher-salinity habitats in the estuarine area. However, additional studies with cultivated isolates or enrichment cultures are needed to fully elucidate this novel group.
Estuaries represent a typical land-sea transitional zone and might harbor mixed populations of both marine and terrestrial AOA (11, 12). This study confirmed that most of the archaeal amoA sequences obtained from the Yangtze Estuary were affiliated with those in marine and other estuarine sediments (20, 21, 54), while the remaining sequences were affiliated with the soil group 1.1b (55). The percentage of sequences in soil cluster decreased from the upstream to the downstream reaches of the Yangtze Estuary. This could be explained by river runoff of soil.
Multiple environmental factors affect the communities of ammonia oxidizers with complicated interactions (18, 20). In this study, both AOB and AOA communities had significant correlation with salinity, pH, ammonium, total phosphorus, and organic carbon. Of all these factors, salinity, which plays a major role in ammonium adsorption in sediments, has been suggested as a key parameter regulating ammonia oxidizer communities (20, 44, 57, 58, 59, 60, 61). Generally, bacterial amoA clone libraries acquired from the higher-salinity sites of the Yangtze Estuary were significantly different from communities obtained from lower-salinity sites. Also, the diversity of AOB was positively (but not significantly) correlated with salinity (r = 0.441, P = 0.114, n = 14). Likewise, in the Plum Island Sound estuary, a slight increase in diversity was also observed associated with salinity increasing from low (0.5 to 8.7 PSU) to medium (6.3 to 24.7 PSU), which was in the same range of salinity in this study (57). The medium-salinity sites might be more heterogeneous to provide diverse niche differentiation. Along the salinity gradient of the Yangtze Estuary, spatial variations of archaeal amoA gene libraries were also observed, as well as in the Bahía del Tóbari Estuary, where AOA communities differ remarkably between the interior and the mouths of the estuary (11).
Consistent with our results, it has been reported that sediment pH might affect the growth and community compositions of amoA-containing microorganisms by affecting the chemical form, concentration, and availability of the substrates (62). Also, the impact of pH on the distribution of amoA-containing prokaryotes has been proposed (55). In this study, the concentration of ammonium also had significant correlation with AOB and AOA communities. Ammonium as the primary energy source might promote the activity of ammonia oxidizers, and previous studies also reported that high concentrations of ammonium partially inhibited the activity of ammonia-oxidizing prokaryotes (63). The detected correlation between the community structure of ammonia-oxidizing bacteria and organic carbon was unexpected, since AOB are obligate chemolithotrophic bacteria (9). However, reports showed that the growth of certain AOB groups could be enhanced by organic compounds, while other species had low tolerance to organic matter (64). Further work is required to examine this result and explore the underlying mechanism, including the complicated interaction between AOA community and organic carbon. Unprecedentedly, both AOB and AOA communities correlated significantly with total phosphorus in this study. This correlation was likely attributed to the changes of nutrient ratios caused mainly by different levels of phosphorus in sediments which may define habitats of ammonia oxidizers. Sediment clay content was observed to have a significant correlation with AOA communities in this study. Grain size of sediment was important because it might control many physicochemical characteristics of sediment as it was related to in situ hydrological conditions, such as tides, river runoff, water mixing, and the intensity and dynamics of these activities (56). However, the exact mechanism needs to be determined in future studies.
Bacterial amoA gene libraries at all sampling sites and archaeal amoA gene libraries at 6 out of 7 sampling sites in the Yangtze Estuary were statistically indistinguishable between summer and winter. Likewise, AOB communities in the Plum Island Sound Estuary showed little seasonal variability over 3 years (57), and AOA communities sampled 9 months apart were virtually indistinguishable at 4 out of 5 sites in the Bahía del Tóbari Estuary (11). The absence of apparent seasonal shifts in the community compositions of ammonia oxidizers at most of the sampling sites suggests that their population distribution in the Yangtze Estuary reflects an adaptation to site-specific characteristics (57).
In contrast to previous reports that AOA were more abundant than AOB in both terrestrial and marine systems (19, 22, 24, 32, 65), our qPCR results indicated that bacterial amoA gene copies were more numerous (1.1 to 47.1 times) than archaeal amoA gene copies in most of the sampling habitats. This was consistent with the results in the Weeks Bay estuary and the San Francisco Bay estuary, where the AOB amoA copy numbers were also greater than AOA amoA copy numbers at most of the sampling sites (14, 20). Salinity was considered to be a key factor causing higher abundance of AOB amoA genes than AOA amoA genes in the high- (22- to 31-PSU) and medium-salinity (6- to 9-PSU) estuarine regions (20). However, this conclusion was challenged by the report that the abundance of AOA was greater than that of AOB along an estuarine salinity gradient (0.5 to 31.7 PSU) (31). Thus, it remains unclear whether salinity is a controlling variable (23, 31). Although we did not detect a linear relationship between salinity and archaeal amoA gene abundance, there was a pattern of higher AOA abundance at the low-salinity sites. This result was consistent with the previous study in the Plum Island Sound estuary (31). In this study, the ratio of AOB to AOA amoA copy numbers was negatively correlated with total phosphorus, and researchers found that AOA amoA gene abundance was positively correlated with total phosphorus in Matsushima Bay sediment (66). Ammonium concentration is another factor that might affect the relative abundance of bacterial and archaeal amoA genes. AOA may better adapt to oligotrophic (low-ammonium) environments since the half-saturation constant (Km) for ammonia oxidation by Thaumarchaeota is lower than those in other microbes (67). However, no significant correlation was found between the ratios of AOB to AOA amoA copy numbers and the concentrations of ammonium in this study.
Abundance of ammonia oxidizers may be reflected by the potential nitrification rates (68), so we expected these two parameters to correlate significantly. In this study, significant linear relationships were observed between potential nitrification rates and archaeal amoA gene abundance. However, no significant correlations were found between AOB amoA gene abundance and potential nitrification rates in the Yangtze Estuary. Interestingly, no significant decreases of potential nitrification rates were also observed after ammonia-oxidizing bacteria were inhibited by ampicillin, though the bacterial amoA gene copies were more numerous than archaeal amoA gene copies in most of the samples. These results implied that archaea might play a more important role in mediating the oxidation of ammonia to nitrite at the study area. Although recent studies have revealed that some Thaumarchaeotes with expression of ammonia monooxygenase genes are not obligate autotrophic ammonia oxidizers (13), the observed results in this study suggested that amoA-carrying Thaumarchaeotes detected in the collected samples indeed made a significant contribution to ammonia oxidation in the intertidal sediments of the Yangtze Estuary. In addition, AOA in the marine group 1.1a might make more of a contribution to the oxidation of ammonia to nitrite, as potential nitrification rates were found to be closely related only to the abundance of this group.
Nitrification rates can be affected by many environmental factors, including light, salinity, temperature, ammonium concentrations, and dissolved oxygen concentrations (69, 70). Strong negative correlations were found between the potential nitrification rates and salinity in the Yangtze Estuary, showing that the metabolic activity of ammonia oxidizers might be inhibited by high levels of salinity. In addition, there were significantly higher potential nitrification rates in summer, compared to winter. This seasonality could be attributed to enhanced activity of ammonia oxidizers due to high temperature in the warm season. However, it has been documented that potential nitrification rates were higher in winter than in summer in the coastal sediments of the Arctic Ocean (69). The difference might result from the concentration of ammonium, which was a limiting factor for nitrification in the Arctic Ocean. Thus, nitrification rates were higher in winter because of the lack of competition with phytoplankton and other microbes for ammonium (69). In contrast, the potential nitrification rates at the study area were likely not limited by ammonium availability, because ammonium concentrations were always relatively high (36).
Taken together, in the intertidal sediments of the Yangtze Estuary, AOB had a lower diversity of amoA genes than did AOA. Spatial variations of both AOB and AOA communities were observed, while all the AOB communities and 6 out of 7 AOA communities in this study were statistically indistinguishable between summer and winter. Community composition of ammonia oxidizers significantly correlated with salinity, pH, ammonium, total phosphorus, and organic carbon. AOA communities were also significantly correlated with sediment clay content. Bacterial amoA gene copies were more abundant than archaeal amoA gene copies in most of the sampling habitats of the Yangtze Estuary. Potential nitrification rates were strongly correlated with salinity and were significantly greater in summer than in winter. In addition, potential nitrification rates had a significant correlation with archaeal amoA gene abundance, and no significant differences were observed between rates measured with and without ampicillin, showing that archaea might play a more important role in ammonia oxidation at the estuarine area.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the National Natural Science Foundations of China (grants 41130525, 41322002, 41071135, 41271114, and 41021064), the Academic Doctoral Prize of East China Normal University (XRZZ2013024), the State Key Laboratory of Estuarine and Coastal Research (SKLEC, no. 2010RCDW07), and the SKLEC Fostering Project for Top Doctoral Dissertations. It was also supported by the Program for New Century Excellent Talents in University (NCET) and the Fundamental Research Funds for the Central Universities and the Marine Scientific Research Project for Public Interest (no. 200905007).
We thank Jun Gong and Xiaoli Zhang for sharing their analytical expertise on measurements of ammonia oxidizers. The anonymous reviewers are thanked for their constructive comments.
Footnotes
Published ahead of print 1 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03035-13.
REFERENCES
- 1.Galloway JN, Townsend AR, Erisman JW, Bekunda M, Cai ZC, Freney JR, Martinelli LA, Seitzinger SP, Sutton MA. 2008. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320:889–892. 10.1126/science.1136674 [DOI] [PubMed] [Google Scholar]
- 2.Munn T, Whyte A, Timmerman P. 1999. Emerging environmental issues: a global perspective of SCOPE. Ambio 28:464–471 [Google Scholar]
- 3.Seitzinger SP. 2008. Nitrogen cycle: out of reach. Nature 452:162–163. 10.1038/452162a [DOI] [PubMed] [Google Scholar]
- 4.Jickells TD. 1998. Nutrient biogeochemistry of the coastal zone. Science 281:217–222. 10.1126/science.281.5374.217 [DOI] [PubMed] [Google Scholar]
- 5.Gruber N, Galloway JN. 2008. An Earth-system perspective of the global nitrogen cycle. Nature 451:293–296. 10.1038/nature06592 [DOI] [PubMed] [Google Scholar]
- 6.Hou LJ, Liu M, Carini SA, Gardner WS. 2012. Transformation and fate of nitrate near the sediment-water interface of Copano Bay. Cont. Shelf Res. 35:86–94. 10.1016/j.csr.2012.01.004 [DOI] [Google Scholar]
- 7.Seitzinger SP. 1988. Denitrification in fresh water and coastal marine ecosystems: ecological and geochemical significance. Limnol. Oceanogr. 33:702–724. 10.4319/lo.1988.33.4_part_2.0702 [DOI] [Google Scholar]
- 8.Jensen K, Sloth NP, Risgaard-Petersen N, Rysgaard S, Revsbech NP. 1994. Estimation of nitrification and denitrification from microprofiles of oxygen and nitrate in model sediment systems. Appl. Environ. Microbiol. 60:2094–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kowalchuk GA, Stephen JR. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55:485–529. 10.1146/annurev.micro.55.1.485 [DOI] [PubMed] [Google Scholar]
- 10.Purkhold U, Pommerening-Röser A, Juretschko S, Schmid MC, Koops HP, Wagner M. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368–5382. 10.1128/AEM.66.12.5368-5382.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beman JM, Francis CA. 2006. Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hyper-nitrified subtropical estuary: Bahia del Tobari, Mexico. Appl. Environ. Microbiol. 72:7767–7777. 10.1128/AEM.00946-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683–14688. 10.1073/pnas.0506625102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mussmann M, Brito I, Pitcher A, Sinninghe Damsté JS, Hatzenpichler R, Richter A, Nielsen JL, Halkjaer Nielsen P, Müller A, Daims H, Wagner M, Head IM. 2011. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc. Natl. Acad. Sci. U. S. A. 108:16771–16776. 10.1073/pnas.1106427108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caffrey JM, Bano N, Kalanetra K, Hollibaugh JT. 2007. Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J. 1:660–662. 10.1038/ismej.2007.79 [DOI] [PubMed] [Google Scholar]
- 15.Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809. 10.1038/nature04983 [DOI] [PubMed] [Google Scholar]
- 16.Li M, Cao H, Hong Y, Gu JD. 2011. Spatial distribution and abundances of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in mangrove sediments. Appl. Microbiol. Biotechnol. 89:1243–1254. 10.1007/s00253-010-2929-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang SY, Wang Y, Feng XJ, Zhai LM, Zhu GB. 2011. Quantitative analyses of ammonia-oxidizing Archaea and bacteria in the sediments of four nitrogen-rich wetlands in China. Appl. Microbiol. Biotechnol. 90:779–787. 10.1007/s00253-011-3090-0 [DOI] [PubMed] [Google Scholar]
- 18.Dang H, Zhang X, Sun J, Li T, Zhang Z, Yang G. 2008. Diversity and spatial distribution of sediment ammonia-oxidizing crenarchaeota in response to estuarine and environmental gradients in the Changjiang Estuary and East China Sea. Microbiology 154:2084–2095. 10.1099/mic.0.2007/013581-0 [DOI] [PubMed] [Google Scholar]
- 19.Jin T, Zhang T, Ye L, Lee OO, Wong YH, Qian PY. 2011. Diversity and quantity of ammonia-oxidizing Archaea and Bacteria in sediment of the Pearl River Estuary, China. Appl. Microbiol. Biotechnol. 90:1137–1145. 10.1007/s00253-011-3107-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mosier AC, Francis CA. 2008. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ. Microbiol. 10:3002–3016. 10.1111/j.1462-2920.2008.01764.x [DOI] [PubMed] [Google Scholar]
- 21.Santoro AE, Francis CA, de Sieyes NR, Boehm AB. 2008. Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ. Microbiol. 10:1068–1079. 10.1111/j.1462-2920.2007.01547.x [DOI] [PubMed] [Google Scholar]
- 22.Beman JM, Popp BN, Francis CA. 2008. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2:429–441. 10.1038/ismej.2007.118 [DOI] [PubMed] [Google Scholar]
- 23.Bouskill NJ, Eveillard D, Chien D, Jayakumar A, Ward BB. 2012. Environmental factors determining ammonia-oxidizing organism distribution and diversity in marine environments. Environ. Microbiol. 14:714–729. 10.1111/j.1462-2920.2011.02623.x [DOI] [PubMed] [Google Scholar]
- 24.Newell SE, Babbin AR, Jayakumar A, Ward BB. 2011. Ammonia oxidation rates and nitrification in the Arabian Sea. Global Biogeochem. Cycles 25:GB4016. 10.1029/2010GB003940 [DOI] [Google Scholar]
- 25.Mohamed NM, Saito K, Tal Y, Hill RT. 2010. Diversity of aerobic and anaerobic ammonia-oxidizing bacteria in marine sponges. ISME J. 4:38–48. 10.1038/ismej.2009.84 [DOI] [PubMed] [Google Scholar]
- 26.Beman JM, Roberts KJ, Wegley L, Rohwer F, Francis CA. 2007. Distribution and diversity of archaeal ammonia monooxygenase genes associated with corals. Appl. Environ. Microbiol. 73:5642–5647. 10.1128/AEM.00461-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park HD, Wells GF, Bae H, Criddle CS, Francis CA. 2006. Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl. Environ. Microbiol. 72:5643–5647. 10.1128/AEM.00402-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ouverney CC, Fuhrman JA. 2000. Marine planktonic archaea take up amino acids. Appl. Environ. Microbiol. 66:4829–4833. 10.1128/AEM.66.11.4829-4833.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker BJ, Lesniewski RA, Dick G. 2012. Genome-enabled transcriptomics reveals archaeal populations that drive nitrification in a deep-sea hydrothermal plume. ISME J. 6:2269–2279. 10.1038/ismej.2012.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P, Strous M, Teira E, Herndl GJ, Middelburg JJ, Schouten S, Sinninghe Damste JS. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. U. S. A. 103:12317–12322. 10.1073/pnas.0600756103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernhard AE, Landry ZC, Blevins A, de la Torre JR, Giblin AE, Stahl DA. 2010. Abundance of ammonia-oxidizing archaea and bacteria along an estuarine salinity gradient in relation to potential nitrification rates. Appl. Environ. Microbiol. 76:1285–1289. 10.1128/AEM.02018-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam P, Jensen MM, Lavik G, McGinnis DF, Muller B, Schubert CJ, Amann R, Thamdrup B, Kuypers MMM. 2007. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc. Natl. Acad. Sci. U. S. A. 104:7104–7109. 10.1073/pnas.0611081104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu JR, Wang JH, Shen HT, Wu H. 2005. Observation and analysis of the diluted water and red tide in the sea off the Changjiang River mouth in middle and late June 2003. Chin. Sci. Bull. 50:240–247. 10.1360/03wd0107 [DOI] [Google Scholar]
- 34.Chai C, Yu ZM, Song XX, Gao XH. 2006. The status and characteristics of eutrophication in the Yangtze River (Changjiang) Estuary and the adjacent East China Sea, China. Hydrobiologia 563:313–328. 10.1007/s10750-006-0021-7 [DOI] [Google Scholar]
- 35.Liu SM, Zhang J, Chen HT, Wu Y, Xiong H, Zhang EF. 2003. Nutrients in the Changjiang and its tributaries. Biogeochemistry. 62:1–18. 10.1023/A:1021162214304 [DOI] [Google Scholar]
- 36.Hou LJ, Liu M, Xu SY, Ou DN, Yu J, Cheng SB, Lin X, Yang Y. 2007. The effects of semi-lunar spring and neap tidal change on nitrification, denitrification, and N2O vertical distribution in the intertidal sediments of the Yangtze estuary, China. Estuar. Coast. Shelf Sci. 73:607–616. 10.1016/j.ecss.2007.03.002 [DOI] [Google Scholar]
- 37.Murphy J, Riley JP. 1962. A modified single solution method for the determination of phosphate in natural water. Anal. Chim. Acta 27:31–36. 10.1016/S0003-2670(00)88444-5 [DOI] [Google Scholar]
- 38.Rotthauwe JH, Witzel KP, Liesack W. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maidak BL, Cole JR, Parker CT, Jr, Garrity GM, Larsen N, Li B, Lilburn TG, McCaughey MJ, Olsen MJ, Overbeek R, Pramanik S, Schmidt TM, Tiedje JM, Woese CR. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171–173. 10.1093/nar/27.1.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150–163. 10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- 43.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- 44.Bernhard AE, Tucker J, Giblin AE, Stahl DA. 2007. Functionally distinct communities of ammonia-oxidizing bacteria along an estuarine salinity gradient. Environ. Microbiol. 9:1439–1447. 10.1111/j.1462-2920.2007.01260.x [DOI] [PubMed] [Google Scholar]
- 45.van de Graaf AA, Mulder A, de Bruijn P, Jetten MS, Robertson LA, Kuenen JG. 1995. Anaerobic oxidation of ammonium is a biologically mediated process. Appl. Environ. Microbiol. 61:1246–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Souza MP, Chu D, Zhao M, Zayed AM, Ruzin SE, Schichnes D, Terry N. 1999. Rhizosphere bacteria enhance selenium accumulation and volatilization by Indian mustard. Plant Physiol. 119:565–573. 10.1104/pp.119.2.565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.ter Braak CJF, Smilauer P. 2002. CANOCO reference manual and CanoDraw for Windows user's guide: software for canonical community ordination (version 4.5). Microcomputer Power, Ithaca, NY [Google Scholar]
- 48.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. 2011. UniFrac: an effective distance metric for microbial community comparison. ISME J. 5:169–172. 10.1038/ismej.2010.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aakra A, Utaker JB, Nes IF. 2001. Comparative phylogeny of the ammonia monooxygenase subunit A and 16S rRNA genes of ammonia-oxidizing bacteria. FEMS Microbiol. Lett. 205:237–242. 10.1111/j.1574-6968.2001.tb10954.x [DOI] [PubMed] [Google Scholar]
- 51.Purkhold U, Wagner M, Timmermann G, Pommerening-Roser A, Koops HP. 2003. 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the nitrosomonads. Int. J. Syst. Evol. Microbiol. 53:1485–1494. 10.1099/ijs.0.02638-0 [DOI] [PubMed] [Google Scholar]
- 52.Urakawa H, Kurata S, Fujiwara T, Kuroiwa D, Maki H, Kawabata S, Hiwatari T, Ando H, Kawai T, Watanabe M, Kohata K. 2006. Characterization and quantification of ammonia-oxidizing bacteria in eutrophic coastal marine sediments using polyphasic molecular approaches and immunofluorescence staining. Environ. Microbiol. 8:787–803. 10.1111/j.1462-2920.2005.00962.x [DOI] [PubMed] [Google Scholar]
- 53.Avrahami S, Conrad R. 2003. Patterns of community change among ammonia oxidizers in meadow soils upon long-term incubation at different temperatures. Appl. Environ. Microbiol. 69:6152–6164. 10.1128/AEM.69.10.6152-6164.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao H, Hong Y, Li M, Gu JD. 2011. Diversity and abundance of ammonia-oxidizing prokaryotes in sediments from the coastal Pearl River estuary to the South China Sea. Antonie Van Leeuwenhoek 100:545–556. 10.1007/s10482-011-9610-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia Z, Conrad R. 2009. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11:1658–1671. 10.1111/j.1462-2920.2009.01891.x [DOI] [PubMed] [Google Scholar]
- 56.Dang H, Li J, Chen R, Wang L, Guo L, Zhang Z, Klotz MG. 2010. Diversity, abundance, and spatial distribution of sediment ammonia-oxidizing Betaproteobacteria in response to environmental gradients and coastal eutrophication in Jiaozhou Bay, China. Appl. Environ. Microbiol. 76:4691–4702. 10.1128/AEM.02563-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernhard AE, Donn T, Giblin AE, Stahl DA. 2005. Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ. Microbiol. 7:1289–1297. 10.1111/j.1462-2920.2005.00808.x [DOI] [PubMed] [Google Scholar]
- 58.Francis CA, O'Mullan GD, Ward BB. 2003. Diversity of ammonia monooxygenase (amoA) genes across environmental gradients in Chesapeake Bay sediments. Geobiology 1:129–140. 10.1046/j.1472-4669.2003.00010.x [DOI] [Google Scholar]
- 59.Bollmann A, Laanbroek HJ. 2002. Influence of oxygen partial pressure and salinity on the community composition of ammonia-oxidizing bacteria in the Schelde Estuary. Aquat. Microb. Ecol. 28:239–247. 10.3354/ame028239 [DOI] [Google Scholar]
- 60.Cébron A, Coci M, Garnier J, Laanbroek HJ. 2004. Denaturing gradient gel electrophoretic analysis of ammonia-oxidizing bacterial community structure in the lower Seine River: impact of Paris wastewater effluent. Appl. Environ. Microbiol. 70:6726–6737. 10.1128/AEM.70.11.6726-6737.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Bie MJM, Speksnijder A, Kowalchuk GA, Schuurman T, Zwart G, Stephen JR, Diekmann OE, Laanbroek HJ. 2001. Shifts in the dominant populations of ammonia-oxidizing beta-subclass proteobacteria along the eutrophic Schelde Estuary. Aquat. Microb. Ecol. 23:225–236. 10.3354/ame023225 [DOI] [Google Scholar]
- 62.Nicol GW, Leininger S, Schleper C, Prosser JI. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10:2966–2978. 10.1111/j.1462-2920.2008.01701.x [DOI] [PubMed] [Google Scholar]
- 63.Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. U. S. A. 105:2134–2139. 10.1073/pnas.0708857105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Antje K, Heinz H. 1982. Effect of organic matter on growth and cell yield of ammonia-oxidizing bacteria. Arch. Microbiol. 133:50–54. 10.1007/BF00943769 [DOI] [Google Scholar]
- 65.Yao HY, Gao YM, Nicol GW, Campbell CD, Prosser J, Zhang LM, Han WY, Singh BK. 2011. Links between ammonia oxidizer community structure, abundance, and nitrification potential in acidic soils. Appl. Environ. Microbiol. 77:4618–4625. 10.1128/AEM.00136-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakami T. 2012. Distribution of ammonia-oxidizing archaea and bacteria in the surface sediments of Matsushima Bay in relation to environmental variables. Microbes Environ. 27:61–66. 10.1264/jsme2.ME11218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976–979. 10.1038/nature08465 [DOI] [PubMed] [Google Scholar]
- 68.Henriksen K. 1980. Measurement of in situ rates of nitrification in sediment. Microb. Ecol. 6:329–337. 10.1007/BF02010495 [DOI] [PubMed] [Google Scholar]
- 69.Christman GD, Cottrell MT, Popp BN, Gier E, Kirchman DL. 2011. Abundance, diversity, and activity of ammonia-oxidizing prokaryotes in the coastal arctic ocean in summer and winter. Appl. Environ. Microbiol. 77:2026–2034. 10.1128/AEM.01907-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henriksen K, Kemp WM. 1986. Nitrification in estuarine and coastal marine sediments: methods, patterns and regulating factors, p 207–250 In Blackburn TH, Sørensen J. (ed), Nitrogen cycling in coastal marine environments. John Wiley and Sons, New York, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.