Abstract
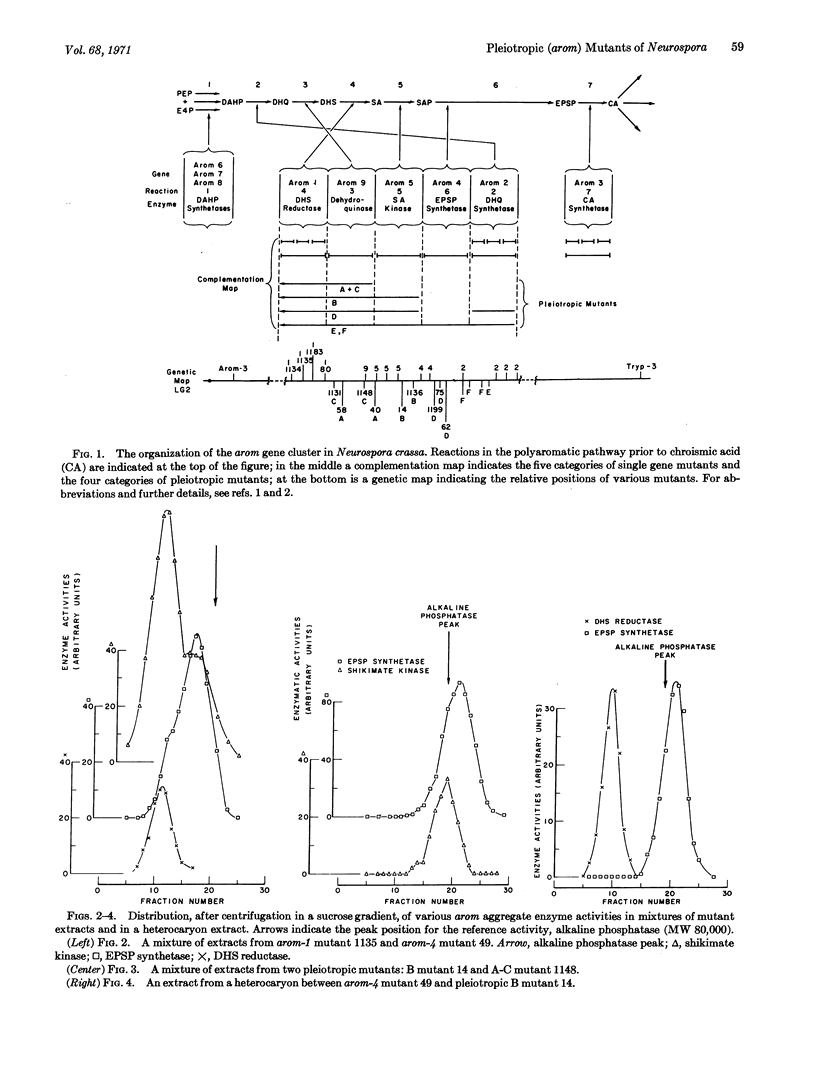
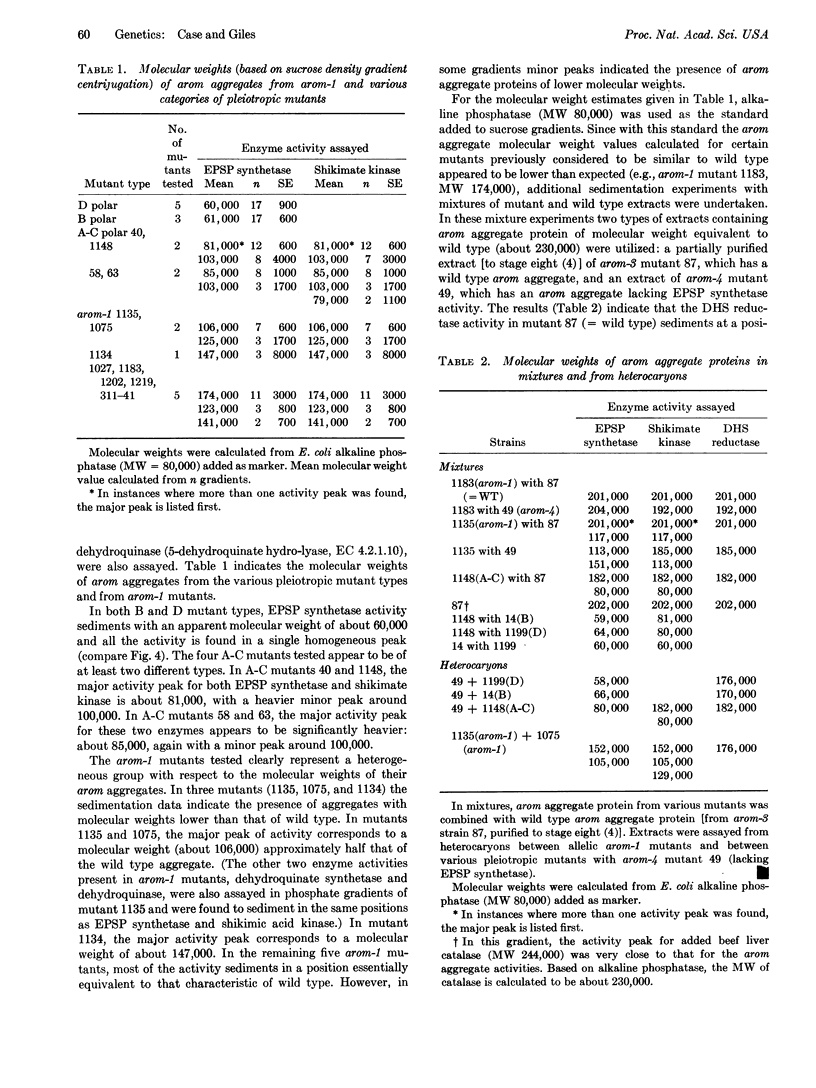
Molecular weights of enzymically active arom aggregates produced by pleiotropic and various arom-1 mutants of Neurospora crassa have been estimated by sucrose density gradient centrifugation. In contrast to most single-gene mutants (which produce intact arom multienzyme aggregates of normal molecular weight— about 230,000), pleiotropic mutants that lack two or more of the five enzyme activities in the arom aggregate produce partial arom aggregates of molecular weights ranging from about 60,000 to about 85,000. In addition, certain arom-1 mutants are pleiotropic in producing arom aggregates of about half the normal molecular weight. Experiments with heterocaryons, as well as with mixtures of extracts from various mutants, have provided evidence concerning the presence or absence of interactions (as detectable molecular hybridization) between different arom aggregates. An evaluation has been made of the relation between the genetic location of particular mutants within the arom gene cluster and the size and enzymic content of the arom aggregates they produce. Interpretations concerning the molecular organization of the arom multienzyme aggregate in N. crassa are presented.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed S. I., Giles N. H. Organization of enzymes in the common aromatic synthetic pathway: evidence for aggregation in fungi. J Bacteriol. 1969 Jul;99(1):231–237. doi: 10.1128/jb.99.1.231-237.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlyn M. B., Giles N. H. Organization of enzymes in the polyaromatic synthetic pathway: separability in bacteria. J Bacteriol. 1969 Jul;99(1):222–230. doi: 10.1128/jb.99.1.222-230.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne L., Case M. E., Giles N. H. Purification and properties of the aromatic (arom) synthetic enzyme aggregate of Neurospora crassa. Biochim Biophys Acta. 1969 Nov 4;191(2):452–462. doi: 10.1016/0005-2744(69)90264-2. [DOI] [PubMed] [Google Scholar]
- Case M. E., Burgoyne L., Giles N. H. In vivo and in vitro complementation between DHQ synthetase mutants in the arom gene cluster of Neurospora crassa. Genetics. 1969 Nov;63(3):581–588. doi: 10.1093/genetics/63.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case M. E., Giles N. H. Evidence for nonsense mutations in the arom gene cluster of Neurospora crassa. Genetics. 1968 Sep;60(1):49–58. doi: 10.1093/genetics/60.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles N. H., Case M. E., Partridge C. W., Ahmed S. I. A gene cluster in Nuerospora crassa coding for an aggregate of five aromatic synthetic enzymes. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1453–1460. doi: 10.1073/pnas.58.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- Rines H. W., Case M. E., Giles N. H. Mutants in the arom gene cluster of Neurospora crassa specific for biosynthetic dehydroquinase. Genetics. 1969 Apr;61(4):789–800. doi: 10.1093/genetics/61.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]



