Abstract
Mussels attach to virtually all types of inorganic and organic surfaces in aqueous environments, and catecholamines composed of 3,4-dihydroxy-l-phenylalanine (DOPA), lysine, and histidine in mussel adhesive proteins play a key role in the robust adhesion. DOPA is an unusual catecholic amino acid, and its side chain is called catechol. In this study, we displayed the adhesive moiety of DOPA-histidine on Escherichia coli surfaces using outer membrane protein W as an anchoring motif for the first time. Localization of catecholamines on the cell surface was confirmed by Western blot and immunofluorescence microscopy. Furthermore, cell-to-cell cohesion (i.e., cellular aggregation) induced by the displayed catecholamine and synthesis of gold nanoparticles on the cell surface support functional display of adhesive catecholamines. The engineered E. coli exhibited significant adhesion onto various material surfaces, including silica and glass microparticles, gold, titanium, silicon, poly(ethylene terephthalate), poly(urethane), and poly(dimethylsiloxane). The uniqueness of this approach utilizing the engineered sticky E. coli is that no chemistry for cell attachment are necessary, and the ability of spontaneous E. coli attachment allows one to immobilize the cells on challenging material surfaces such as synthetic polymers. Therefore, we envision that mussel-inspired catecholamine yielded sticky E. coli that can be used as a new type of engineered microbe for various emerging fields, such as whole living cell attachment on versatile material surfaces, cell-to-cell communication systems, and many others.
INTRODUCTION
Microbial cell attachment is a crucial process in the overall efficiency of a variety of systems that exploits the whole microbial cell, such as biocatalysts (1–9) or biosensors (10). Existing methods of attaching microbial cells to surfaces utilize various chemistries, such as adsorption (11), covalent attachment (12, 13), and entrapment within matrices (13, 14). However, adsorption often results in the detachment of bacteria due to the weak bond with the substrates, and covalent coupling requires cross-linking agents which results in the loss of cell activity and viability. The entrapment process requires harsh conditions in some cases, which jeopardizes cellular activity and viability. Another important issue is that the types of materials that the conventional methods can be applied to are largely limited due to the available surface chemistry. Therefore, cell attachment to variety of material surfaces without surface modification or harsh chemical treatment remains a great challenge. The facile cell attachment would circumvent the complex chemical modifications and reduce the loss of cellular activity and viability of the whole cell.
As an emerging method for cell attachment, engineered cells expressing binding affinity tags on the cell surfaces through display technology have been utilized, although the approach is not prevalent. Cell surface display allows peptides and proteins to be displayed on the surfaces of microbial cells by fusing them with the anchoring motif through recombinant DNA techniques. A wide range of applications of the display technology include bioadsorption (15–20), whole-cell biocatalyst (21–27), peptide library screening (28, 29), live vaccine development (30–32), antibody production (33), and biosensors (34, 35). Although cell attachment through cell surface display is not a well-known application, cell attachment through displaying peptides with binding affinity has been reported on chitin, cellulose, and gold substrates. The first report of displaying affinity tags for cell attachment was recombinant Escherichia coli expressing Cex exoglucanase and cellulose-binding domain on the cell surface (36). The engineered E. coli was found to bind tightly and rapidly to cellulosic materials; it was later shown that this binding is very specific and stable (36, 37). Also, recombinant E. coli with chitin-binding domain expressed on the surface exhibited highly stable immobilization on the chitin substrate (38). Cell attachment for whole-cell biosensor was also reported on the gold substrate with recombinant E. coli expressing gold-binding peptides on the cell surface (10). Limitation of the aforementioned cell attachments is that the engineered recombinant E. coli can only bind to a specific substrate depending on the displayed peptide motif on the cell surface. In order to overcome this limitation related to material versatility, we hypothesized that not only can introduction of the adhesive property of mussels extend the range of materials for cell attachment but the adhesive E. coli can also spontaneously bind to surfaces without chemical process.
Adhesive properties are unique in that mussels can attach to virtually all types of inorganic and organic surfaces in wet conditions. Therefore, the production of mussel adhesive proteins (MAPs) and its mimetic peptides have been investigated in depth to be used as bioadhesives. The expression systems of MAPs and hybrid fusion MAPs in E. coli have been explored through recombinant DNA technology (39–43), but there are as yet no reports of displaying mussel adhesive protein or adhesive mimetic motifs of mussels on the surface to exploit its adhesive property. The major component of the specialized mussel adhesive proteins, Mytilus edulis foot protein 5 (Mefp-5), located at a distal side of adhesive pads, was found to have extensive repeats of 3,4-dihydroxy-l-phenylalanine (DOPA) and lysine (44). Also, when the amino acid sequence of the Mefp-5 is carefully observed, repeats of DOPA and histidine are present in the protein (Fig. 1). Based on this observation, the coexistence of catechol and cationic amine functional group was hypothesized to be a key factor in achieving adhesion to a wide spectrum of materials. Therefore, histidine and DOPA, although they have not been tested for surface adhesion, are expected to exhibit adhesive property.
FIG 1.
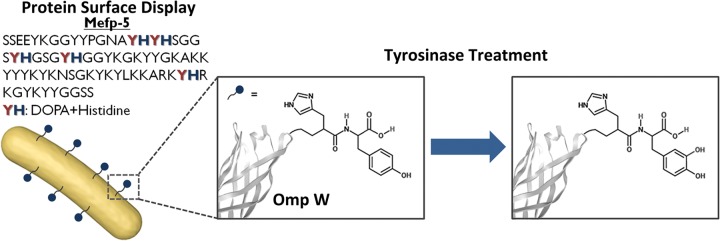
Schematic illustration of engineering mussel-inspired catecholamine-displayed E. coli. DOPA and histidine repeats found in the mussel adhesive protein, Mefp-5, are displayed on the E. coli surface using outer membrane protein W (OmpW) as the anchoring motif. Since DOPA is an unnatural amino acid, tyrosine was displayed first and converted to DOPA using tyrosinase.
In the present study, we combined cell surface display techniques and the mussel-inspired adhesive catecholamine moiety, histidine and DOPA, to prepare sticky E. coli to achieve spontaneous adhesion onto various material surfaces. The engineered sticky E. coli exhibited unprecedented adhesion to various material surfaces, including silica and glass beads, titanium, and gold substrates. Furthermore, the sticky E. coli even adhered to synthetic substrates that are typically incompatible with immobilization chemistry, such as polyethylene terephthalate (PET), polydimethylsiloxane (PDMS), and polyurethane (PU). Our findings suggest that the display of catecholamine on living bacterial cells can be a new approach for facile cell attachment, bypassing complicated, harsh chemical processes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strain XL10-Gold Kanr {Tetr Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [F′ proAB lacIqZΔM15 Tn10 (Tetr) Tn5 (Kanr) Amy]} (Stratagene, United Kingdom) was used as a host strain for all recombinant plasmid work in the present study. All recombinant strains were cultivated in Luria-Bertani medium (10 g of Bacto tryptone/liter, 5 g of Bacto yeast extract/liter, and 10 g of NaCl/liter) supplemented with 50 g of ampicillin/ml. Control strains with no recombinant plasmids were cultured in Luria-Bertani medium supplemented with 40 g of kanamycin/ml. The recombinant cells were all cultivated at 30°C and induced at an optical density at 600 nm (OD600) of 0.4 to 0.6 by addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 0.2 mM. After induction, the cells were grown for an additional 4 h and harvested for further experiments.
Plasmids and DNA manipulation.
PCR was performed using the primers from Table 1 with a PCR 5000 thermal cycler (Bio-Rad) using an ExPrime Taq premix kit (GeNet Bio, Inc.). All DNA manipulation and cloning procedures, including restriction enzyme digestion, ligation, and agarose gel electrophoresis, were carried out according to standard procedures (45). The primers and plasmids used in the present study are listed in Tables 1 and 2. Truncated E. coli OmpW was amplified from the genomic DNA of the E. coli W3110 strain, and Pseudomonas aeruginosa perhydrolase genes were amplified from the genomic DNA of P. aeruginosa using a primer design based on the reported DNA sequences.
TABLE 1.
Primers used in PCR experiments
| Primer sequence (5′–3′)a | Gene to be amplified | Template |
|---|---|---|
| GAATTCATGAAAAAGTTAACAGTGGCG | Truncated OmpW_forward | E. coli W3110 gDNA |
| TCTAGAGCCATGATCGTTAAATCCATT | Truncated OmpW (aa 139)_reverse | E. coli W3110 gDNA |
| TCTAGATGCACCGCCCAGCTTATAAT | Truncated OmpW (aa 191)_reverse | E. coli W3110 gDNA |
| TCTAGAATGGGTTACGTGACAACGAAGG | Perhydrolase_forward | P. aeruginosa PAO1 gDNA |
| AAGCTTTCAGCCGCGGATGAAGGCC | Perhydrolase_reverse | P. aeruginosa PAO1 gDNA |
| AAGCTTTTAATGGTGATGATGGTGATGGCCGCGGATGAAGGC | Perhydrolase + 6-His_reverse | P. aeruginosa PAO1 gDNA |
| AAGCTTTTAATAATGGTGATGATGGTGATG | Perhydrolase + 6-His + 1Tyr_reverse | Perhydrolase + 6-His PCR product |
| AAGCTTTTAATAATAATGGTGATGATGGTGATG | Perhydrolase + 6-His + 2Tyr_reverse | Perhydrolase + 6-His PCR product |
Restriction enzyme sites are indicated in boldface.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristicsa | Reference |
|---|---|---|
| pKK223-3 | Amp resistant; tac promoter; 4.5 kb | Brosius and Holy (56) |
| pKK223-18MCS | pKK223 derivative; containing additional 18 multicloning site; 4.6 kb | Hernan et al. (57) |
| pKK223-pOW13F | pKK223 derivative; containing 417-bp fragment of truncated OmpW at aa 139 of E. coli; 5.0 kb | This study |
| pKK223-pOW19F | pKK223 derivative; containing 573-bp fragment of truncated OmpW at aa 191 of E. coli; 5.1 kb | This study |
| pOW13F-PerPA1 | pKK223-pOW13F derivative; P. aeruginosa perhydrolase; 5.8 kb | This study |
| pOW13F-PerPA2 | pKK223-pOW13F derivative; P. aeruginosa perhydrolase + 6-His; 5.8 kb | This study |
| pOW13F-PerPA3 | pKK223-pOW13F derivative; P. aeruginosa perhydrolase + 6-His + 1Tyr; 5.8 kb | This study |
| pOW13F-PerPA4 | pKK223-pOW13F derivative; P. aeruginosa perhydrolase + 6-His + 2Tyr; 5.8 kb | This study |
| pOW19F-PerPA1 | pKK223-pOW19F derivative; P. aeruginosa perhydrolase; 6.0 kb | This study |
| pOW19F-PerPA2 | pKK223-pOW19F derivative; P. aeruginosa perhydrolase + 6-His; 6.0 kb | This study |
| pOW19F-PerPA3 | pKK223-pOW19F derivative; P. aeruginosa perhydrolase + 6-His + 1Tyr; 6.0 kb | This study |
| pOW19F-PerPA4 | pKK223-pOW19F derivative; P. aeruginosa perhydrolase + 6-His + 2Tyr; 6.0 kb | This study |
aa, amino acid.
Construction of surface display system.
Outer membrane protein W (OmpW) was used as the protein platform, and perhydrolase from P. aeruginosa (PerPA) was used as a spacer protein to display the mussel-inspired sticky catecholamine moiety of histidine and DOPA. OmpW is an outer membrane protein that belongs to a family of small outer membrane beta-barrel proteins that are widespread in Gram-negative bacteria (46, 47). The X-ray structure of OmpW showed that it forms an eight-stranded beta-barrel with a hydrophobic channel (48), and this transmembrane structure made it possible for the OmpW to be used as a stable anchoring motif for microbial cell surface display. To develop a strategy for displaying PerPA with DOPA and histidine residues using OmpW as an anchoring motif, we first searched for the potential fusion site in OmpW. Based on the predicted secondary structure of OmpW, two sites were chosen—at amino acids 139 and 191—to display the PerPA fusion protein. The truncated OmpW genes encoding the amino acids 139 and 191 from the N terminus were amplified by PCR using the primer sets shown in Table 1 and were cloned into the EcoRI and XbaI sites of pKK223-18MCS to generate pOW13F and pOW19F, respectively. A variety of PerPA gene-containing inserts, such as PerPA (PerPA1), PerPA+His (PerPA2), PerPA+His+1-DOPA (PerPA3), and PerPA+His+2-DOPA (PerPA4), were amplified and cloned into the XbaI and HindIII sites of pOW13F and pOW19F. Thus, the resulting construct designated pOW13F-PerPA1 is a perhydrolase-OmpW fusion protein-encoding gene, and the fusion site is amino acid 139 in OmpW. Similar constructs with different fusion proteins include pOW13F-PerPA2, pOW13F-PerPA3, and pOW13F-PerPA4. Similarly, the pOW19F-PerPA1 construct is a perhydrolase-OmpW fusion protein-encoding gene, and the fusion site is amino acid 191 in OmpW. Thus, we constructed pOW19F-PerPA2, pOW19F-PerPA3, and pOW19F-PerPA4 (Table 2). Recombinant XL10-Gold cells harboring the recombinant DNA were cultivated at 30°C and induced with IPTG.
SDS-PAGE and Western blot analysis.
Proteins in the whole-cell lysate and outer membrane fractions were analyzed by 12% (wt/vol) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Outer membrane proteins were prepared as follows: cells were harvested by centrifuging the cell broth (3 ml) at 6,000 rpm for 5 min at 4°C. The cell pellet was washed with 1 ml of 10 mM Na2HPO4 buffer (pH 7.2), centrifuged at 6,000 rpm for 5 min at 4°C, and resuspended in 0.2 ml of 10 mM Na2HPO4 buffer (pH 7.2). The resuspended cells underwent three cycles of sonication (pulse on 5 s, pulse off 10 s at 20% of maximum output). The sonicated cells were then centrifuged at 10,000 rpm for 2 min at room temperature to remove the partially disrupted cells, and the supernatant was collected. The collected supernatant was centrifuged again at 10,000 rpm for 30 min at 18°C. After centrifugation, the cell pellet was resuspended in 0.5 ml of 10 mM Na2HPO4 buffer (pH 7.2) containing 0.5% (wt/vol) sarcosyl and incubated for 30 min in a water bath at 37°C. After incubation, the membrane proteins were obtained by washing the insoluble pellet with 10 mM Na2HPO4 buffer (pH 7.2), followed by resuspension in 40 μl of Tris-EDTA buffer (pH 8.0). The whole-cell lysate and outer membrane protein samples were prepared by heating the protein with protein sample buffer (5×; Elpis Biotech) for 10 min at 100°C. Western blot analysis was carried out according to standard procedures (45). Mouse monoclonal anti-6×His tag antibody was used as the primary antibody (Abcam), and goat anti-mouse IgG–alkaline phosphatase conjugate (Sigma-Aldrich) was used as the secondary antibody for immunodetection of the fusion protein. The enzyme-catalyzed chromogenic reaction between nitroblue tetrazolium and BCIP (5-bromo-4-chloro-3-indolylphosphate; Roche) was used for Western blotting signal detection.
Immunofluorescence microscopy.
Cells were harvested by centrifugation at 6,000 rpm for 5 min at 4°C for analysis by using immunofluorescence microscopy. The cells were then washed and resuspended in 3% (wt/vol) bovine serum albumin (BSA; Sigma-Aldrich) in phosphate-buffered saline (PBS). The cells were incubated with mouse monoclonal anti-6×His tag antibody (Abcam) diluted 1:1,000 in PBS buffer containing 3% (wt/vol) BSA for 4 h at 4°C. After five washes with PBS buffer, the cell-antibody complex was incubated overnight at 4°C with goat polyclonal secondary antibody to mouse IgG conjugated with fluorescein isothiocyanate (FITC) diluted 1:3,000 in PBS buffer containing 3% (wt/vol) BSA. After overnight incubation, the cells were washed five times with PBS buffer prior to microscopic observation to remove any unbound goat anti-mouse IgG conjugated with FITC. The cells were mounted on microscopic glass slides and examined by fluorescence microscopy. Photographs were taken with a Nikon fluorescence microscope.
Tyrosinase treatment of recombinant E. coli and gold reduction.
Tyrosinase from mushroom (Sigma-Aldrich) was used throughout the experiment to convert tyrosine residues to DOPA. Tyrosinase was dissolved in distilled water to a concentration of 500 U/ml, where one unit represents ΔA280 of 0.001 per min at pH 6.5 at 25°C in a 3-ml reaction mixture containing l-tyrosine. The recombinant E. coli used for the tyrosinase treatment experiment was resuspended in 10 mM phosphate buffer (pH 7.0) to an OD600 of 2. An equal amount of tyrosinase was added to the solution, and the mixture was reacted at 25°C overnight. A solution of 3 mM HAuCl4 (Sigma-Aldrich) was incubated with tyrosinase (500 U/ml) for the gold reduction experiment. Recombinant E. coli cells were resuspended in 10 mM phosphate buffer (pH 7.0) to a cell OD600 of 2 and then incubated with tyrosinase and gold solution for 1 h to confirm the successful conversion of tyrosine to DOPA. After 1 h of incubation, the cells were washed and resuspended in 10 mM phosphate buffer (pH 7.0). Transmission electron microscopy (TEM) was used to analyze the surface of the recombinant E. coli. The resuspended cells were placed on a TEM grid, washed, and dried for TEM analysis.
Adhesion test of recombinant E. coli on various substrates.
Recombinant E. coli cells were resuspended in 10 mM phosphate buffer (pH 7.0) to an OD600 of 2 and incubated with tyrosinase (500 U/ml) and Silnos 290 silica beads (200 mg/liter; ABC Nanotech) or glass beads (200 mg/liter; Polyscience, Inc.) overnight at 25°C with shaking in a Thermomixer (Eppendorf) at 1,000 rpm. After incubation, the beads were dried on a silicon wafer and washed three times with 10 mM phosphate buffer (pH 7.0). Scanning electron microscopy (SEM) was used to analyze the surfaces of the beads. A further cell adhesion experiment was performed using silica (Si), titanium (Ti), and gold (Au) substrates as the metallic substrates and PET, PU, and PDMS as the polymeric substrates. For the Si substrates, an Si wafer was used, and the Ti and Au substrates were prepared by thermal evaporation (BAK641; Evatec, Switzerland) onto an Si wafer (thickness = 100 nm). PET and PU films were purchased from Hanmi, Inc. (South Korea). PDMS substrates were prepared using a Sylgard 184 silicone elastomer kit (Dow Corning, USA). Recombinant E. coli cells were resuspended in 10 mM phosphate buffer (pH 7.0) to an OD600 of 2 and then incubated overnight with tyrosinase (500 U/ml) on various surfaces at 25°C with shaking (220 rpm) to examine the adhesive properties of the sticky E. coli. As a control experiment, recombinant E. coli cells were not treated with tyrosinase. After incubation of recombinant E. coli on metallic surfaces, the surfaces were washed three times with 10 mM phosphate buffer (pH 7.0) and dried. SEM was then used to analyze the surfaces. After the recombinant E. coli was incubated on polymeric surfaces, the surfaces were washed three times with 10 mM phosphate buffer (pH 7.0) and dried. The surfaces were then dyed with acridine orange (Sigma-Aldrich) for 10 min to stain the E. coli and washed again with 10 mM phosphate buffer (pH 7.0). The surfaces were then analyzed by fluorescence microscopy.
RESULTS
Construction of surface display system.
In the present study, outer membrane protein W was used as the anchoring motif and perhydrolase from P. aeruginosa (PerPA) as a functional spacer protein to display the mussel-inspired catecholamine moiety. Perhydrolase was chosen as a spacer protein that can provide an additional function to the sticky E. coli, based on the fact that it can provide a valuable platform for developing a biocatalyst exploiting its practical reactions such as epoxidation of carbon-carbon double bonds of alkene or the reversible formation of peroxyacids from carboxylic acid and peroxide. To develop a strategy for displaying PerPA with 6-histidine and tyrosine residues using the OmpW as an anchoring motif, we first searched for the potential fusion sites in the OmpW. Based on the predicted secondary structure of OmpW, two fusion sites were chosen—at amino acid 139 and amino acid 191—for displaying PerPA fusion protein. The truncated OmpW genes encoding amino acids 139 and 191 from the N terminus were amplified by PCR using the primer sets shown in Table S1 in the supplemental material and were cloned into EcoRI and XbaI sites of pKK223-18MCS to make pOW13F and pOW19F. The inserts containing PerPA genes—PerPA (PerPA1), PerPA+6-His (PerPA2), PerPA+6-His+2Tyr (PerPA3), PerPA+6His+1Tyr (PerPA4)—were amplified and cloned into the XbaI and HindIII sites of pOW13F and pOW19F each to make pOW13F-PerPA1, pOW13F-PerPA2, pOW13F-PerPA3, pOW13F-PerPA4, pOW19F-PerPA1, pOW19F-PerPA2, pOW19F-PerPA3, and pOW19F-PerPA4, which were used to display PerPA genes on the E. coli cell surface. Recombinant XL10-Gold cells harboring the recombinant DNA were cultivated at 30°C and induced with IPTG.
Confirmation of OmpW-PerPA fusion protein display on the E. coli cell surface.
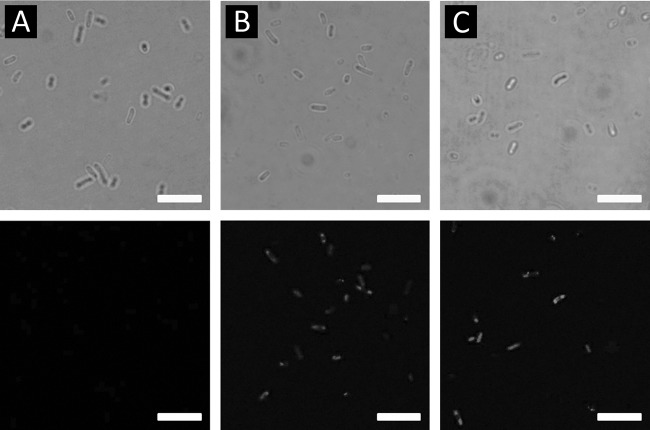
After transforming the E. coli XL10-Gold with the recombinant plasmids, for the confirmation of the fusion protein display on the cell surface, the whole-cell lysates and outer membrane proteins of recombinant XL10-Gold cells were analyzed by SDS-PAGE. The fusion protein could hardly be detected by the Coomassie blue staining due to the low expression level, therefore, Western blot analysis was carried out using 6×His tag antibody as the primary antibody (Abcam) and goat anti-mouse IgG–alkaline phosphatase conjugate (Sigma) was used as the secondary antibody for the immunodetection of the fusion protein (Fig. 2 and see Fig. S1 in the supplemental material). The outer membrane protein of truncated OmpW at amino acid 139 and the PerPA fusion protein have estimated sizes of ∼45.3 kDa calculated from the amino acid sequence. From the Western blot result, a protein band around that size was not detected on the Western blot results; instead, a protein band of ∼30 kDa was observed (see Fig. S1 in the supplemental material). The band with a size of 30 kDa seems to correspond to the PerPA, which is 30.44 kDa; therefore, we can infer that the OmpW truncated at amino acid 139 was eliminated or excised as the recombinant DNA was translated and expressed. The fact that a band at ∼30 kDa was observed in the whole-cell lysate and in the soluble fractions of recombinant E. coli XL10-Gold harboring pOW13F-PerPA2, pOW13F-PerPA3, and pOW13F-PerPA4 and not in the outer membrane fraction supports our theory. The fact that a protein band of ∼45.4 kDa was not detected shows that the Omp W-PerPA fusion protein was not successfully displayed on the surface of E. coli. The outer membrane protein size of truncated OmpW at amino acid 191 and the PerPA fusion protein have estimated sizes of ca. 51.0 kDa calculated from the amino acid sequence. The bands corresponding to the 51.0-kDa fusion protein were detected in whole-cell lysates and the outer membrane fraction of the recombinant XL10-Gold harboring pOW19F-PerPA2, pOW19F-PerPA3, and pOW19F-PerPA4, suggesting a successful display of fusion proteins on the cell surface (Fig. 2). Therefore, recombinant XL10-Gold harboring pOW19F-PerPA1, pOW19F-PerPA2, pOW19F-PerPA3, and pOW19F-PerPA4 were used in further studies. As another method to confirm the successful display of the OmpW-PerPA fusion protein on the surface of the E. coli cell surface, immunofluorescence microscopy was used. As shown in Fig. 3, recombinant E. coli harboring pOW19F-PerPA3 and pOW19F-PerPA4 became fluorescent due to the binding of the anti-6×His antibody, followed by binding of FITC-conjugated secondary antibody, indicating that the OmpW-PerPA fusion protein was successfully displayed on the surface of the E. coli. On the other hand, control E. coli XL10-Gold cells did not exhibit any fluorescence.
FIG 2.
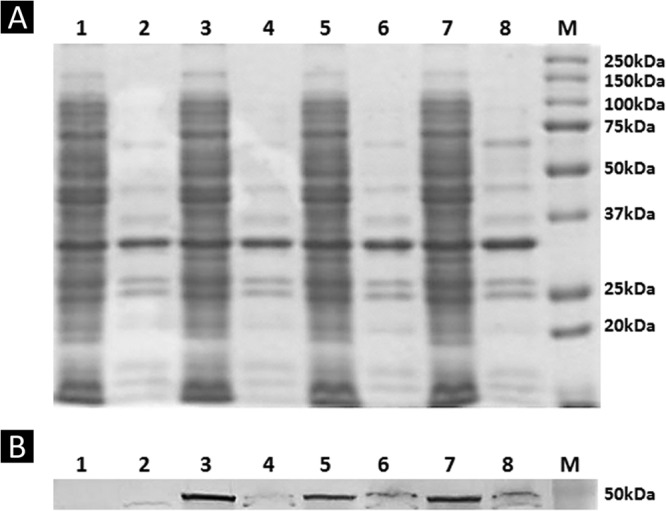
SDS-PAGE analysis and immunoblotting analysis. SDS-PAGE analysis (A) and Western blot analysis (B) of recombinant E. coli XL10-Gold cells expressing OmpW-PerPA fusion proteins were performed. Lane 1, whole-cell lysate of XL10-Gold harboring pOW19F-PerPA1; lane 2, outer membrane fraction of XL10-Gold harboring pOW19F-PerPA1; lane 3, whole-cell lysate of XL10-Gold harboring pOW19F-PerPA2; lane 4, outer membrane fraction of XL10-Gold harboring pOW19F-PerPA2; lane 5, whole-cell lysate of XL10-Gold harboring pOW19F-PerPA3; lane 6, outer membrane fraction of XL10-Gold harboring pOW19F-PerPA3; lane 7, whole-cell lysate of XL10-Gold harboring pOW19F-PerPA4; lane 8, outer membrane fraction of XL10-Gold harboring pOW19F-PerPA4; lane M, molecular mass standards.
FIG 3.
Immunofluorescence analysis using differential interference micrographs (upper row) and immunofluorescence micrographs (lower row) of XL10-Gold cells (A) and XL10-Gold cells harboring pOW19F-PerPA3 (B) or pOW19F-PerPA4 (C). Cells were incubated with mouse anti-His probe antibody, followed by probing with goat anti-mouse IgG-FITC conjugate. Scale bars, 5 μm.
Tyrosinase treatment and confirmation of tyrosine conversion to DOPA.
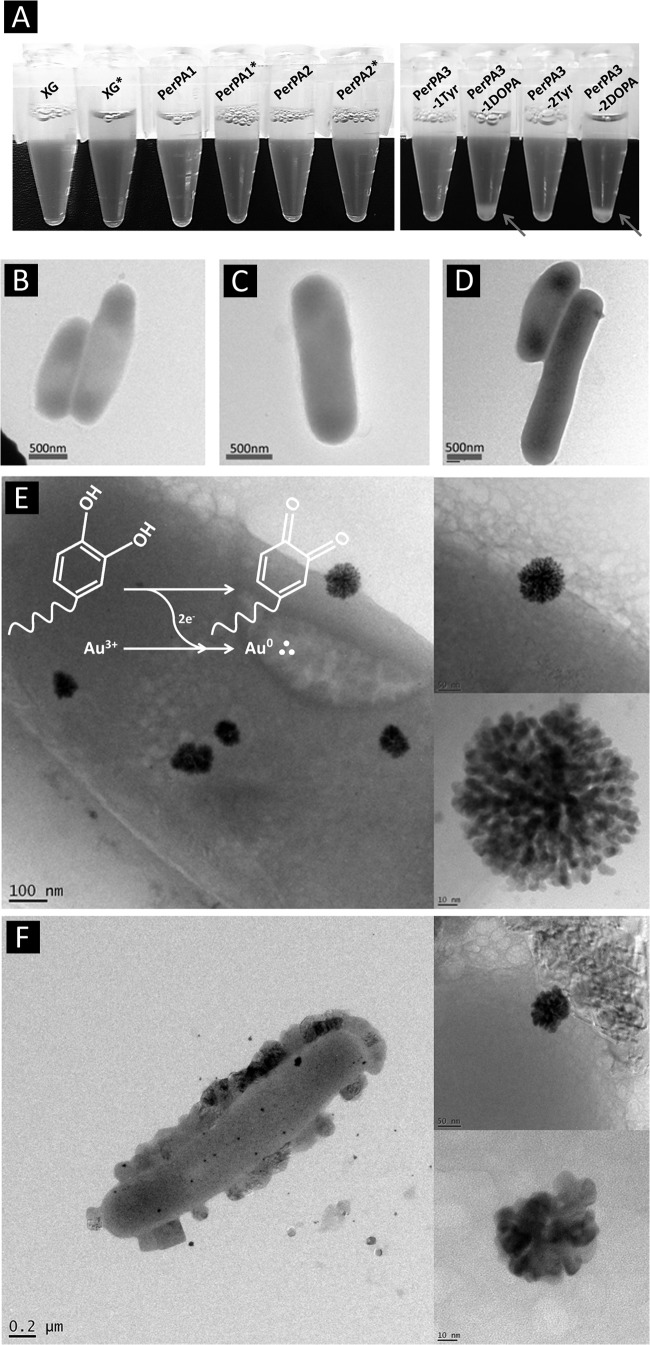
After we confirmed the successful display, the recombinant E. coli cells were treated with tyrosinase to convert the tyrosine reside on the surface to DOPA since DOPA is an unnatural amino acid and thus requires postmodification. XL10-Gold and recombinant E. coli XL10-Gold harboring pOW19F-PerPA1, pOW19F-PerPA2, pOW19F-PerPA3, and pOW19F-PerPA4 were treated with tyrosinase (500 U/ml). After the recombinant cells were treated with tyrosinase, cell aggregation occurred only with the recombinant E. coli harboring pOW19F-PerPA3 and pOW19F-PerPA4 (Fig. 4A) that have tyrosine residue on the surface, which indirectly confirms the conversion of the tyrosine residue to DOPA since molecules with a DOPA moiety tend to aggregate due to their specific cohesive interactions. In the other recombinant E. coli strains, cells that did not have tyrosine on the surface did not show any aggregation after tyrosinase treatment (Fig. 4A). As another method to confirm the successful conversion of tyrosine residue on the surface of recombinant E. coli XL10-Gold harboring plasmid pOW19F-PerPA3 and pOW19F-PerPA4 to DOPA, recombinant E. coli cells were incubated with tyrosinase with 10 mM HAuCl4 solution. It is well known that the DOPA moiety has a metal-reducing ability that reduces Au3+ to Au0+. Therefore, we hypothesize that gold nanoparticles will be reduced on the surface of E. coli where the DOPA moiety is present. E. coli XL10-Gold and recombinant E. coli XL10-Gold harboring pOW19F-PerPA1, pOW19F-PerPA2, pOW19F-PerPA3, and pOW19F-PerPA4 were incubated with tyrosinase and HAuCl4 for 1 h at 25°C with mild agitation. After the incubation, the cells were observed for the gold nanoparticle formation on the surfaces of the recombinant E. coli cells using TEM. In the controls, XL10-Gold, and recombinant E. coli strains harboring pOW19F-PerPA1 and pOW19F-PerPA2, no gold nanoparticle formation was observed on the surface (Fig. 4B to D). Also, when the gold nanoparticle formation experiments were performed with the recombinant E. coli harboring pOW19-PerPA3 and pOW19-PerPA4 without the tyrosinase treatment, no gold nanoparticle formation was observed (see Fig. S2 in the supplemental material). Gold nanoparticles with sizes ranging from 50 to 120 nm were only observed on the surfaces of the tyrosinase-treated recombinant E. coli XL10-Gold harboring pOW19F-PerPA3 and pOW19F-PerPA4, which had tyrosine residues on the surface before the tyrosinase treatment (Fig. 4E and F). The gold nanoparticle was confirmed by the energy dispersive X-ray spectrum taken by TEM (see Fig. S3 in the supplemental material). The gold nanoparticle formation therefore can be used to confirm the successful conversion of tyrosine residue on the cell surface to DOPA residue in the recombinant E. coli XL10-Gold harboring pOW19F-PerPA3 and pOW19F-PerPA4. Tyrosinase could not be responsible for the gold nanoparticle formation since all of the controls were also treated with tyrosinase. Hereafter, tyrosinase-treated recombinant E. coli XL10-Gold harboring pOW19F-PerPA3 and pOW19F-PerPA4 will be referred to as recombinant E. coli PerPA3-DOPA and PerPA4-DOPA, respectively.
FIG 4.
Confirmation of tyrosine-to-DOPA conversion. (A) Tyrosinase was added to E. coli XL10-Gold and recombinant E. coli cells harboring pOW19F-PerPA1, pOW19F-PerPA2, pOW19F-PerPA3, and pOW19F-PerPA4. Aggregation was observed only for tyrosinase-treated recombinant E. coli harboring pOW19F-PerPA3(PerPA3-1DOPA) and pOW19F-PerPA4 (PerPA4-2DOPA). ∗, tyrosinase treatment. (B to F) TEM analysis of XL10-Gold cells (B) and XL10-Gold cells harboring pOW19-PerPA1 (C), pOW19F-PerPA2 (D), pOW19F-PerpA3 (E), and pOW19F-PerPA4 (F) after incubation with tyrosinase and gold solution (HAuCl4). Gold nanoparticle formation was only observed in the PerPA3-DOPA and PerPA4-DOPA E. coli cells.
Adhesion property of mussel-inspired catecholamine-displayed engineered E. coli.
After we confirmed the successful display of the fusion protein on the surface and the successful conversion of tyrosine to DOPA, we explored the adhesive property of the engineered E. coli. In order to observe the adhesive property of our mussel-inspired catecholamine surface-displayed recombinant E. coli, cells were incubated with a variety of substrates and surfaces. First, microparticles were chosen as a substrate for recombinant E. coli attachment. Silnos 290, a silica microparticle with an average size of 9 μm, and glass beads with sizes between 30 and 50 μm were used. Recombinant E. coli cells were incubated with tyrosinase and the beads overnight. After incubation of XL10-Gold and recombinant E. coli XL10-Gold harboring pOW19F-PerPA1, pOW19F-PerPA2, pOW19F-PerPA3, and pOW19F-PerPA4 with tyrosinase and beads, the incubated beads were observed by SEM to examine the degree of adhesion to the beads. In the case of adhesion test to the Silnos 290 silica bead, tyrosinase-treated recombinant E. coli harboring pOW19F-PerPA3 and pOW19F-PerPA4 showed significant adhesion to the Silnos 290 silica beads (Fig. 5B and D). As shown in Fig. 5, recombinant E. coli XL-Gold harboring pOW19F-PerPA3 and pOW19F-PerPA4 without tyrosinase treatment (Fig. 5A and C) did not exhibit immobilization to the microparticle, confirming the adhesive property of the catecholamine-displayed recombinant E. coli PerPA3-DOPA and PerPA4-DOPA. In the other control experiments, where tyrosinase was treated to XL10-Gold, recombinant E. coli XL10-Gold harboring pOW19F-PerPA1 and pOW19F-PerPA2, no or only a few E. coli cells adhered to the surfaces of the Silnos 290 silica beads (see Fig. S4 in the supplemental material). Based on this observation, it is reasonable to conclude that the catecholamine-displayed recombinant E. coli has an adhesive property. As a different microparticle with different size for the adhesion test, glass bead of size of 30 to 50 μm was used. In the case of the adhesion test to the glass bead, recombinant E. coli PerPA3-DOPA and PerPA4-DOPA showed significant adhesion to the glass beads (Fig. 5F and H). As can be seen in Fig. 5, control recombinant E. coli harboring pOW19F-PerPA3 and pOW19F-PerPA4 without tyrosinase treatment showed little or no cell adhesion to the microparticles. In the other control experiments, in which tyrosinase was treated with XL10-Gold and recombinant E. coli harboring pOW19F-PerPA1 and pOW19F-PerPA2, some cells adhered to the surfaces of the glass beads, but the degree of adhesion was still significantly low compared to recombinant E. coli PerPA3-DOPA and PerPA4-DOPA (see Fig. S4 in the supplemental material). These results also confirm the adhesive property of mussel-inspired catecholamine-displayed recombinant E. coli. The engineered E. coli cells that adhered to various microparticles were counted (cells per surface area of the microparticles), as shown in Fig. 5I. When the adhesion levels of PerPA3-DOPA and PerPA4-DOPA were compared using the quantified data, no significant differences were observed. Because of this, we used recombinant E. coli PerPA4-DOPA throughout this study.
FIG 5.
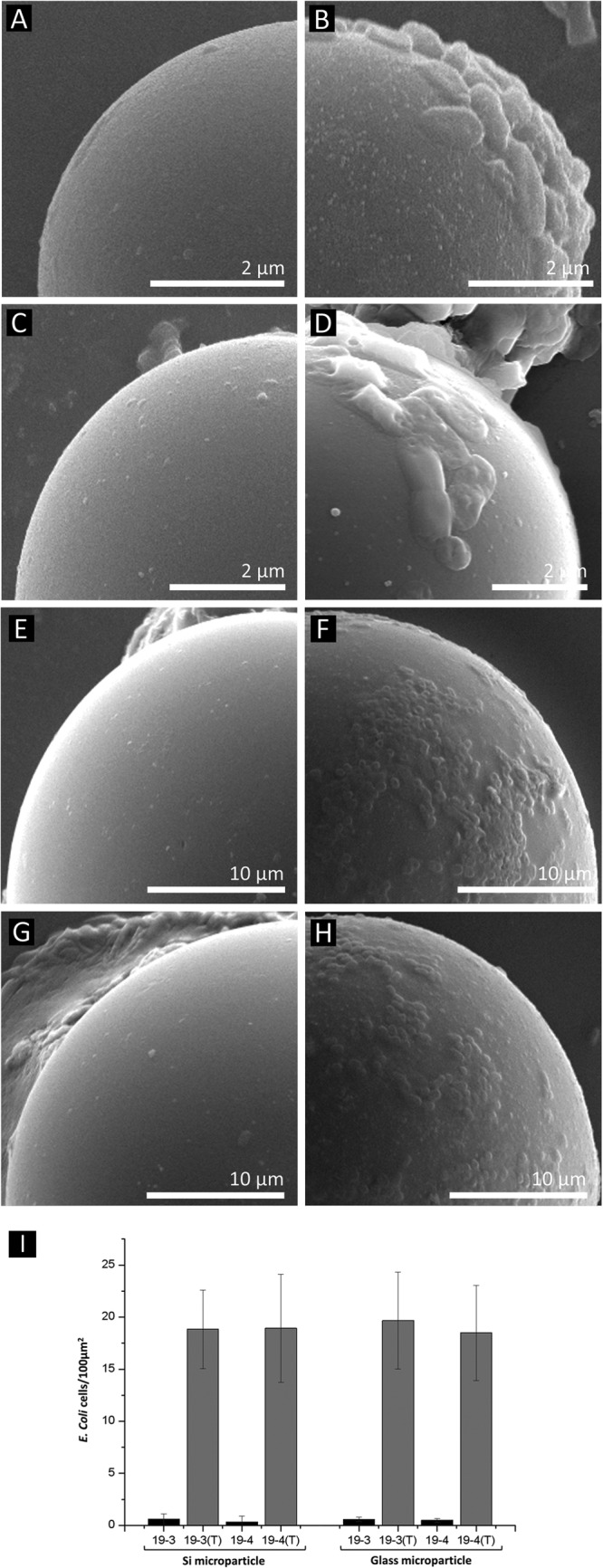
Cell adhesion test. A cell adhesion test was performed on silica microparticles (A, B, C, and D) and glass microparticles (E, F, G, and H). SEM analysis was used to observe the surfaces of the microparticles. The SEM images on the left are the control XL10-Gold harboring pOW19F-PerPA3 (A and E) and pOW19F-PerPA4 (C and G) without tyrosinase treatment. The SEM images on the right are tyrosinase-treated recombinant E. coli PerPA3-DOPA (B and F) and PerPA4-DOPA (D and H). Only catecholamine-displayed E. coli exhibited adhesive properties. (I) The cells adhered onto the microparticles were quantified and expressed as cells per area in the bar graph. 19-3 and 19-4 denote recombinant E. coli strains harboring pOW19F-PerPA3 and pOW19F-PerPA4, respectively. Tyrosinase-treated cells are labeled with a “(T)”.
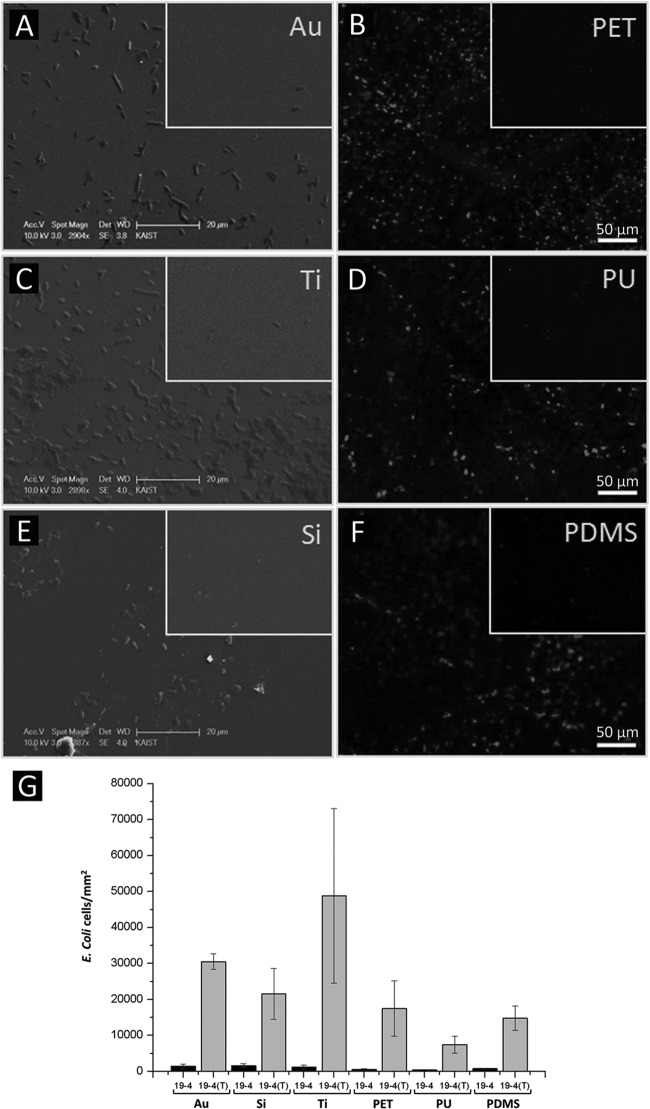
Two different types of surface substrates, metallic and polymeric, were used for the further adhesion test of the engineered recombinant E. coli. Au, Ti, and Si surfaces were chosen as the metallic surfaces for the cell adhesion experiment. These metallic surfaces were incubated with the recombinant E. coli XL10-Gold harboring pOW19F-PerPA4 with or without tyrosinase. In the control group without tyrosinase-treated recombinant E. coli, equal amounts of 10 mM phosphate buffer (pH 7.0) were added in order to counteract the addition of tyrosinase. After the recombinant E. coli XL10-Gold harboring pOW19F-PerPA4 with or without tyrosinase was incubated on various metallic surfaces overnight, SEM was used to observe the adhesion of recombinant E. coli on the surfaces. SEM images of surfaces incubated with tyrosinase-treated and untreated recombinant E. coli cells harboring pOW19F-PerPA4 were compared (Fig. 6A, C, and E). As shown in Fig. 6, recombinant E. coli PerPA4-DOPA, significantly adhered more to the metallic surfaces, which demonstrates the adhesive property of the engineered E. coli that has mussel-inspired catecholamine on the surface. To test the adhesion of catecholamine-displayed recombinant E. coli on different surfaces, PET, PU, and PDMS surfaces were selected as the polymeric surfaces for the cell adhesion experiment. After recombinant E. coli XL10-Gold harboring pOW19F-PerPA4 with or without tyrosinase was incubated on PET, PU, and PDMS surfaces, the surfaces were stained with acridine orange dye, and fluorescence microscopy was used to observe the adhesion of E. coli on the surfaces. Fluorescence images of surfaces incubated with tyrosinase-treated and untreated recombinant E. coli harboring pOW19F-PerPA4 were compared (Fig. 6B, D, and F). As shown in Fig. 6, recombinant E. coli PerPA4-DOPA adhered significantly more often to the PET, PU, and PDMS surfaces, which demonstrates the adhesive property of the catecholamine cell surface-displayed recombinant E. coli. The E. coli cells that adhered to various substrates were quantified per unit area (Fig. 6G). The number of cells that adhered to metallic surfaces can be directly counted from the SEM images. For the polymeric surfaces, we obtained a linear relationship between the fluorescence intensity and the cell number using many sampling areas, which was subsequently utilized to convert the fluorescence intensity into the number of cells. The quantification result showed a greater degree of adhesion to metallic surfaces than to polymeric surfaces. The numbers of attached E. coli cells per mm2 area were as follows: Ti (48,812 ± 24,232) > Au (30,541 ± 2,203) > Si (21,583 ± 7,042) > PET (17,490 ± 7,696) > PDMS (14,793 ± 3,363) > PU (7,457 ± 2,398), i.e., in descending order.
FIG 6.
Cell adhesion test on various substrates. A cell adhesion test was performed on the metallic surfaces Au (A), Ti (C), and Si (E) and on the polymeric surfaces PET (B), PU (D), and PDMS (F) using PerPA4-DOPA recombinant cells. Inset images show the control XL10-Gold harboring pOW19F-PerP4. (G) The adhered cells were quantified and expressed in cells per area in the bar graph. 19-4 denotes recombinant E. coli harboring pOW19F-PerPA4. Tyrosinase-treated cells are labeled with “(T)”. The scale bar for the inlet controls are the same as for the main images.
DISCUSSION
Cell attachment through microbial cell surface display is a fairly new method, and a few studies have been reported for the attachment of a limited range of material substrates such as cellulose, chitin, or gold (10, 36–38). Such limitation in surface adhesion is largely due to adhesion peptide identification by a screening process for a given material. In order to expand the type of materials to which the engineered E. coli is able to bind without labor-intensive, time-consuming screening processes, we turned our attention to interesting adhesive features of the mussel adhesive proteins. It is well known that the catecholamine group in MAPs plays a crucial role in the adhesion of mussels. For example, Mefp-5, which is responsible for robust, material-independent adhesion, shows extensive repeats of lysine and DOPA (44). Furthermore, synthetic versions of catecholamine polymers, such as poly(dopamine) (49), poly(norepinephrine) (50), poly(ethylenimine)-catechol (51), and chitosan-catechol (52, 53), have demonstrated material-independent adhesion in aqueous conditions. In the present study, the histidine-DOPA moiety was chosen for the preparation of sticky E. coli for the following two reasons: (i) the histidine-DOPA moiety is found in Mefp-5, which is expected to exhibit adhesive property similar to the catecholamines mentioned earlier, and (ii) the histidine-DOPA moiety was chosen for the ease of purification and detection of the surface-displayed peptide, since polyhistidine has been widely used for the affinity purification of genetically modified proteins. The catecholamine moiety exhibits various unique properties; it can form stable complexes with metal ions and undergo redox reactions to form covalent catechol-catechol/-amine/-thiol cross-links. One of the important properties of catechol moieties is the oxidative chemical conversion to quinone, and the quinone is highly reactive with amine and thiol compounds via Michael-type addition or Schiff base formation (49). This mechanism is able to achieve robust irreversible adhesion on organic substrates (54). Thus, the display of histidine-DOPA on the surface of E. coli can convert nonadhesive E. coli into adhesive bacteria that can adhere to the surfaces of virtually all types of materials. The aforementioned covalent-bond-forming mechanism can explain the result of E. coli aggregation shown in Fig. 4. The DOPA displayed on the cell surface can undergo catechol-catechol cross-linking with other displayed DOPA molecules and catechol-amine cross-linking between the displayed DOPA and other amine groups that might be present in intrinsic proteins on the E. coli surface. Another way to confirm the display of the sticky catecholamine moiety was to exploit the redox activity of catechol. As the catechol moiety is oxidized to catechol-quinone, electrons are released from the catechol, and gold and silver ions are spontaneously reduced by the released electrons, as shown in the scheme of Fig. 4E. Therefore, gold reduction occurred on the surface of the sticky E. coli can be expected. Successfully, gold nanoparticle formation was observed on the surface only when the catecholamine, histidine-DOPA, was displayed (PerPA3-DOPA and PerPA4-DOPA).
After the preparation of sticky E. coli, the spontaneous attachment of the cells was studied using various types of material surfaces. Since cell aggregation was already observed for the sticky E. coli displaying histidine-DOPA, E. coli is expected to have enhanced ability attaching spontaneously onto surfaces. In fact, microbials with a certain tendency of aggregation have been shown to attach to surfaces, although the adhesive force might not be as strong as the one used by mussel adhesive molecules (55). The mussel-inspired sticky E. coli exhibited significant adhesion to unprecedented various substrates, including silica, glass microparticles, and Au, Ti, Si, PET, PU, and PDMS surfaces compared to the control microbes. This observation is significant because a new type of a catecholamine amino acid moiety, DOPA-His, was shown to exhibit wet-resistant adhesive properties that can potentially be applied to any living organisms when adhesive properties are needed. Also, it is noteworthy that the sticky E. coli was attached to various substrates without applying any surface chemistry (e.g., chemical agents for coupling). In particular, glass and PDMA are challenging materials for adhesion. Catecholamine moieties adhere to virtually any substrates via coordination bonds, π-π stacking, electrostatics, and/or hydrogen bonding (49, 54). For surfaces such as Si and Ti wafers, their oxide forms, SiO2 and TiO2, are presented, onto which catechol forms a coordination bond with the −OH groups, and electrostatic interactions can be established by the interactions between amine groups along the catecholamine and the hydroxyl ions from the substrates. Catecholamine is stacked onto the gold substrates via the interactions between pi (π) electrons in catechol and the electrons in the conduction bands of the Au surfaces. PET and PU are composed of chains that contain phenyl rings that enable the displayed catecholamine moiety to interact with those substrates by π-π interactions. The adhesion of recombinant E. coli onto the PDMS can be attributed to the electrostatic interactions between the hydroxyl ions present on the surfaces of PDMS and the amine groups of catecholamine. Also, it has been suggested that the catechol also contributes to the surface adhesion, but this remains unclear. Thus, our results indicate that the sticky E. coli is likely to attach to a wide range of material surfaces. In contrast, previous experiments regarding cell immobilization achieved by surface-engineered peptide display showed that the engineered E. coli adhered only to limited types of materials that have binding affinity with the displayed peptide. Regarding the robustness of the sticky E. coli, our experimental condition was typically under agitation (1,000 rpm for microparticles and 220 rpm for other substrates), and washing steps were performed in which the cells remained attached, demonstrating the robustness of the sticky chemistry indirectly. These findings suggest that E. coli displaying mussel-inspired catecholamine can be used as a new method for cell immobilization on a broader range of substrates.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the a National Research Foundation of Korea (NRF) grant funded by the South Korean government (MSIP; no. 20090083525), the Biomedical Technology Development Program (2012-0006085 [H.L.]), and the Technology Development Program To Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012-C1AAA001 and NRF-2012-M1A2A2026556 [S.H.L.]) of the National Research Foundation of Korea. The World Premier Material Program of the Ministry of Knowledge and Economy also provided support.
Footnotes
Published ahead of print 11 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02223-13.
REFERENCES
- 1.Nigam JN. 2000. Continuous ethanol production from pineapple cannery waste using immobilized yeast cells. J. Biotechnol. 80:189–193. 10.1016/S0168-1656(00)00246-7 [DOI] [PubMed] [Google Scholar]
- 2.LeBaron HM, Mumma RO. 1999. Honeycutt. Continuous degradation of phenol at low concentration using immobilized Pseudomonas putida. Enzyme Microb. Technol. 25:530–536 [Google Scholar]
- 3.Chibata I, Tosa T. 1980. Immobilized microbial cells and their application. Trends Biochem. Sci. 5:88–90. 10.1016/0968-0004(80)90254-6 [DOI] [Google Scholar]
- 4.Chibata I, Tosa T, Sato T. 1986. Continuous production of l-aspartic acid. Appl. Biochem. Biotechnol. 13:231–240. 10.1007/BF02798461 [DOI] [Google Scholar]
- 5.Cheetham PSJ, Garrett C, Clark J. 1985. Isomaltulose production using immobilized cells. Biotechnol. Bioeng. 27:471–481. 10.1002/bit.260270412 [DOI] [PubMed] [Google Scholar]
- 6.Carvalho W, Silva SS, Converti A, Vitolo M. 2002. Metabolic behavior of immobilized Candida guilliermondii cells during batch xylitol production from sugarcane bagasse acid hydrolysate. Biotechnol. Bioeng. 79:165–169. 10.1002/bit.10319 [DOI] [PubMed] [Google Scholar]
- 7.Sato T, Nishida Y, Tosa T, Chibata I. 1979. Immobilization of Escherichia coli cells containing aspartase activity with κ-carrageenan. Enzymic properties and application for l-aspartic acid production. Biochim. Biophys. Acta 570:179–186. 10.1016/0005-2744(79)90212-2 [DOI] [PubMed] [Google Scholar]
- 8.Yun JW, Jung KH, Oh JW, Lee JH. 1990. Semibatch production of fructo-oligosaccharides from sucrose by immobilized cells of Aureobasidium pullulans. Appl. Biochem. Biotechnol. 25:299–308 [Google Scholar]
- 9.Yun JW, Jung KH, Jeon YJ, Lee JH. 1992. Continuous production of fructo-oligosaccharides from sucrose by immobilized cells of Aureobasidium pullulans. J. Microbiol. Biotechnol. 2:298–301 [Google Scholar]
- 10.Park TJ, Zheng S, Kang YJ, Lee SY. 2009. Development of a whole-cell biosensor by cell surface display of gold-binding polypeptide on the gold surface. FEMS Microbiol. Lett. 293:141–147. 10.1111/j.1574-6968.2009.01525.x [DOI] [PubMed] [Google Scholar]
- 11.Klein J, Ziehr H. 1990. Immobilization of microbial cells by adsorption. J. Biotechnol. 16:1–16. 10.1016/0168-1656(90)90061-F [DOI] [PubMed] [Google Scholar]
- 12.Barié N, Rapp M. 2001. Covalent bound sensing layers on surface acoustic wave (SAW). Biosens. Bioelectron. 16:979–987. 10.1016/S0956-5663(01)00198-1 [DOI] [PubMed] [Google Scholar]
- 13.Bickerstaff GF. 1997. Immobilization of enzymes and cells. Methods Biotechnol. 1:1–11 [Google Scholar]
- 14.Pilkington PH, Margaritis A, Mensour NA. 1998. Mass transfer characteristics of immobilized cells used in fermentation processes. Crit. Rev. Biotechnol. 18:237–255. 10.1080/0738-859891224239 [DOI] [Google Scholar]
- 15.Bae W, Chen W, Mulchandani A, Mehra RK. 2000. Enhanced bioaccumulation of heavy metals by bacterial cells displaying synthetic phytochelatins. Biotechnol. Bioeng. 70:518–524. [DOI] [PubMed] [Google Scholar]
- 16.Bae W, Mulchandani A, Chen W. 2002. Cell surface display of synthetic phytochelatins using ice nucleation protein for enhanced heavy metal bioaccumulation. J. Inorg. Biochem. 88:223–227. 10.1016/S0162-0134(01)00392-0 [DOI] [PubMed] [Google Scholar]
- 17.Wu CH, Mulchandani A, Chen W. 2008. Versatile microbial surface-display for environmental remediation and biofuels production. Trends Microbiol. 16:181–188. 10.1016/j.tim.2008.01.003 [DOI] [PubMed] [Google Scholar]
- 18.Xu Z, Lee SY. 1999. Display of polyhistidine peptides on the Escherichia coli cell surface by using outer membrane protein C as an anchoring motif. Appl. Environ. Microbiol. 65:5142–5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biondo R, da Silva FA, Vicente EJ, Souza Sarkis JE, Schenberg ACG. 2012. Synthetic phytochelatin surface display in Cupriavidus metallidurans CH34 for enhanced metals bioremediation. Environ. Sci. Technol. 46:8325–8332. 10.1021/es3006207 [DOI] [PubMed] [Google Scholar]
- 20.Kuroda K, Ueda M. 2011. Molecular design of the microbial cell surface toward the recovery of metal ions. Curr. Opin. Biotechnol. 22:427–433. 10.1016/j.copbio.2010.12.006 [DOI] [PubMed] [Google Scholar]
- 21.Richins RD, Kaneva I, Mulchandani A, Chen W. 1997. Biodegradation of organophosphorus pesticides by surface-expressed organophosphorus hydrolase. Nat. Biotechnol. 15:984–987. 10.1038/nbt1097-984 [DOI] [PubMed] [Google Scholar]
- 22.Lee SH, Choi J, Han M, Choi JH, Lee SY. 2005. Display of lipase on the cell surface of Escherichia coli using OprF as an anchor and its applications to enantioselective resolution in organic solvent. Biotechnol. Bioeng. 90:223–230. 10.1002/bit.20399 [DOI] [PubMed] [Google Scholar]
- 23.Schumacher SD, Hannemann F, Teese MG, Bernhardt R, Jose J. 2012. Autodisplay of functional CYP106A2 in Escherichia coli. J. Biotechnol. 161:104–112. 10.1016/j.jbiotec.2012.02.018 [DOI] [PubMed] [Google Scholar]
- 24.Schumacher SD, Jose J. 2012. Expression of active human P450 3A4 on the cell surface of Escherichia coli by autodisplay. J. Biotechnol. 161:113–120. 10.1016/j.jbiotec.2012.01.031 [DOI] [PubMed] [Google Scholar]
- 25.Yang TH, Kwon MA, Song JK, Pan JG, Rhee JS. 2010. Functional display of Pseudomonas and Burkholderia lipases using a translocator domain of EstA autotransporter on the cell surface of Escherichia coli. J. Biotechnol. 146:126–129. 10.1016/j.jbiotec.2010.01.022 [DOI] [PubMed] [Google Scholar]
- 26.Hasunuma T, Kondo A. 2012. Development of yeast cell factories for consolidated bioprocessing of lignocellulose to bioethanol through cell surface engineering. Biotechnol. Adv. 30:1207–1218. 10.1016/j.biotechadv.2011.10.011 [DOI] [PubMed] [Google Scholar]
- 27.Baek SH, Kim S, Lee K, Lee JK, Hahn JS. 2012. Cellulosic ethanol production by combination of cellulase-displaying yeast cells. Enzyme Microb. Technol. 51:366–372. 10.1016/j.enzmictec.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 28.Boder ET, Wittrup KD. 1997. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15:553–557. 10.1038/nbt0697-553 [DOI] [PubMed] [Google Scholar]
- 29.Westerlund-Wikström B, Tanskanen J, Virkola R, Hacker J, Lindberg M, Skurnik M, Korhonen TK. 1997. Functional expression of adhesive peptide as functions to Escherichia coli flagellin. Protein Eng. 10:1319–1326. 10.1093/protein/10.11.1319 [DOI] [PubMed] [Google Scholar]
- 30.Lee JS, Shin KS, Pan JG, Kim CJ. 2000. Surface-displayed viral antigens on Salmonella carrier vaccine. Nat. Biotechnol. 18:645–648. 10.1038/76494 [DOI] [PubMed] [Google Scholar]
- 31.Liljeqvist S, Samuelson P, Hansson M, Nguyen TN, Binz H, Stahl S. 1997. Surface display of the cholera toxin B subunit on Staphylococcus xylosus and Staphylococcus carnosus. Appl. Environ. Microbiol. 63:2481–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahbarizadeh F, Nouri M, Ahmadvand D, Nourollahi H, Iri-Sofla FJ, Farokhimanesh S. 2011. Cell surface display of Salmonella outer membrane protein A on Lactobacillus salivarius: a first step towards food-grade live vaccine against Salmonella infections. Food Biotechnol. 25:151–164. 10.1080/08905436.2011.576569 [DOI] [Google Scholar]
- 33.Martineau P, Charbit A, Leclerc C, Werts C, O'Callaghan D, Hofnung M. 1991. A genetic system to elicit and monitor antipeptide antibodies without peptide synthesis. Biotechnology 9:170–172. 10.1038/nbt0291-170 [DOI] [PubMed] [Google Scholar]
- 34.Dhillon JK, Drew PD, Porter AJ. 1999. Bacterial surface display of an anti-pollutant antibody fragment. Lett. Appl. Microbiol. 28:350–354. 10.1046/j.1365-2672.1999.00552.x [DOI] [PubMed] [Google Scholar]
- 35.Shibasaki S, Ueda M, Ye K, Shimizu K, Kamasawa N, Osumi M, Tanaka A. 2001. Creation of cell surface-engineered yeast that display different fluorescent proteins in response to the glucose concentration. Appl. Microbiol. Biotechnol. 57:528–533. 10.1007/s002530100767 [DOI] [PubMed] [Google Scholar]
- 36.Francisco JA, Stathopoulos C, Warren RAJ, Kilburn DG, Georgiou G. 1993. Specific adhesion and hydrolysis of cellulose by intact Escherichia coli cells expressing surface-anchored cellulase or cellulose binding domains. Nat. Biotechnol. 11:491–495. 10.1038/nbt0493-491 [DOI] [PubMed] [Google Scholar]
- 37.Wang AA, Mulchandani A, Chen W. 2001. Whole-cell immobilization using cell surface-exposed cellulose-binding domain. Biotechnol. Prog. 17:407–411. 10.1021/bp0100225 [DOI] [PubMed] [Google Scholar]
- 38.Wang J-Y, Chao Y-P. 2006. Immobilization of cells with surface-displayed chitin-binding domain. Appl. Environ. Microbiol. 72:927–931. 10.1128/AEM.72.1.927-931.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strausberg RL, Andersen DM, Filpula DR, Finkelman M, Link RP, McCandliss R, Orndorff SA, Strausberg SL, Wei T. 1989. Development of a microbial system for production of mussel adhesive protein, p 453–464 In Hemingway RW, Conner AH, Branham SJ. (ed), Adhesives from renewable resources. ACS Symp. Ser., vol 385 American Chemical Society, Washington, DC [Google Scholar]
- 40.Kitamura M, Kawakami K, Nakamura N, Tsumoto K, Uchiyama H, Ueda Y, Kumagai I, Nakaya T. 1999. Expression of a model peptide of a marine mussel adhesive protein in Escherichia coli and characterization of its structural and functional properties. J. Polym. Sci. Part A Polym. Chem. 37:729–736 [Google Scholar]
- 41.Hwang DS, Yoo HJ, Jun JH, Moon WK, Cha HJ. 2004. Expression of functional recombinant mussel adhesive protein Mgfp-5 in Escherichia coli. Appl. Environ. Microbiol. 70:3352–3359. 10.1128/AEM.70.6.3352-3359.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang DS, Gim Y, Cha HJ. 2005. Expression of functional recombinant mussel adhesive protein type 3A in Escherichia coli. Biotechnol. Prog. 21:965–970. 10.1021/bp050014e [DOI] [PubMed] [Google Scholar]
- 43.Hwang DS, Gim Y, Yoo HJ, Cha HJ. 2007. Practical recombinant hybrid mussel bioadhesive fp-151. Biomaterials 28:3560–3568. 10.1016/j.biomaterials.2007.04.039 [DOI] [PubMed] [Google Scholar]
- 44.Waite JH, Qin X. 2001. Polyphosphoprotein from the adhesive pads of Mytilus edulis. Biochemistry 40:2887–2893. 10.1021/bi002718x [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 46.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593–656. 10.1128/MMBR.67.4.593-656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Busch W, Saier MH. 2004. The IUBMB-endorsed transporter classification system. Mol. Biotechnol. 27:253–262. 10.1385/MB:27:3:253 [DOI] [PubMed] [Google Scholar]
- 48.Hong H, Patel DR, Tamm LK, van den Berg B. 2006. The outer membrane protein OmpW forms an eight-stranded β-barrel with a hydrophobic channel. J. Biol. Chem. 281:7568–7577. 10.1074/jbc.M512365200 [DOI] [PubMed] [Google Scholar]
- 49.Lee H, Dellatore SM, Miller WM, Messersmith PB. 2007. Mussel-inspired surface chemistry for multifunctional coatings. Science 318:426–430. 10.1126/science.1147241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang SM, Rho J, Choi IS, Messersmith PB, Lee H. 2009. Norepinephrine: material-independent, multifunctional surface modification reagent. J. Am. Chem. Soc. 131:13224–13225. 10.1021/ja905183k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim E, Song IT, Lee S, Kim JS, Lee H, Jang JH. 2012. Drawing sticky adeno-associated viruses on surfaces for spatially patterned gene expression. Angew. Chem. Int. Ed. Engl. 51:5598–5601. 10.1002/anie.201201495 [DOI] [PubMed] [Google Scholar]
- 52.Ryu JH, Lee Y, Kong WH, Kim TG, Park TG, Lee H. 2011. Catechol-functionalized chitosan/pluronic hydrogels for tissue adhesives and hemostatic materials. Biomacromolecules 12:2653–2659. 10.1021/bm200464x [DOI] [PubMed] [Google Scholar]
- 53.Kim K, Ryu JH, Lee DY, Lee H. 2013. Bio-inspired catechol conjugation converts water-insoluble chitosan into a highly water-soluble, adhesive chitosan derivative for hydrogels and LBL assembly. Biomater. Sci. 1:783–790. 10.1039/c3bm00004d [DOI] [PubMed] [Google Scholar]
- 54.Lee H, Scherer NF, Messersmith PB. 2006. Single-molecule mechanics of mussel adhesion. Proc. Natl. Acad. Sci. U. S. A. 103:12999–13003. 10.1073/pnas.0605552103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burdman S, Okon Y, Jurkevitch E. 2000. Surface characteristics of Azospirillum brasilense in relation to cell aggregation and attachment to plant roots. Crit. Rev. Microbiol. 26:91–110. 10.1080/10408410091154200 [DOI] [PubMed] [Google Scholar]
- 56.Brosius J, Holy A. 1984. Regulation of ribosomal promoters with a synthetic lac operator. Proc. Natl. Acad. Sci. U. S. A. 81:6929–6933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hernan RA, Hui HL, Andracki ME, Noble RW, Sligar SG, Walder JA, Walder RY. 1992. Human hemoglobin expression in Escherichia coli: importance of optimal codon usage. Biochemistry 31:8619–8628 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.