Abstract
In order to survive a temperature downshift, bacteria have to sense the changing environment and adjust their metabolism and structure. Two-component signal transduction systems (TCSs) play a central role in sensing and responding to many different environmental stimuli. Although the nonproteolytic (group II) Clostridium botulinum represents a major hazard in chilled foods, the cold adaption mechanisms of group II C. botulinum organisms are not known. Here, we show that the CLO3403/CLO3404 TCS of C. botulinum E1 Beluga is involved in the cold shock response and growth at 12°C. Cold shock induced the expression of the genes encoding the histidine kinase (clo3403) and the response regulator (clo3404) by more than 100-fold after 5 h relative to their expression in a nonshocked culture at the corresponding time point. The involvement of CLO3403/CLO3404 in growth at low temperature was demonstrated by impaired growth of the insertional clo3403 and clo3404 knockout mutants at 12°C compared to the growth of the wild-type culture. Additionally, the inactivation of clo3403 had a negative effect on motility. The growth efficiency at 12°C of the TCS mutants and the motility of the kinase mutants were restored by introducing a plasmid harboring the operon of the CLO3403/CLO3404 TCS. The results suggest that the CLO3403/CLO3404 TCS is important for the cold tolerance of C. botulinum E1 Beluga.
INTRODUCTION
Group II (nonproteolytic) Clostridium botulinum type E is a notorious food-borne pathogen which is mainly found in aquatic environments in cold regions of the world (e.g., Northern Europe, Japan, Canada, Alaska, and Greenland) (1, 2), with a high prevalence of spores in the Baltic Sea (3). Type E botulism is usually caused through consumption of contaminated fish or seafood products such as vacuum-packed smoked fish, salted fish, and fermented marine mammals (4–11). Concerning modern food processing, C. botulinum type E is the principal food safety hazard in refrigerated, minimally processed packaged foods of aquatic origin (12). The mild heat treatments used in the production of these foods are not sufficient to eliminate all spores, and the limited use of salt and preservatives, as well as vacuum packaging, may support growth and toxin production by C. botulinum. Most importantly, thermal control is often not sufficient to prevent food poisoning, as some strains of group II C. botulinum are able to grow and produce toxin at temperatures as low as 3°C (13–15).
The mechanisms of cold shock response and growth at low temperature have been thoroughly studied in the model organisms Escherichia coli and Bacillus subtilis (16–18) but are still unknown in the psychrotrophic C. botulinum type E. The cold shock proteins (Csps) are a universal group of cold-induced proteins occurring in a wide range of bacteria and other organisms (17, 19). While group I C. botulinum strains carry Csp genes in their genome (20), none were found in the genomes of C. botulinum type E strains (21), which raises the question of alternative strategies for coping with cold by C. botulinum type E. It is known that two-component signal transduction systems (TCSs) play a role in the cold tolerance and cold shock response of various bacteria (22–26), and recent studies have shown the importance of two TCSs in the cold adaption of the mesophilic group I C. botulinum (27, 28). Typical TCSs consist of a sensory histidine kinase and a response regulator. The histidine kinase is often embedded in the cell membrane, sensing a designated stimulus with its N-terminal sensor domain. When stimulated, the C-terminal histidine residue in the cytoplasmic kinase domain becomes autophosphorylated and the phosphoryl group is transferred to the N-terminal aspartate residue in the receiver domain of the cognate intracellular response regulator. Furthermore, the phosphoryl group is transferred to the C-terminal output domain. Response regulators control the transcription of their target genes through specific DNA-binding activity. TCSs respond to many environmental stimuli, including pH, osmolarity, oxidative stress, and temperature (29–31). The best-characterized bacterial TCS contributing to cold tolerance is the DesK/DesR of B. subtilis. DesK/DesR responds to decreased membrane fluidity caused by a temperature downshift and induces the transcription of des. The des-encoded desaturase restores membrane fluidity at low temperatures by increasing the desaturation of the acyl chains of membrane phospholipids (22, 32–37). Whether and how TCSs in group II C. botulinum organisms respond to low temperatures is not known.
Here, we show that the CLO3403/CLO3404 TCS of C. botulinum E1 Beluga is involved in the cold shock response and growth at 12°C. The relative mRNA levels of clo3403 and clo3404 were induced after a cold shock (temperature downshift from 30°C to 12°C), and insertional knockout mutants with disrupted clo3403 and clo3404 showed a cold-sensitive phenotype. The mutations were successfully complemented by introducing a plasmid harboring the operon of the CLO3403/CLO3404 TCS.
MATERIALS AND METHODS
Strains, plasmids, and culture.
The bacterial strains and plasmids used in this study are listed in Table 1. C. botulinum E1 Beluga was used as a wild-type (WT) strain and for constructing insertional clo3403 and clo3404 knockout mutants. C. botulinum E1 Beluga was initially isolated from a food-borne botulism outbreak in Alaska that was associated with beluga flippers (38). C. botulinum strains were grown at 30°C or 12°C in tryptone-peptone-glucose-yeast extract (TPGY) broth (50 g/liter tryptone, 5 g/liter peptone, 4 g/liter glucose, and 20 g/liter yeast extract [Difco, Becton Dickinson, Sparks, MD] plus 1 g/liter sodium thioglycolate [Merck KGaA, Darmstadt, Germany]) or on TPGY agar (1%) supplemented with thiamphenicol (15 μg/ml), cycloserine (250 μg/ml), and erythromycin (2.5 μg/ml) when appropriate (Sigma-Aldrich, St. Louis, MO). Culturing was performed under strictly anaerobic conditions in an anaerobic work station (MG1000 anaerobic work station; Don Whitley Scientific Ltd., Shipley, United Kingdom) with an atmosphere of 85% N2, 10% CO2 and 5% H2. Agar plates and broth were deoxygenated before use by anaerobic storage for 48 h or by boiling for 15 min, respectively. E. coli was grown aerobically in Luria-Bertani (LB) broth (Difco) or on LB agar plates at 37°C. When appropriate, chloramphenicol (25 μg/ml) and kanamycin (30 μg/ml) were used for selection (Sigma-Aldrich).
TABLE 1.
Strains and plasmids used in this study
| Strain description or plasmid | Description | Source or reference |
|---|---|---|
| C. botulinum strains | ||
| E1 Beluga | Wild-type strain (WT) | IFRa |
| clo3403s | Insertional mutation of clo3403 in sense direction at position 630-631 | This study |
| clo3403a | Insertional mutation of clo3403 in antisense direction at position 227-228 | This study |
| clo3404s | Insertional mutation of clo3404 in sense direction at position 384-385 | This study |
| clo3404a | Insertional mutation of clo3404 in antisense direction at position 505-506 | This study |
| WT-pMTL82151 | WT strain with empty plasmid | This study |
| clo3403s-pMTL82151 | clo3403s mutant with empty plasmid | This study |
| clo3403a-pMTL82151 | clo3403a mutant with empty plasmid | This study |
| clo3404s-pMTL82151 | clo3404s mutant with empty plasmid | This study |
| clo3404a-pMTL82151 | clo3404a mutant with empty plasmid | This study |
| WT-pMTL82151::clo3404-clo3401 | WT strain with complementation plasmid | This study |
| clo3403s-pMTL82151::clo3404-clo3401 | clo3403s mutant with complementation plasmid | This study |
| clo3403a-pMTL82151::clo3404-clo3401 | clo3403a mutant with complementation plasmid | This study |
| clo3404s-pMTL82151::clo3404-clo3401 | clo3404s mutant with complementation plasmid | This study |
| clo3404a-pMTL82151::clo3404-clo3401 | clo3404a mutant with complementation plasmid | This study |
| E. coli strains | ||
| CA434 | Conjugation donor | 44 |
| NEB 5-alpha competent | Cloning strain | New England BioLabs |
| Plasmids | ||
| pMTL007C-E2 | ClosTron vector for mutagenesis | 42 |
| pMTL007C-E2::clo3403-631s | pMTL007C-E2 targeting clo3403 in sense direction | This study (DNA2.0) |
| pMTL007C-E2::clo3403-228a | pMTL007C-E2 targeting clo3403 in antisense direction | This study (DNA2.0) |
| pMTL007C-E2::clo3404-385s | pMTL007C-E2 targeting clo3404 in sense direction | This study (DNA2.0) |
| pMTL007C-E2::clo3404-506a | pMTL007C-E2 targeting clo3404 in antisense direction | This study (DNA2.0) |
| pMTL82151 | Plasmid vector | 45 |
| pMTL82151::clo3404-clo3401 | Complementation plasmid containing the operon of the CLO3403/CLO3404 TCS | This study |
Culture Collection of the Institute of Food Research, Norwich, United Kingdom.
All experiments were performed in triplicate. Three single colonies of each strain (biological replicates) were separately inoculated into 10 ml TPGY broth and incubated at 30°C for 24 h. The cultures were diluted (1:100) into fresh TPGY (10 ml) and incubated at 30°C for 16 h. The second overnight culture (ONC) was used as the starting culture for the experiments.
Cold shock.
The C. botulinum E1 Beluga ONC was diluted (1:100) into 200 ml of fresh TPGY broth and incubated at 30°C. Growth was tracked by measurement of the optical density at 600 nm (OD600). At an OD600 of 1.0 to 1.2, the culture was divided into two 100-ml cultures and one culture was transferred to an ice-water bath. The temperature of the culture was followed by mixing the culture with a sterile thermometer. Reaching 12°C implied cold shock. The culture was incubated at 12°C for 24 h. The other culture represented the nonshocked control and was incubated at 30°C over the same time period. Samples for RNA isolation were taken from cold-shocked and nonshocked cultures immediately before (time zero [t0]) and 1 min, 30 min, 1 h, 2 h, 5 h, and 24 h after the cold shock (Fig. 1). A volume of 250 μl of an ethanol (99%)–phenol (95%) solution (9:1) was added to 1,250 μl of each sample. The mixture was incubated for 30 min at 4°C, and the cells were pelleted by centrifugation (5 min and 8,000 rpm at 4°C). The cell pellets were stored at −70°C. RNA isolation was performed within 30 days.
FIG 1.
Cold shock of C. botulinum E1 Beluga. The C. botulinum E1 Beluga ONC was diluted (1:100) into 200 ml of fresh TPGY broth and incubated at 30°C (solid line). During mid-exponential growth phase, half of the culture was cold shocked (indicated by arrow) and growth was continued at 12°C (dotted line). Samples for RT-qPCR analysis were collected from both cultures before (t0) and 1 min, 30 min, 1 h, 2 h, 5 h, and 24 h after the cold shock (vertical dashed lines). Error bars represent standard deviations of three biological replicates.
RNA isolation.
Frozen cell pellets were resuspended in 250 μl of lysis buffer containing lysozyme (50 mg/ml; Sigma-Aldrich), mutanolysin (1 U/ml; Sigma-Aldrich), and Tris-EDTA (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) at a ratio of 2:1:1. The mixture was incubated at 37°C for 2 h. RNA was isolated using the RNeasy minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Genomic DNA was removed by DNase treatment (RNase-free DNase set; Qiagen) and, after the final RNA elution, by a second DNase treatment (DNA-free; Ambion, Life Technologies Corporation, Carlsbad, CA) (39). The RNA concentration was determined using the NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA). The RNA quality was checked with the Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA).
RT.
The cDNA was synthesized using the DyNAmo cDNA synthesis kit (Thermo Scientific). A total of 800 ng of RNA was reverse transcribed according to the manufacturer's instructions, with the exception of a predenaturation step for 5 min at 65°C to reduce secondary structures of the RNA template. For each RNA sample, two reactions were performed. DNA contamination was controlled by reactions with no reverse transcriptase. Synthesized cDNA was stored at −20°C before use in quantitative real-time reverse transcription-PCR (RT-qPCR).
RT-qPCR.
For RT-qPCR, the Maxima SYBR green qPCR master mix (2×; Thermo Scientific) was used according to the manufacturer's instructions. The PCR mixtures consisted of 1× Maxima SYBR green qPCR master mix, 0.3 μM each primer (Table 2), and 4 μl of 10−2-fold-diluted cDNA (for clo3403 and clo3404) or 10−6-fold-diluted cDNA (for 16S rrn) in a total volume of 25 μl. Two PCRs for each cDNA sample (PCR replicates) were performed in the Rotor-Gene 3000 real-time thermal cycler (Qiagen). The run comprised one cycle at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s, and a final cycle at 60°C for 60 s. The PCR efficiencies were determined for each primer pair with serially diluted cDNA pools as the templates. The efficiencies were 0.97 for 16S rrn, 0.98 for clo3403, and 0.92 for clo3404. Melting curve analysis was performed after each run from 60°C to 98°C in 0.5°C steps for 10 s to confirm the specificity of the PCR products. Gene expression was normalized to the expression of 16S rrn, and the relative gene expression values were calculated as described by Pfaffl (40). The two-tailed Student's t test was used to compare fold changes of relative gene expression.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′→3′) | Use |
|---|---|---|
| clo3403-F | GACAAGGCATTCCGAAAGAA | RT-qPCR |
| clo3403-R | CAGTAGTTCCATTCAAACTGCTTA | RT-qPCR |
| clo3404-F | AGCAGAATGTGGGATAGATGC | RT-qPCR |
| clo3404-R | CAGCACCAAGCTGAAGACCT | RT-qPCR |
| 16S rrn-F | AGGAGCAATCCGCTATGAGA | RT-qPCR |
| 16S rrn-R | GTGCAATATTCCCCACTGCT | RT-qPCR |
| clo3403-F2 | AGAAAACGTTTTGAATGAGATGAG | Screening for mutants |
| clo3403-R2 | CCTAAAATTGCTGCTAATGGAG | Screening for mutants |
| clo3404-F2 | GTCTTCAGCTTGGTGCTGAC | Screening for mutants |
| clo3404-R2 | ACTATATCCCCATATCTTGTCCAA | Screening for mutants |
| EBS Universal | CGAAATTAGAAACTTGCGTTCAGTAAAC | Screening for mutants |
| clo3404-F-NotI | NNNNNNGCGGCCGCGGAAATCGCATTCCTTTCAT | Construction of complementation plasmid |
| clo3401-R-NdeI | NNNNNNCATATGTGTATTTTATTGTTTGGTTGTGTTTTT | Construction of complementation plasmid |
| Intron-F | TGGCAATGATAGCGAAACAA | Southern blotting |
| Intron-R | GGTACCGCCTTGTTCACATT | Southern blotting |
Construction of insertional clo3403 and clo3404 knockout mutants with the ClosTron.
The TCS genes clo3403 and clo3404 were disrupted by inserting a mobile group II intron from Lactococcus lactis (Ll.ltrB) using the ClosTron system (41, 42). Target sites were designed using the intron design tool, based on the Perutka algorithm (43), at ClosTron.com (42). For mutagenesis of each gene, two target sites were designed for inserting the intron in the sense orientation (s) of the target gene and in the antisense orientation (a), resulting in two mutants (sense and antisense) for each gene. For clo3403, the insertions were targeted in sense orientation to position 630-631 (clo3403s) and in antisense orientation to position 227-228 (clo3403a). The insertions to clo3404 were targeted at position 384-385 (clo3404s) and at position 505-506 (clo3404a). Retargeted pMTL007C-E2 plasmids were ordered from DNA2.0, Inc. (Menlo Park, CA), and transformed by a heat shock into chemically competent E. coli CA434 (44). Transformants were grown in 5 ml of LB broth supplemented with kanamycin (30 μg/ml) and chloramphenicol (25 μg/ml). Retargeted plasmids were then conjugated into C. botulinum E1 Beluga on TPGY agar for 6 h. Transconjugants were screened on TPGY agar containing thiamphenicol (15 μg/ml) and cycloserine (250 μg/ml) and streaked on TPGY agar containing erythromycin (2.5 μg/ml) for the selection of integrants. Insertion in the correct site and orientation were confirmed by PCR using target-specific intron-flanking primers and the intron-binding EBS Universal primer. Single intron insertion was verified by Southern blotting as described previously (23).
Construction of the complementation plasmid.
A 4,660-bp fragment containing clo3404, clo3403, clo3402, and clo3401, 457 bp upstream from clo3404 and 154 bp downstream from clo3401, was amplified. This fragment represents the operon containing the TCS genes (clo3403 and clo3404), an ABC transporter gene (clo3401), a gene encoding a conserved hypothetical protein with a predicted transmembrane helix (clo3402), and the putative natural promoter of the operon. The fragment was purified with the Gene JET PCR purification kit (Thermo Scientific), digested with NotI and NdeI, and ligated into pMTL82151 (45, 46). The resulting pMTL82151::clo3404-clo3401 was sequenced, transformed into E. coli CA434, and further conjugated into the mutants and the WT strain. As a vector control, the empty pMTL82151 plasmid was conjugated into the mutants and the WT.
Growth experiments at 12°C and 30°C.
The C. botulinum ONC was diluted (1:100) into 10 ml of anaerobic TPGY broth. Volumes of 350 μl of this dilution were pipetted into four wells (technical replicates) of a 100-well microtiter plate and incubated at 12°C for 10 days or at 30°C for 2 days in the Bioscreen C microbiology reader (Oy Growth Curves AB, Helsinki, Finland) in an anaerobic workstation under continuous shaking (47, 48). The OD600 values were measured automatically at 15-min intervals, and growth curves were constructed by plotting the OD600 values against time. The maximum growth rate (OD600 value per hour) of each culture was calculated by fitting the growth curve to the Baranyi and Roberts model (49) using the DMFit Microsoft Excel add-in program (Institute of Food Research, Norwich, United Kingdom). The mean maximum growth rate of the four technical replicates was determined. Three biological replicates were used for each strain. The two-tailed Student's t test was used to compare maximum growth rates.
Motility assay.
Single colonies, grown on TPGY agar, were stab inoculated with a sterile inoculating loop into tubes containing TPGY agar (0.3%). The tubes were incubated at 12°C or 30°C under anaerobic conditions. Growth and motility were followed over 7 days and 2 days, respectively. Three biological replicates were used for each strain.
Electron microscopy.
The C. botulinum ONCs were diluted (1:100) into 50 ml of fresh TPGY broth in duplicate and incubated at 12°C or 30°C. One ml of each mid-exponential-phase culture was sampled and diluted (1:1) with 5% glutaraldehyde (Sigma-Aldrich) for 2 h at room temperature to fix the cells. The samples were washed with sterile water, and 3.5-μl amounts were incubated on carbon-coated grids (Electron Microscopy Sciences, Hatfield, PA) for 2 min. The liquid was carefully removed. Cells were stained by adding 3.5 μl phosphotungstic acid (1%) on the grid. Negative stain was incubated for 15 s and carefully removed. The grids were analyzed with the Tecnai 12 transmission electron microscope (Philips Electron Optics, Holland) in the electron microscopy unit of the Institute of Biotechnology, University of Helsinki.
RESULTS
Expression of clo3403 and clo3404 is induced after a cold shock.
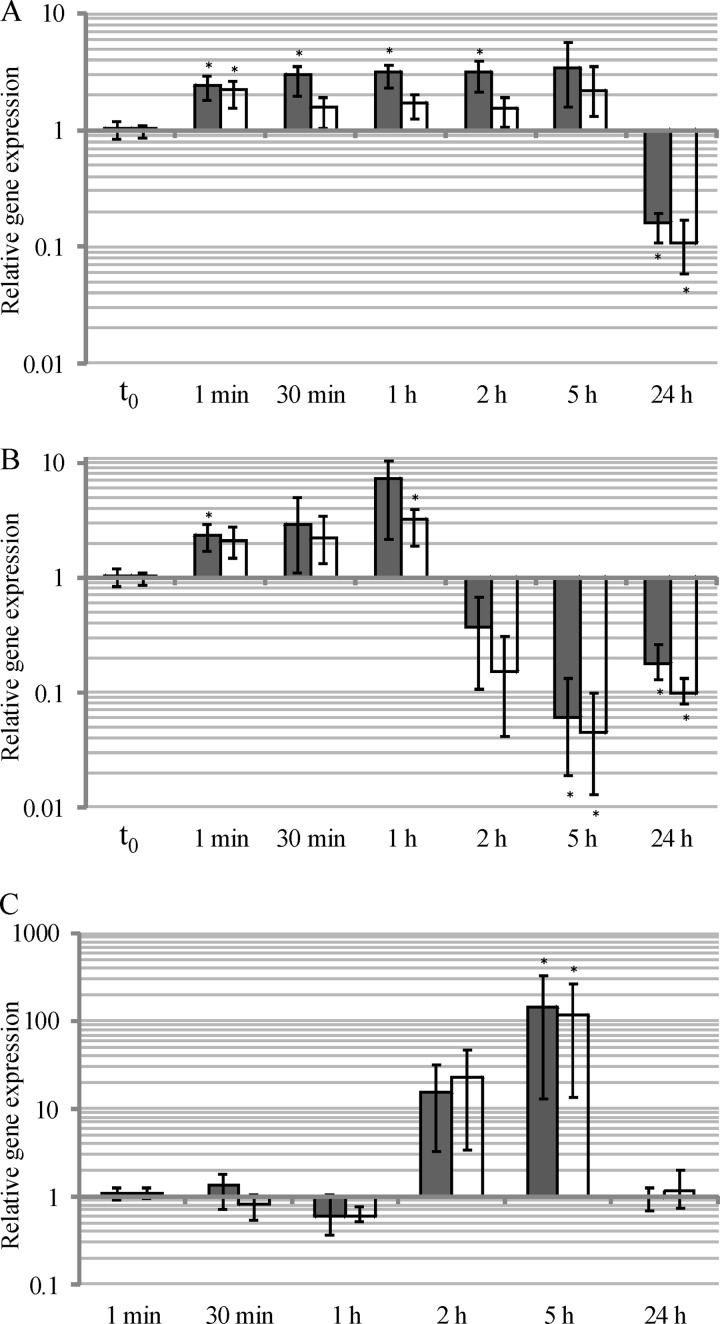
The expression of the CLO3403/CLO3404 TCS in the cold shock response of C. botulinum E1 Beluga was studied by measuring the relative mRNA levels of clo3403 and clo3404 immediately before (t0) and 1 min, 30 min, 1 h, 2 h, 5 h, and 24 h after a cold shock (temperature downshift from 30°C to 12°C) using RT-qPCR. A nonshocked culture served as a control (Fig. 1). The cold shock induced the expression of the histidine kinase (clo3403) and response regulator (clo3404) genes 1.5- to 3.4-fold in the first 5 h relative to their expression at time point t0 (Fig. 2A). At 24 h after the cold shock, the expression of both genes (clo3403 and clo3404) was significantly downregulated relative to their expression at t0 (P < 0.05; the expression ratios were 0.16 and 0.11, respectively). The nonshocked control culture showed downregulation of the TCS genes already at 2 h and 5 h (expression ratios of 0.37 to 0.04) (Fig. 2B). Calibrating the results for the cold-shocked culture to the results for the nonshocked culture revealed more than 10-fold (not significant) and more than 100-fold (P < 0.05) induction of clo3403 and clo3404 mRNA expression in the cold-shocked culture at 2 h and 5 h, respectively (Fig. 2C).
FIG 2.
Expression of clo3403 (dark gray) and clo3404 (white) is induced after a cold shock. Relative expression levels of clo3403 and clo3404 in cold-shocked culture calibrated to t0 (A), in nonshocked culture calibrated to t0 (B), and in cold-shocked culture calibrated to nonshocked culture at corresponding time points (C). Gene expression was normalized to 16S rrn. Error bars indicate the minimum and maximum values of three biological replicates. *, P < 0.05.
Insertional inactivation of clo3403 and clo3404.
To examine the role of the CLO3403/CLO3404 TCS in growth at low temperature (12°C), insertional knockout mutants were constructed. The genes encoding clo3403 and clo3404 were disrupted by inserting a mobile group II intron from Lactococcus lactis (Ll.ltrB) using the ClosTron system. Insertion in the correct site was confirmed by PCR using primers flanking the intron insertion site (Fig. 3A). The resulting clo3403s (sense) and clo3403a (antisense) and clo3404s and clo3404a mutants were confirmed by amplifying a 2,615-bp and a 2,311-bp fragment, respectively. Correct orientation of the insertion was analyzed by PCR using a gene-specific primer and the intron-binding EBS Universal primer (Fig. 3B). The clo3403s and clo3403a mutants were confirmed by obtaining a 679-bp and an 837-bp fragment, respectively, and the clo3404s and clo3404a mutants by obtaining a 365-bp and a 323-bp fragment, respectively. Single intron insertion was shown by Southern blotting using an intron-specific probe (Fig. 3C). HindIII-digested genomic DNA of each mutant gave one band on the Southern blot. Digested WT DNA was the negative control. To our knowledge, the clo3403 and clo3404 mutants constructed are the first confirmed and reported insertional knockout mutants of group II C. botulinum.
FIG 3.

Insertional inactivation of clo3403 and clo3404 in sense (s) and antisense (a) orientation using the ClosTron. (A and B) Insertion in the correct site was confirmed by PCR using primers flanking the intron insertion site (A), and insertion in correct orientation was confirmed by PCR using a gene-specific primer and the intron-binding EBS Universal primer (B). (C) Southern blot analysis of HindIII-digested genomic DNA from the WT and clo3403 and clo3404 mutants using an intron-specific probe.
The clo3403 and clo3404 mutants showed impaired growth at 12°C.
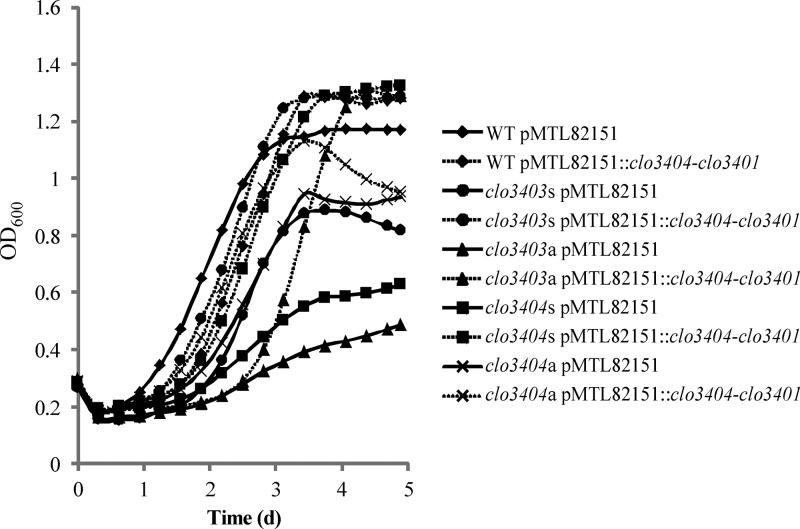
The TCS mutants showed impaired growth at 12°C compared to the growth of the WT strain (Fig. 4). Both clo3403 mutants exhibited an extended lag phase, and the clo3403s mutant was unable to reach the WT level of cell density. The clo3404s mutant showed a prolonged lag phase but reached the WT level of cell density, whereas the growth of the clo3404a mutant was slightly impaired compared to that of the WT, with a delayed (∼8 h) entry into stationary growth phase and slightly reduced maximum cell density. Determining the maximum growth rates revealed significantly (P < 0.01) reduced maximum growth rates of all four TCS mutants relative to the growth of the WT at 12°C (Table 3). These negative effects on growth were not observed in cultures growing at the optimum temperature (30°C). Additionally, the role of the CLO3403/CLO3404 TCS in growth at low temperature was confirmed by successful complementation of the mutations (Fig. 5). The mutants harboring pMTL82151::clo3404-clo3401 showed improved growth (increased maximum cell densities and increased maximum growth rates) at 12°C compared to the growth of the vector control strains, and their maximum growth rates were fully restored to WT levels. The results suggest that the functional CLO3403/CLO3404 TCS is needed for efficient growth at low temperature.
FIG 4.
The clo3403 and clo3404 mutants show impaired growth at 12°C. Growth curves of WT and clo3403s, clo3403a, clo3404s, and clo3404a strains at 12°C and 30°C. Volumes of 350 μl of diluted ONCs (1:100 diluted into fresh TPGY broth) were incubated at 12°C or 30°C in the Bioscreen C microbiology reader in an anaerobic workstation. OD600 values were measured automatically at 15-min intervals. Error bars represent standard deviations of three biological replicates.
TABLE 3.
Maximum growth rates of WT, complementation, and vector control strains at optimum and/or low temperaturea
| Strain description | Mean maximum growth rate (OD600/h) ± SD at: |
|
|---|---|---|
| 12°C | 30°C | |
| WT | 0.045 ± 0.000 | 0.313 ± 0.028 |
| clo3403s | 0.019 ± 0.009b | 0.279 ± 0.001 |
| clo3403a | 0.041 ± 0.001b | 0.315 ± 0.013 |
| clo3404s | 0.033 ± 0.003b | 0.341 ± 0.029 |
| clo3404a | 0.033 ± 0.004b | 0.322 ± 0.017 |
| WT-pMTL82151 | 0.026 ± 0.005 | |
| WT-pMTL82151::clo3404-clo3401 | 0.026 ± 0.001 | |
| clo3403s-pMTL82151 | 0.021 ± 0.006 | |
| clo3403s-pMTL82151::clo3404-clo3401 | 0.028 ± 0.009 | |
| clo3403a-pMTL82151 | 0.005 ± 0.002 | |
| clo3403a-pMTL82151::clo3404-clo3401 | 0.032 ± 0.001c | |
| clo3404s-pMTL82151 | 0.008 ± 0.004 | |
| clo3404s-pMTL82151::clo3404-clo3401 | 0.026 ± 0.003c | |
| clo3404a-pMTL82151 | 0.018 ± 0.000 | |
| clo3404a-pMTL82151::clo3404-clo3401 | 0.024 ± 0.002c | |
WT, clo3403s, clo3403a, clo3404s, clo3404a, and respective complementation and vector control strains were grown at 12°C and/or 30°C.
Result is significantly (P < 0.01) different from the value for the WT.
Value is significantly (P < 0.01) increased compared to the value for the respective vector control.
FIG 5.
Improved growth of TCS mutants at 12°C by introducing pMTL82151::clo3404-clo3401. Growth curves of WT, clo3403s, clo3403a, clo3404s, and clo3404a strains harboring the empty vector control (pMTL82151) or the complementation plasmid (pMTL82151::clo3404-clo3401) at 12°C. Volumes of 350 μl of diluted ONCs (1:100 diluted into fresh TPGY broth) were incubated at 12°C in the Bioscreen C microbiology reader in an anaerobic workstation. OD600 values were measured automatically at 15-min intervals. Curves represent the means of three biological replicates.
The clo3403s mutant is not motile.
The motility of C. botulinum E1 Beluga and the TCS mutants was studied by stab inoculation into TPGY agar (0.3%) and culture at 12°C and 30°C. The WT, clo3404s, and clo3404a strains showed clear motility at 12°C and 30°C after 2 days of incubation. The clo3403s mutant was not motile at all at either temperature, and clo3403a showed reduced motility at 12°C compared to that of the WT (Fig. 6). Hence, C. botulinum E1 Beluga is motile at optimum and low temperature, whereas inactivation of clo3403 has a negative effect on motility. Inactivation of clo3404 does not affect motility.
FIG 6.
The clo3403s mutant is nonmotile, and its motility is restored by introducing pMTL82151::clo3404-clo3401. Motility assays of WT, clo3403s, clo3403a, clo3404s, and clo3404a (left) strains at 12°C and 30°C and of WT, clo3403s, and clo3403a strains harboring pMTL82151 or pMTL82151::clo3404-clo3401 (right) at 12°C and 30°C. Single colonies were stab inoculated into tubes containing TPGY agar (0.3%) and incubated at 12°C or 30°C. Images were taken after 2 days of incubation, with the exception of the image showing complementation at 12°C taken after 4 days of incubation.
The motility of the clo3403s mutant was fully restored at 12°C and 30°C by introducing the complementation plasmid pMTL82151::clo3404-clo3401 (Fig. 6), and the motility of the clo3403a mutant at 12°C was restored by introducing pMTL82151::clo3404-clo3401. Surprisingly, the WT carrying the empty vector control was nonmotile at 12°C, but this could be an effect of antibiotic pressure and/or plasmid replication effort under cold stress, as the WT was shown to be motile in the absence of the antibiotic and the vector. Nevertheless, all strains carrying the complementation plasmid were fully motile at both temperatures, affirming the possible role of the CLO3403/CLO3404 TCS, or at least the histidine kinase CLO3403, in the motility of C. botulinum E1 Beluga.
The clo3403s mutant does not form flagella.
To study whether disturbed flagellum synthesis could explain the nonmotility of the clo3403s mutant, we performed electron microscopy analysis. The clo3403s mutant did not form flagella during mid-exponential growth phase at 12°C and 30°C, whereas the WT formed flagella at both temperatures (Fig. 7). Similar to the WT, the clo3404s, clo3404a, and clo3403a mutants were motile and produced flagella at 12°C and 30°C (data not shown). Thus, the nonmotile phenotype of clo3403s is most probably linked to its missing flagella. The complemented clo3403s mutant was motile and produced flagella (Fig. 8).
FIG 7.
The clo3403s mutant does not form flagella. Electron microscopy images of WT and clo3403s strains. The C. botulinum ONCs were diluted (1:100) into 50 ml of fresh TPGY broth in duplicate and incubated at 12°C or 30°C. Samples for transmission electron microscopy were taken during mid-exponential growth phase at 12°C and 30°C.
FIG 8.
The flagellum formation of the clo3403s mutant is restored by introducing pMTL82151::clo3404-clo3401. Electron microscopy images of the clo3403s mutant harboring pMTL82151::clo3404-clo3401 and the clo3403s mutant harboring pMTL82151. The C. botulinum ONCs were diluted (1:100) into 50 ml of fresh TPGY broth and incubated at 30°C. Samples for transmission electron microscopy were taken during mid-exponential growth phase.
Electron microscopy analysis also revealed that the cell size (3 to 5 μm) and cell shape are very similar among the tested strains during mid-exponential growth phase at both temperatures, confirming that the differential optical densities measured between the WT and mutant cultures at 12°C were growth related and not caused by varying cell sizes.
DISCUSSION
Determining the relative expression levels of the TCS genes clo3403 and clo3404 revealed induction of expression for at least 5 h after a cold shock, whereas in the nonshocked culture grown at the optimum temperature (30°C), the relative mRNA levels of clo3403 and clo3404 were downregulated at 2 h and 5 h. The prolonged induction of the TCS genes after the cold shock is suggested to be a response to the temperature downshift. TCSs responding to low temperature on the transcriptional level have been reported in group I C. botulinum organisms (27, 28). Similar to the current study, the previous works showed that the relative expression levels of cbo0366/cbo0365 and cbo2306/cbo2307 in group I C. botulinum organisms was most increased 5 h after the cold shock when calibrating the cold-shocked culture to the nonshocked culture. Thus, we suggest that TCSs in group I and group II C. botulinum are responding to low temperature in a similar time range. In Yersinia pseudotuberculosis, 44 TCS genes were significantly induced under cold stress (23). It is thus plausible that C. botulinum is also using multiple TCSs to respond to cold. At 24 h after the cold shock, the expression of the TCS genes clo3403 and clo3404 was downregulated to similar levels in cold-shocked and nonshocked cultures, which is most likely a growth-phase-dependent effect.
The involvement of the CLO3403/CLO3404 TCS in growth at low temperature was demonstrated by impaired growth (significantly reduced maximum growth rates) of the clo3403s, clo3403a, clo3404s, and clo3404a mutants at 12°C compared to the WT growth. At the optimum growth temperature, no growth difference was observed between the mutants and the WT. Comparison between the mutants with sense and antisense mutations of clo3403 or clo3404 showed that the insertion orientation had an effect on the intensity of the cold-sensitive phenotype and motility in the clo3403 mutants. As discussed earlier, these differences may be linked to polar effects of the strong erm promoter within the intron or, more likely, to the presence of antisense RNA (28). Insertion of the intron in the sense orientation within clo3403 could lead to antisense RNA production against the upstream regulator gene, clo3404, thus not only disrupting clo3403 but also attenuating clo3404 and thereby knocking out the entire TCS. We hypothesize that the more-drastic phenotypes observed for clo3403s than for clo3403a were due to such a total silencing of this TCS. A disturbed flagellum synthesis most probably affected the cold tolerance of this mutant. Another explanation for the phenotypic differences between the sense and antisense mutants could be the different intron insertion sites used for constructing the mutants. In the clo3403s mutant, the insertion site was in the C-terminal HAMP domain, whereas the antisense mutant had the insertion in the N-terminal sensing domain. Therefore, insertion site-dependent functional differences cannot be excluded. Nevertheless, all of the mutants grew less efficiently than the WT at 12°C, and this finding was further confirmed by successful complementation of the mutations. Loss of growth efficiency at low temperature (15°C) was also reported for the group I C. botulinum TCS mutants with mutations of cbo0366, cbo0365, cbo2306, and cbo2307 (27, 28), which affirms the general role of TCSs in the cold tolerance of C. botulinum. A protein blast analysis suggests that neither CBO0366/CBO0365 nor CBO2306/CBO2307 is the closest homologue to CLO3403/CLO3404. The closest homologous proteins to CLO3403/CLO3404 are CLH0370/CLH0369 and CLLA0382/CLLA0381 in C. botulinum Alaska E43 and C. botulinum Eklund 17B, respectively. Whether those TCSs are also important for the cold tolerance of C. botulinum Alaska E43 and C. botulinum Eklund 17B must be tested. More research is needed to solve the mechanism of TCSs behind the cold adaptation of this dangerous pathogen.
The clo3403s histidine kinase mutant, the most cold-sensitive mutant among those tested, appeared nonmotile and nonflagellated, and the clo3403a mutant showed reduced motility at 12°C. In contrast, the clo3404s and clo3404a mutants were fully motile, which suggests that the response regulator CLO3404 alone is not essential for motility or that other regulators may compensate for it. As discussed above, the nonmotile phenotype of the clo3403s mutant could be explained by a combined effect of the disrupted clo3403 and the antisense RNA-attenuated clo3404, silencing the entire TCS. How the blocking of this TCS affects flagellation remains to be characterized. The operon encoding most of the flagellar genes is located 400 kb downstream from the TCS genes, making polar effects on motility highly unlikely. The motility of clo3403s and clo3403a, however, was fully restored by introducing the complementation plasmid, which affirms the role of the histidine kinase in motility. Furthermore, motility seems to be important for the growth of C. botulinum E1 Beluga at the low but not at the optimum temperature. This result is consistent with previous hypotheses that motility and cold growth are linked in bacteria (23, 50, 51). In Y. pseudotuberculosis, the CheA/CheY TCS was shown to regulate motility and to be involved in the cold stress response. cheA and cheY were highly induced at 3°C, and a functional cheA was important for growth at cold temperature (23). Similar findings were made in Listeria monocytogenes, in which the response regulators DegU and GmaR played a role in the temperature-dependent regulation of flagellar genes (52–54) and the flagellar proteins FlhA and MotA were needed for efficient growth at low temperature (50). Moreover, in E. coli, the flagellar genes fliC, fliH, and fliN were highly induced after a temperature downshift (55).
In conclusion, we demonstrate that the CLO3403/CLO3404 TCS of the psychrotrophic food-borne pathogen C. botulinum type E is required for efficient growth at low temperature and that a cold shock induces the expression of the TCS for at least 5 h. We also showed that inactivation of the histidine kinase gene clo3403 has a negative effect on motility. The results were confirmed by successful complementation of the mutations.
ACKNOWLEDGMENTS
The work was performed in the Finnish Centre of Excellence in Microbial Food Safety Research and supported by the Academy of Finland (grants 141140 and 118602), the Finnish Graduate School on Applied Bioscience, the Finnish Foundation of Veterinary Research, the European Community's Seventh Framework Program FP7/2007-2013 (grant 237942), and the Doctoral Program of the Faculty of Veterinary Medicine of the University of Helsinki.
We thank Esa Penttinen, Hanna Korpunen, and Heimo Tasanen for excellent technical assistance.
Footnotes
Published ahead of print 1 November 2013
REFERENCES
- 1.Huss HH. 1980. Distribution of Clostridium botulinum. Appl. Environ. Microbiol. 39:764–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peck MW. 2009. Biology and genomic analysis of Clostridium botulinum. Adv. Microb. Physiol. 55:183–265. 10.1016/S0065-2911(09)05503-9 [DOI] [PubMed] [Google Scholar]
- 3.Hielm S, Hyytiä E, Andersin AB, Korkeala H. 1998. A high prevalence of Clostridium botulinum type E in Finnish freshwater and Baltic Sea sediment samples. J. Appl. Microbiol. 84:133–137. 10.1046/j.1365-2672.1997.00331.x [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention 2001. Botulism outbreak associated with eating fermented food—Alaska, 2001. MMWR Morb. Mortal. Wkly. Rep. 50:680–682 http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5032a2.htm [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention 2003. Outbreak of botulism type E associated with eating a beached whale—Western Alaska, July 2002. MMWR Morb. Mortal. Wkly. Rep. 52:24–26 http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5202a2.htm [PubMed] [Google Scholar]
- 6.Dawar M, Moody L, Martin JD, Fung C, Isaac-Renton J, Patrick DM. 2002. Two outbreaks of botulism associated with fermented salmon roe—British Columbia, August 2001. Can. Commun. Dis. Rep. 28:45–49 [PubMed] [Google Scholar]
- 7.Korkeala H, Stengel G, Hyytiä E, Vogelsang B, Bohl A, Wihlman H, Pakkala P, Hielm S. 1998. Type E botulism associated with vacuum-packaged hot-smoked whitefish. Int. J. Food Microbiol. 43:1–5. 10.1016/S0168-1605(98)00080-4 [DOI] [PubMed] [Google Scholar]
- 8.Leclair D, Fung J, Isaac-Renton JL, Proulx JF, May-Hadford J, Ellis A, Ashton E, Bekal S, Farber JM, Blanchfield B, Austin JW. 2013. Foodborne botulism in Canada, 1985-2005. Emerg. Infect. Dis. 19:961–968. 10.3201/eid1906.120873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Proulx JF, Milor-Roy V, Austin J. 1997. Four outbreaks of botulism in Ungava Bay, Nunavik, Quebec. Can. Commun. Dis. Rep. 23:30–32 [PubMed] [Google Scholar]
- 10.Peck MW. 2006. Clostridium botulinum and the safety of minimally heated, chilled foods: an emerging issue? J. Appl. Microbiol. 101:556–570. 10.1111/j.1365-2672.2006.02987.x [DOI] [PubMed] [Google Scholar]
- 11.Lindström M, Vuorela M, Hinderink K, Korkeala H, Dahlsten E, Raahenmaa M, Kuusi M. 2006. Botulism associated with vacuum-packed smoked whitefish in Finland, June-July 2006. Euro Surveill. 11:E060720.3 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=3004 [DOI] [PubMed] [Google Scholar]
- 12.Lindström M, Kiviniemi K, Korkeala H. 2006. Hazard and control of group II (non-proteolytic) Clostridium botulinum in modern food processing. Int. J. Food Microbiol. 108:92–104. 10.1016/j.ijfoodmicro.2005.11.003 [DOI] [PubMed] [Google Scholar]
- 13.Eklund MW, Wieler DI, Poysky FT. 1967. Outgrowth and toxin production of nonproteolytic type B Clostridium botulinum at 3.3 to 5.6°C. J. Bacteriol. 93:1461–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham AF, Mason DR, Maxwell FJ, Peck MW. 1997. Effect of pH and NaCl on growth from spores of non-proteolytic Clostridium botulinum at chill temperature. Lett. Appl. Microbiol. 24:95–100. 10.1046/j.1472-765X.1997.00348.x [DOI] [PubMed] [Google Scholar]
- 15.Schmidt CF, Lechowich RV, Folinazzo JF. 1961. Growth and toxin production by type E Clostridium Botulinum below 40°F. J. Food Sci. 26:626–630. 10.1111/j.1365-2621.1961.tb00807.x [DOI] [Google Scholar]
- 16.Phadtare S. 2004. Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 6:125–136 [PubMed] [Google Scholar]
- 17.Ermolenko DN, Makhatadze GI. 2002. Bacterial cold-shock proteins. Cell. Mol. Life Sci. 59:1902–1913. 10.1007/PL00012513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palonen E, Lindström M, Korkeala H. 2010. Adaptation of enteropathogenic Yersinia to low growth temperature. Crit. Rev. Microbiol. 36:54–67. 10.3109/10408410903382581 [DOI] [PubMed] [Google Scholar]
- 19.Graumann PL, Marahiel MA. 1998. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 23:286–290. 10.1016/S0968-0004(98)01255-9 [DOI] [PubMed] [Google Scholar]
- 20.Söderholm H, Lindström M, Somervuo P, Heap J, Minton N, Linden J, Korkeala H. 2011. cspB encodes a major cold shock protein in Clostridium botulinum ATCC 3502. Int. J. Food Microbiol. 146:23–30. 10.1016/j.ijfoodmicro.2011.01.033 [DOI] [PubMed] [Google Scholar]
- 21.Söderholm H, Jaakkola K, Somervuo P, Laine P, Auvinen P, Paulin L, Lindström M, Korkeala H. 2013. Comparison of Clostridium botulinum genomes shows the absence of cold shock protein coding genes in type E neurotoxin producing strains. Botulinum J. 2:189–207. 10.1504/TBJ.2013.055662 [DOI] [Google Scholar]
- 22.Aguilar PS, Hernandez-Arriaga AM, Cybulski LE, Erazo AC, de Mendoza D. 2001. Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 20:1681–1691. 10.1093/emboj/20.7.1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palonen E, Lindström M, Karttunen R, Somervuo P, Korkeala H. 2011. Expression of signal transduction system encoding genes of Yersinia pseudotuberculosis IP32953 at 28°C and 3°C. PLoS One 6:e25063. 10.1371/journal.pone.0025063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ullrich M, Peñaloza-Vázquez A, Bailey AM, Bender CL. 1995. A modified two-component regulatory system is involved in temperature-dependent biosynthesis of the Pseudomonas syringae phytotoxin coronatine. J. Bacteriol. 177:6160–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hesami S, Metcalf DS, Lumsden JS, Macinnes JI. 2011. Identification of cold-temperature-regulated genes in Flavobacterium psychrophilum. Appl. Environ. Microbiol. 77:1593–1600. 10.1128/AEM.01717-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan YC, Hu Y, Chaturongakul S, Files KD, Bowen BM, Boor KJ, Wiedmann M. 2008. Contributions of two-component regulatory systems, alternative sigma factors, and negative regulators to Listeria monocytogenes cold adaptation and cold growth. J. Food Prot. 71:420–425 http://www.ingentaconnect.com/content/iafp/jfp/2008/00000071/00000002/art00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindström M, Dahlsten E, Söderholm H, Selby K, Somervuo P, Heap JT, Minton NP, Korkeala H. 2012. Involvement of two-component system CBO0366/CBO0365 in the cold shock response and growth of group I (proteolytic) Clostridium botulinum ATCC 3502 at low temperatures. Appl. Environ. Microbiol. 78:5466–5470. 10.1128/AEM.00555-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derman Y, Isokallio M, Lindström M, Korkeala H. 2013. The two-component system CBO2306/CBO2307 is important for cold adaptation of Clostridium botulinum ATCC 3502. Int. J. Food Microbiol. 167:87–91. 10.1016/j.ijfoodmicro.2013.06.004 [DOI] [PubMed] [Google Scholar]
- 29.Hoch JA. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165–170. 10.1016/S1369-5274(00)00070-9 [DOI] [PubMed] [Google Scholar]
- 30.Laub MT, Goulian M. 2007. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41:121–145. 10.1146/annurev.genet.41.042007.170548 [DOI] [PubMed] [Google Scholar]
- 31.Mitrophanov AY, Groisman EA. 2008. Signal integration in bacterial two-component regulatory systems. Genes Dev. 22:2601–2611. 10.1101/gad.1700308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aguilar PS, Cronan JE, Jr, de Mendoza D. 1998. A Bacillus subtilis gene induced by cold shock encodes a membrane phospholipid desaturase. J. Bacteriol. 180:2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aguilar PS, Lopez P, de Mendoza D. 1999. Transcriptional control of the low-temperature-inducible des gene, encoding the delta5 desaturase of Bacillus subtilis. J. Bacteriol. 181:7028–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beranová J, Mansilla MC, de Mendoza D, Elhottová D, Konopásek I. 2010. Differences in cold adaptation of Bacillus subtilis under anaerobic and aerobic conditions. J. Bacteriol. 192:4164–4171. 10.1128/JB.00384-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cybulski LE, Martín M, Mansilla MC, Fernández A, de Mendoza D. 2010. Membrane thickness cue for cold sensing in a bacterium. Curr. Biol. 20:1539–1544. 10.1016/j.cub.2010.06.074 [DOI] [PubMed] [Google Scholar]
- 36.Mansilla MC, de Mendoza D. 2005. The Bacillus subtilis desaturase: a model to understand phospholipid modification and temperature sensing. Arch. Microbiol. 183:229–235. 10.1007/s00203-005-0759-8 [DOI] [PubMed] [Google Scholar]
- 37.Martín M, de Mendoza D. 2013. Regulation of Bacillus subtilis DesK thermosensor by lipids. Biochem. J. 451:269–275. 10.1042/BJ20121825 [DOI] [PubMed] [Google Scholar]
- 38.Dolman CE, Chang H. 1953. The epidemiology and pathogenesis of type E and fishborne botulism. Can. J. Public Health 44:231–244 [PubMed] [Google Scholar]
- 39.Kirk DG, Dahlsten E, Zhang Z, Korkeala H, Lindström M. 2012. Involvement of Clostridium botulinum ATCC 3502 sigma factor K in early-stage sporulation. Appl. Environ. Microbiol. 78:4590–4596. 10.1128/AEM.00304-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP. 2007. The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 70:452–464. 10.1016/j.mimet.2007.05.021 [DOI] [PubMed] [Google Scholar]
- 42.Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, Scott JC, Minton NP. 2010. The ClosTron: mutagenesis in Clostridium refined and streamlined. J. Microbiol. Methods 80:49–55. 10.1016/j.mimet.2009.10.018 [DOI] [PubMed] [Google Scholar]
- 43.Perutka J, Wang W, Goerlitz D, Lambowitz AM. 2004. Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J. Mol. Biol. 336:421–439. 10.1016/j.jmb.2003.12.009 [DOI] [PubMed] [Google Scholar]
- 44.Purdy D, O'Keeffe TA, Elmore M, Herbert M, McLeod A, Bokori-Brown M, Ostrowski A, Minton NP. 2002. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 46:439–452. 10.1046/j.1365-2958.2002.03134.x [DOI] [PubMed] [Google Scholar]
- 45.Heap JT, Pennington OJ, Cartman ST, Minton NP. 2009. A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods 78:79–85. 10.1016/j.mimet.2009.05.004 [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z, Korkeala H, Dahlsten E, Sahala E, Heap JT, Minton NP, Lindström M. 2013. Two-component signal transduction system CBO0787/CBO0786 represses transcription from botulinum neurotoxin promoters in Clostridium botulinum ATCC 3502. PLoS Pathog. 9:e1003252. 10.1371/journal.ppat.1003252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinderink K, Lindström M, Korkeala H. 2009. Group I Clostridium botulinum strains show significant variation in growth at low and high temperatures. J. Food Prot. 72:375–383 http://www.ingentaconnect.com/content/iafp/jfp/2009/00000072/00000002 [DOI] [PubMed] [Google Scholar]
- 48.Derman Y, Lindström M, Selby K, Korkeala H. 2011. Growth of group II Clostridium botulinum strains at extreme temperatures. J. Food Prot. 74:1797–1804. 10.4315/0362-028X.JFP-11-187 [DOI] [PubMed] [Google Scholar]
- 49.Baranyi J, Roberts TA. 1994. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 23:277–294. 10.1016/0168-1605(94)90157-0 [DOI] [PubMed] [Google Scholar]
- 50.Mattila M, Lindström M, Somervuo P, Markkula A, Korkeala H. 2011. Role of flhA and motA in growth of Listeria monocytogenes at low temperatures. Int. J. Food Microbiol. 148:177–183. 10.1016/j.ijfoodmicro.2011.05.022 [DOI] [PubMed] [Google Scholar]
- 51.Markkula A, Mattila M, Lindström M, Korkeala H. 2012. Genes encoding putative DEAD-box RNA helicases in Listeria monocytogenes EGD-e are needed for growth and motility at 3°C. Environ. Microbiol. 14:2223–2232 [DOI] [PubMed] [Google Scholar]
- 52.Shen A, Kamp HD, Gründling A, Higgins DE. 2006. A bifunctional O-GlcNAc transferase governs flagellar motility through anti-repression. Genes Dev. 20:3283–3295. 10.1101/gad.1492606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mauder N, Williams T, Fritsch F, Kuhn M, Beier D. 2008. Response regulator DegU of Listeria monocytogenes controls temperature-responsive flagellar gene expression in its unphosphorylated state. J. Bacteriol. 190:4777–4781. 10.1128/JB.00258-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gueriri I, Cyncynatus C, Dubrac S, Arana AT, Dussurget O, Msadek T. 2008. The DegU orphan response regulator of Listeria monocytogenes autorepresses its own synthesis and is required for bacterial motility, virulence and biofilm formation. Microbiology 154:2251–2264. 10.1099/mic.0.2008/017590-0 [DOI] [PubMed] [Google Scholar]
- 55.Kim YH, Han KY, Lee K, Lee J. 2005. Proteome response of Escherichia coli fed-batch culture to temperature downshift. Appl. Microbiol. Biotechnol. 68:786–793. 10.1007/s00253-005-0053-3 [DOI] [PubMed] [Google Scholar]