Abstract
Arabinoxylan oligosaccharides (AXOS) are prebiotic carbohydrates with promising health-promoting properties that stimulate the activity of specific colon bacteria, in particular bifidobacteria. However, the mechanisms by which bifidobacterial strains break down these compounds in the colon is still unknown. This study investigates AXOS consumption of a large number of bifidobacterial strains (36), belonging to 11 different species, systematically. To determine their degradation mechanisms, all strains were grown on a mixture of arabinose and xylose, xylo-oligosaccharides, and complex AXOS molecules as the sole added energy sources. Based on principal component and cluster analyses of their different arabinose substituent and/or xylose backbone consumption patterns, five clusters that were species independent could be distinguished among the bifidobacterial strains tested. In parallel, the strains were screened for the presence of genes encoding several putative AXOS-degrading enzymes, but no clear-cut correlation could be made with the different degradation mechanisms. The intra- and interspecies differences in the consumption patterns of AXOS indicate that bifidobacterial strains could avoid competition among each other or even could cooperate jointly to degrade these complex prebiotics. The knowledge gained on the AXOS degradation mechanisms in bifidobacteria can be of importance in the rational design of prebiotics with tailor-made composition and thus increased specificity in the colon.
INTRODUCTION
Arabinoxylan oligosaccharides (AXOS) and arabinoxylans (AX), containing arabinose and xylose substituents, cannot be degraded by human enzymes of the gastrointestinal tract (1). These complex substrates enter the colon intact and represent an energy source for saccharolytic residing bacteria, including species of Bacteroides, Bifidobacterium, Clostridium, Lactobacillus, and Eubacterium (2, 3). Although bifidobacteria represent <5% of the human adult fecal microbiota, they are indispensable for the degradation of these complex molecules and hence for the maintenance of a balanced gut homeostasis. Their formation of partial carbohydrate breakdown products and short-chain fatty acids (SCFA) forms the basis of many complex interactions of cross-feeding with other colon inhabitants (2, 4).
It has already been shown, both in vivo and in vitro, that AXOS and AX selectively increase the abundance of Bifidobacterium spp., a so-called bifidogenic effect exerted by these prebiotics (5–10). Also, the large number of genes possibly coding for AX(OS)-degrading enzymes in for instance the genome of Bifidobacterium longum subsp. longum NCC2705 demonstrates the adaptation of bifidobacterial strains to these substrates important for human health (11). Further, it has been shown that the bifidogenic effect impacts indirectly other genera as well, in particular propionate- and butyrate-producing colon bacteria, possibly through cross-feeding (4, 10, 12).
AX consist of a linear backbone of 1,500 to 15,000 β-(1→4)-linked d-xylopyranosyl (Xylp) units, which can occur unsubstituted, monosubstituted with α-l-arabinofuranosyl (Araf) residues positioned on C-(O)-2 or C-(O)-3, or disubstituted with Araf residues on both C-(O)-2 and C-(O)-3 positions (13). In addition, xylose residues can be substituted with glucuronic acid or acetyl groups, while arabinose residues can be esterified with phenolic acids, such as ferulic acid and p-coumaric acid. Because of the heterogeneous composition of AXOS and AX, their complete hydrolysis requires the action of both debranching and depolymerizing enzymes. The enzymatic cleavage of AX with β-endoxylanase results in AXOS and xylo-oligosaccharides (XOS or unsubstituted backbones of Xylp units) (10). Other AXOS- and AX-degrading enzymes include β-xylosidases and exo-oligoxylanases, which release terminal xylose residues from the nonreducing and reducing ends of the xylose backbones, respectively (5, 14). Debranching enzymes include α-arabinofuranosidases, glucuronidases, acetyl esterases, and feruloyl esterases, which remove arabinose, glucuronic acid, acetic acid, and ferulic acid, respectively (8).
Although several studies have demonstrated the bifidogenic effect of AXOS and AX, the degradation mechanism by which Bifidobacterium spp. break down these compounds is still unknown. Until now, studies of the degradation of AXOS and AX through monoculture fermentations with bifidobacterial strains have been restricted to the monitoring of bacterial growth, pH, and SCFA production (15, 16). Also, fermentations of purified short-chain AXOS standards have been performed (17). However, these studies do not reveal the complete fermentation capacity of bifidobacteria. Moreover, most studies and reviews concerning the prebiotic effect of AXOS and AX consider the bifidobacterial gut population as a whole (5–9, 15). For instance, they do not take into account differences in carbohydrate preferences that exist on species and/or strain level within the genus of Bifidobacterium (4, 18, 19).
We set out to unravel the mechanistic variations in AXOS degradation by a wide range of bifidobacterial strains covering the most common species occurring in the human colon. All bifidobacterial strains were investigated as to their potential to ferment AXOS-derived monosaccharides (arabinose and xylose) and xylose backbones (XOS), as well as the complete AXOS molecules. In parallel, all strains were screened for the presence of putative genes encoding AX(OS)-degrading enzymes based on genome information of B. longum subsp. longum NCC2705 to map the distribution of these genes in the strains studied and to find correlations with their AXOS degradation mechanisms.
MATERIALS AND METHODS
Strains and media.
Thirty-six bifidobacterial strains belonging to 11 different species were used throughout the present study (Table 1). All bifidobacterial strains were grown at 37°C in reinforced clostridial medium (Oxoid, Basingstoke, Hampshire, United Kingdom), except for the B. bifidum strains that were grown in de Man-Rogosa-Sharpe medium (Oxoid), under anaerobic conditions in a modular atmosphere-controlled system (MG anaerobic workstation; Don Whitley Scientific, West Yorkshire, United Kingdom) that was continuously sparged with a mixture of 80% nitrogen gas, 10% carbon dioxide, and 10% hydrogen gas (Air Liquide, Paris, France). For storage of the strains at −80°C, the media were supplemented with 25% (vol/vol) glycerol (Sigma-Aldrich, Stenheim, Germany). Before screening, the authenticity of the strains was confirmed through 16S rRNA gene sequencing using the genus-specific primers Bif164 and Bif662 (20).
TABLE 1.
Bifidobacterium strains used in the present study
| No. | Strain | Origin | Source or referencea |
|---|---|---|---|
| 1 | B. longum subsp. longum BB536 | Infant feces | Morinaga Milk Industry, Tokyo, Japan |
| 2 | B. longum subsp. longum LMG 13197T | Adult intestine | BCCM/LMG, Ghent, Belgium |
| 3 | B. longum subsp. infantis LMG 11570 | Infant intestine | BCCM/LMG, Ghent, Belgium |
| 4 | B. longum subsp. infantis LMG 11588 | Infant feces | BCCM/LMG, Ghent, Belgium |
| 5 | B. longum subsp. longum LMG 11047 | Human | BCCM/LMG, Ghent, Belgium |
| 6 | B. longum subsp. longum 46 | Elderly feces | BENEO-Orafti, Tienen, Belgium |
| 7 | B. longum subsp. longum LMG 13196 | Infant intestine | BCCM/LMG, Ghent, Belgium |
| 8 | B. longum subsp. longum NCC2705 | Infant feces | Nestlé, Lausanne, Switzerland |
| 9 | B. longum subsp. longum CUETM 172 | Not available | Bahaka et al. (62) |
| 10 | B. longum subsp. longum PRO-16-10 | Adult feces | Matamoros et al. (63) |
| 11 | B. longum subsp. longum CUETM 193 | Child feces | Bahaka et al. (62) |
| 12 | B. longum subsp. longum CUETM 239 | Child feces | Bahaka et al. (62) |
| 13 | B. longum subsp. longum CUETM 290 | Child feces | Bahaka et al. (62) |
| 14 | B. longum subsp. longum CUETM 171 | Child feces | Bahaka et al. (62) |
| 15 | B. longum subsp. longum ATCC 51870 | Child feces | ATCC, Manassas, VA, USA |
| 16 | B. pseudolongum subsp. pseudolongum LMG 11594 | Chicken feces | BCCM/LMG, Ghent, Belgium |
| 17 | B. pseudolongum subsp. globosum LMG 11614 | Bovine rumen | BCCM/LMG, Ghent, Belgium |
| 18 | B. bifidum DSM 20082 | Adult intestine | BCCM/LMG, Ghent, Belgium |
| 19 | B. bifidum LMG 11583 | Adult intestine | BCCM/LMG, Ghent, Belgium |
| 20 | B. bifidum LMG 11582 | Adult intestine | BCCM/LMG, Ghent, Belgium |
| 21 | B. bifidum LMG 13195 | Infant intestine | BCCM/LMG, Ghent, Belgium |
| 22 | B. animalis subsp. lactis DN-173 010 | Probiotic drink | Danone Vitapole, Palaiseau, France |
| 23 | B. animalis subsp. lactis R-17143 | Proflora, Chefalo | BCCM/LMG, Ghent, Belgium |
| 24 | B. animalis subsp. lactis BB-12 | Yogurt | Christian Hansen, Hørsholm, Denmark |
| 25 | B. dentium LMG 10507 | Human feces | BCCM/LMG, Ghent, Belgium |
| 26 | B. breve LMG 13194 | Infant intestine | BCCM/LMG, Ghent, Belgium |
| 27 | B. breve LMG 13208 | Infant intestine | BCCM/LMG, Ghent, Belgium |
| 28 | B. breve LMG 11040 | Nursling feces | BCCM/LMG, Ghent, Belgium |
| 29 | B. breve Yakult | Infant feces | Yakult Honsha, Tokyo, Japan |
| 30 | B. thermophilum LMG 11574 | Bovine rumen | BCCM/LMG, Ghent, Belgium |
| 31 | B. catenulatum LMG 11043T | Adult intestine or feces | BCCM/LMG, Ghent, Belgium |
| 32 | B. gallicum LMG 11596T | Adult intestine | BCCM/LMG, Ghent, Belgium |
| 33 | B. angulatum LMG 11039T | Human feces | BCCM/LMG, Ghent, Belgium |
| 34 | B. angulatum LMG 11568 | Sewage | BCCM/LMG, Ghent, Belgium |
| 35 | B. adolescentis LMG 10734 | Adult intestine | BCCM/LMG, Ghent, Belgium |
| 36 | B. adolescentis LMG 10502T | Adult intestine | BCCM/LMG, Ghent, Belgium |
BCCM/LMG, Belgian Co-ordinated Collections of Microorganisms/Laboratory Microbiology Ghent; ATCC, American Type Culture Collection.
Fermentation experiments.
Laboratory fermentations were carried out in glass recipients containing medium for colon bacteria (MCB), which supports the growth of various colon bacteria (21). This medium is composed of the following components (g liter−1): bacteriological peptone (Oxoid), 6.5; soy peptone (Oxoid), 5.0; yeast extract (VWR International, Darmstadt, Germany), 3.0; tryptone (Oxoid), 2.5; NaCl (VWR International), 4.5; KCl (Merck, Darmstadt, Germany), 2.0; MgSO4·7H2O (Merck), 0.5; CaCl2·2H2O (Merck), 0.45; cysteine-HCl (Merck), 0.4; NaHCO3 (VWR International), 0.2; MnSO4.H2O (VWR International), 0.2; FeSO4·7H2O (Merck), 0.005; ZnSO4·7H2O (VWR International), 0.005; hemin (Sigma-Aldrich), 0.005; menadione (Sigma-Aldrich), 0.005; H3PO4 (Merck), 0.5 ml liter−1; and Tween 80 (Merck), 2 ml liter−1. All components were dissolved in ultrapure water, and the medium was adjusted to pH 6.3 by means of 37% HCl (VWR International) and 10 M NaOH solutions (VWR International). After autoclaving at 121°C and 1.1 bar overpressure for 20 min, the recipients were put immediately under anaerobic conditions (MG anaerobic workstation). An arabinose-xylose mixture (further referred to as A+X) consisting of arabinose (1 g liter−1; Sigma-Aldrich) plus xylose (4 g liter−1; Sigma-Aldrich) in a ratio (A/X) of 0.2 typical for AXOS, XOS 95P (5 g liter−1; Shandong Longlive Bio-Technology, Shandong, China), and AXOS-5-0.27 (5 g liter−1; Fugeia, Leuven, Belgium) were used as the sole added energy sources. XOS 95P (further referred to as XOS) mainly consists of XOS with a degree of polymerization (DP) of 2 to 7 (XOS2-7, ≥95.0% [mass/mass]), XOS2-4 (≥65.0% [mass/mass]), and minor amounts of xylose, arabinose, and dextrose (≤0.5% [mass/mass]). Wheat AXOS-5-0.27 has an average DP of 5, an A/X of 0.27, and contains 1% (mass/mass) ferulic acid. Since it has been shown previously that AXOS-5-0.27 contains XOS as well (22), the complete substrate will be further referred to as (A)XOS and the individual components as AXOS and XOS(A)XOS. Solutions of the energy sources were sterilized separately, either by autoclaving (A+X) or by membrane filtration, using Minisart filters (0.2-μm pores; Sartorius AG, Göttingen, Germany) in the case of XOS or MediaKap-50 Plus high-performance hollow fiber membrane filters (0.2-μm pores; Microgon, Rancho Dominguez, CA) in the case of AXOS, and added to the MCB-containing recipients aseptically. There was no partial loss of the samples during filtration, as the chromatograms of high-performance anion-exchange chromatography (HPAEC) with pulsed amperometric detection (PAD) were identical to the chromatograms of the nonfiltrated reference XOS and (A)XOS solutions.
Fermentations with A+X as the added energy source were carried out in duplicate in test tubes containing 10 ml of MCB with A+X. The inocula (5% [vol/vol]) were obtained after transfer of stock culture cells into 10 ml of the appropriate medium, followed by anaerobic incubation at 37°C for 12 h.
Fermentations with XOS or (A)XOS as the added energy sources were carried out in duplicate in Schott flasks containing 100 ml of MCB with XOS or (A)XOS. The inocula (5% [vol/vol]) were prepared through two subcultures. The stock culture cells were first inoculated into 10 ml of the appropriate medium and incubated anaerobically at 37°C for 12 h. Next, 5% (vol/vol) of these subcultures was grown in 10 ml of MCB with A+X, followed by anaerobic incubation at 37°C for 12 h.
Kinetic analysis of bacterial growth and carbohydrate consumption.
Fermentations were sampled at 0 and 24 h for those with A+X and at 0, 4, 8, 12, 24, and 48 h for those with XOS and (A)XOS.
(i) Determination of bacterial growth.
The optical density at 600 nm (OD600) was measured against ultrapure water as blank with a VIS spectrophotometer (Genesys 20; Thermo Scientific, Waltham, MA). Each measurement was performed in triplicate. The errors of the measurements are represented as standard deviations.
(ii) Determination of arabinose and xylose concentrations.
For the fermentations performed in MCB with A+X, residual concentrations of arabinose and xylose were determined by HPAEC-PAD, using an ICS-3000 chromatograph equipped with an ED PAD, a column oven at a constant temperature of 30°C, an AS autosampler, a DP-1 dual pump, a CarboPac PA10 guard column (50 by 4 mm), and a CarboPac PA10 analytical column (250 by 4 mm), all from Thermo Scientific (Waltham, MA). Chromeleon software 6.7 (Thermo Scientific) was used for system control and data analysis. For the mobile phase, three eluent solutions were used at a flow rate of 1 ml min−1: ultrapure water (eluent A), 167 mM NaOH (eluent B), and 500 mM NaOH (eluent C). The following gradient was applied: 0.0 to 20.0 min, 87% A, 13% B, and 0% C; 21.0 to 27.0 min, 0% A, 0% B, and 100% C; and 27.2 to 30.0 min, 87% A, 13% B, and 0% C. NaOH solution (50%) was purchased from Boom (Meppel, The Netherlands). Quantifications were carried out through standard addition. Four standard solutions were prepared (g liter−1): ultrapure water (solution A), 0.010 arabinose and 0.040 xylose (solution B), 0.020 arabinose and 0.080 xylose (solution C), and 0.030 arabinose and 0.120 xylose (solution D). Sample preparation for HPAEC-PAD analysis involved microcentrifugation (14,000 rpm for 15 min) to remove the cells, followed by mixing 300 μl of a 50-fold-diluted cell-free culture supernatant with 300 μl of solutions A, B, C, or D. To remove proteins, 300 μl of Carrez A reagent [36 g of K4Fe(CN)6·3H2O liter−1] and 300 μl of Carrez B reagent (72 g of ZnSO4·7H2O liter−1) were added. After microcentrifugation (14,000 rpm for 15 min), the supernatant was filtered (0.2-μm pores; Uniflo RC filters; Whatman, Dassel, Germany) and injected (10 μl) into the column. The original sample concentrations with corresponding errors were calculated as described previously (23).
(iii) Determination of XOS and XOS(A)XOS concentrations.
Quantitative analysis of arabinose, xylose, XOS, and XOS(A)XOS breakdown products during fermentations of XOS or (A)XOS was performed using HPAEC-PAD. To this end, an ICS-3000 chromatograph at controlled room temperature (18°C) was used, equipped with an ED40 PAD, an AS50 autosampler, a SP-1 single pump, a CarboPac PA100 guard column (50 by 4 mm), and a CarboPac PA100 analytical column (250 by 4 mm), all from Thermo Scientific. Chromeleon software 6.7 (Thermo Scientific) was used for system control and data analysis. The mobile phase, at a flow rate of 1.0 ml min−1, consisted of ultrapure water (eluent A), 0.2 M NaOH (eluent B), and 0.1 M NaOH with 0.4 M NaCH3COOH (eluent C), with the following gradient: 0.0 to 7.0 min, 95% A, 5% B, and 0% C; 7.1 to 9.0 min, 50% A, 50% B, and 0% C; 25.0 min, 38% A, 32% B, and 30% C; 25.1 to 29.0 min, 0% A, 0% B, and 100% C; and 29.1 to 34.0 min, 95% A, 5% B, and 0% C. Anhydrous NaCH3COOH (≥99.0%) was purchased from Sigma-Aldrich. Before injection (10 μl) into the column, samples were deproteinized by adding 300 μl of Carrez A reagent and 300 μl of Carrez B reagent to 600 μl of a 16-fold-diluted XOS or (A)XOS cell-free culture supernatant, obtained through centrifugation at 4,618 × g for 20 min at 4°C. After microcentrifugation (14,000 rpm for 15 min), the supernatant was filtered (pore size, 0.2 μm; Uniflo RC filters; Whatman). A mixture of arabinose (Sigma-Aldrich), xylose (Sigma-Aldrich), and xylobiose, xylotriose, xylotetraose, xylopentaose, and xylohexaose (all from Megazyme International, Bray, Ireland) was used as an external standard to perform quantifications. Ten standard solutions (0.0030, 0.0075, 0.0150, 0.0200, 0.0250, 0.0300, 0.0600, 0.0750, 0.1000, and 0.1500 g liter−1) were prepared by diluting stock solutions starting from 0.150 g of the components liter−1. Samples were analyzed in triplicate and the errors of the measurements are represented as standard deviations.
(iv) Qualitative fingerprinting of AXOS degradation.
Qualitative analysis of AXOS breakdown during fermentations with (A)XOS as the added energy source was performed through HPAEC-PAD, using the equipment described above for XOS and XOS(A)XOS analysis, with a CarboPac PA200 guard column (50 by 3 mm) and a CarboPac PA200 analytical column (250 by 3 mm). Sample preparation was performed as described by Rivière et al. (22). The mobile phase, at a flow rate of 0.5 ml min−1, consisted of ultrapure water (eluent A), 0.2 M NaOH (eluent B), and 0.1 M NaOH with 0.4 M NaCH3COOH (eluent C), with the following gradient: 0.0 to 10.0 min, 90% A, 10% B, and 0% C; 10.1 to 15.0 min, 50% A, 50% B, and 0% C; 55.0 min, 34% A, 34% B, and 32% C; 55.1 to 60.0 min, 0% A, 0% B, and 100% C; and 60.1 to 66 min, 90% A, 10% B, and 0% C. To interpret the chromatograms, enzymatic reference degradation chromatograms that were generated through degradation of (A)XOS in solution by enzymes with known specificities were used, as described previously (22).
Multivariate data analysis.
All data of the A+X, XOS, and (A)XOS fermentations were combined and processed by principal component analysis followed by cluster analysis, using the software package SPSS 20.0 (SPSS, Inc., Chicago, IL), as follows. For all 36 bifidobacterial strains, the data were displayed in a matrix consisting of 32 fermentation variables and 36 objects. Subsequently, they were converted to binary format prior to statistical analysis, except for the quantitative data of growth on XOS and (A)XOS. Concentrations of arabinose, xylose, xylobiose, xylotriose, xylotetraose, xylopentaose, and xylohexaose were given the binary number 1 if >10% of the initial concentration of the component was consumed at the end of the fermentation or 0 if this was not the case. For AXOS substituent degradation, the binary number 1 was used if the peak corresponding with an AXOS molecule in the HPAEC-PAD chromatogram disappeared and 0 if the peak did not disappear. These data were complemented with the results concerning the nature of the backbone degradation (0 or 1 if the xylose concentration did not increase or increased, respectively) and the nature of (A)XOS degradation (0 or 1 if the arabinose concentration did not increase or increased, respectively, and 0 or 1 if the xylose backbone concentrations did not increase or increased, respectively). To determine the number (n) of principal components (PCs), a scree plot was constructed. In the n-dimensional space of the retained PCs, a Varimax with Kaiser normalization rotation method was applied to maximize the sum of the squares of the correlations between the original variables and the rotated PCs (factor loadings) to make the factor loadings more interpretable. In this way, all factor loadings will have a value near one or near zero, with a few intermediate values. A score plot of the fermentation data along the loading vectors was constructed, and the clusters were identified. A three-dimensional score plot was constructed with Matlab (Matworks, Natick, MA). Finally, a K-means cluster analysis based on the squared Euclidian distance was performed using the between-groups linkage.
Genetic screening for AX(OS)-degrading enzymes.
For the isolation of genomic DNA, cell pellets of overnight cultures in the appropriate medium were collected by microcentrifugation at 14,000 rpm for 15 min at 4°C and stored at −20°C for at least 1 h. DNA was extracted from the cell pellets as described before (24).
The strains were screened for the presence of genes putatively encoding AX(OS)-degrading enzymes as those present in the genome of B. longum subsp. longum NCC2705, i.e., an extracellular β-endoxylanase (BL1543), an extracellular β-xylosidase (BL1544), an α-arabinofuranosidase type I (BL0181), four α-arabinofuranosidases type A (BL0544, BL1611, BL1138, and BL1166), and a xylan esterase (BL0682) (11). This was done to map the distribution of these genes in the strains studied. For each of the enzymes, homologous protein sequences within the bifidobacterial genus were retrieved from the nonredundant protein sequence database (February 2012) (25) using the BLASTp algorithm (26). Next, the corresponding gene sequences were aligned to construct a consensus sequence that was used to design primer sets with Primer3 (27) (see Table S1 in the supplemental material). For the four α-arabinofuranosidase type A genes, three primer sets were designed, since the genes BL0544 and BL1611 showed high nucleotide sequence identity (81%), and thus the same primer set could be used. In addition to determine the complete sequence of the extracellular β-xylosidase genes, six primer pairs (see Table S2 in the supplemental material) were designed based on the gene sequence of BL1544 (RefSeq no. NC_004307.2; from positions 1946700 to 1950117) using the SNPbox program (28), resulting in overlapping PCR amplicons. PCR assays were performed in a total volume of 50 μl, containing 100 ng of genomic DNA, 200 μM each deoxynucleoside triphosphate (Fermentas GmbH, St. Leon-Rot, Germany), 20 pmol of each primer, 5 μl of 10× PCR buffer (Roche, Mannheim, Germany), 1.25 U of Taq DNA polymerase (Roche), and ultrapure water. The following conditions were used: (i) initial denaturation at 95°C for 5 min; (ii) a 30-cycle reaction of denaturation at 95°C for 30 s, an annealing step with a temperature of 51.9°C (BL0181, BL0544/BL1611, and BL1166), 58.7°C (BL1544_1), 53.2°C (BL1544_2 to BL1544_6), or 54.2°C (BL1543, BL1544, BL0420, BL1138, and BL0682), and an extension step at 72°C for 1 min; and (iii) a final extension at 72°C for 7 min. After amplification, PCR product sizes were controlled on a 1.0% (mass/vol) agarose gel (Invitrogen, Paisley, United Kingdom). Amplicons resulting from the genetic screening were sequenced by the VIB Genetic Service Facility (Antwerp, Belgium). The sequences were assembled using the BioEdit software (29), and their identities were confirmed with the nucleotide collection database (May 2012) (25) using the BLASTn algorithm (26). The sequences obtained for the complete extracellular β-xylosidase gene were assembled using the Phred and Phrap programs (30) and aligned using ClustalW (31). Finally, conserved protein domains were predicted in the translated protein sequences using SignalP (32), TMHMM (33), CAZy (34), and the Conserved Domain Database (35) (February 2013).
Nucleotide sequence accession numbers.
The complete gene sequences of the extracellular β-xylosidases were submitted to the EMBL database (25) with accession numbers HG316485 to HG316495.
RESULTS
Fermentation experiments and multivariate data analysis.
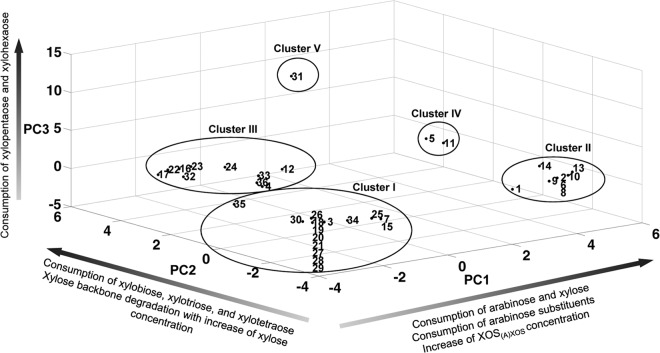
Principal component analysis reduced the original 32 variables to three independent rotated PCs, accounting for 78% of the total variance after Varimax Kaiser rotation (PC1, 34%; PC2, 27%; PC3, 17%). High factor loadings (>0.7) for PC1 corresponded with the consumption of the monosaccharides arabinose and xylose, the consumption of arabinose substituents of AXOS (both mono- and disubstituted), and an increase of the XOS(A)XOS concentration in the medium (Table 2). High factor loadings for the consumption of xylobiose, xylotriose, and xylotetraose, together with the nature of the xylose backbone degradation, namely, with or without an increase of the xylose concentration in the medium, were found for PC2. The consumption of xylopentaose and xylohexaose contributed to PC3. Since the other factor loadings were not as high, it can be suggested that these variables did not contribute to the rotated PCs. Based on their different arabinose substituent and/or xylose backbone consumption patterns, five different clusters could be distinguished among the bifidobacterial strains tested in the three-dimensional score plot based on the three PCs (Fig. 1 and Table 3).
TABLE 2.
Rotated principal component (PC) matrix of 32 fermentation variablesa
| Fermentation variable | Factor loading |
||
|---|---|---|---|
| PC1 | PC2 | PC3 | |
| Arabinose plus xylose fermentations | |||
| Arabinose | 0.74 | 0.15 | –0.33 |
| Xylose | 0.68 | 0.08 | –0.32 |
| XOS 95P fermentations | |||
| Xylose | 0.82 | –0.06 | –0.25 |
| Xylobiose | –0.03 | 0.89 | 0.11 |
| Xylotriose | –0.07 | 0.95 | 0.11 |
| Xylotetraose | –0.07 | 0.95 | 0.11 |
| Xylopentaose | –0.01 | 0.13 | 0.96 |
| Xylohexaose | –0.01 | 0.13 | 0.96 |
| Xylose increase | –0.04 | 0.89 | 0.15 |
| Maximal growth on XOS | 0.04 | 0.64 | –0.30 |
| AXOS-5-0.27 fermentations | |||
| M1 | 0.66 | 0.57 | 0.16 |
| M2 | 0.91 | –0.13 | 0.32 |
| M3 | 0.91 | –0.13 | 0.32 |
| M4 | 0.59 | 0.18 | –0.03 |
| M5 | 0.91 | –0.13 | 0.32 |
| M6 | 0.91 | –0.13 | 0.32 |
| M7 | 0.94 | –0.18 | –0.02 |
| D1 | 0.69 | 0.43 | 0.19 |
| D2 | 0.84 | –0.03 | 0.33 |
| D3 | 0.82 | –0.06 | 0.35 |
| D4 | –0.17 | 0.68 | –0.16 |
| Arabinose | 0.72 | 0.14 | –0.32 |
| Xylose | 0.82 | –0.06 | –0.25 |
| Xylobiose | 0.08 | 0.93 | 0.11 |
| Xylotriose | 0.08 | 0.93 | 0.11 |
| Xylotetraose | –0.02 | 0.96 | 0.12 |
| Xylopentaose | –0.01 | 0.13 | 0.96 |
| Xylohexaose | –0.01 | 0.13 | 0.96 |
| XOS increase | 0.93 | –0.11 | –0.05 |
| Xylose increase | –0.09 | 0.82 | 0.21 |
| Arabinose increase | 0.17 | 0.11 | 0.56 |
| Maximal growth on AXOS | 0.43 | 0.57 | 0.13 |
Factor loadings of >0.7 are indicated in boldface. M1 to M7 represent AXOS peaks containing at least one monosubstituted Xylp residue. D1 to D4 represent AXOS peaks containing at least one doubly substituted Xylp residue.
FIG 1.
Three-dimensional score plot of a principal component analysis of the fermentation data of 36 bifidobacterial strains. The numbers correspond to the strains listed in Table 1.
TABLE 3.
Overview of different clusters of bifidobacterial strains representing different degradation strategies of XOS and (A)XOS
| Cluster | Bifidobacterium representative strain | XOS consumption | Arabinose substituent consumption |
|---|---|---|---|
| I | B. bifidum LMG 11583 | – | – |
| II | B. longum subsp. longum NCC2705 | – | + (broad degradation) |
| III | B. longum subsp. infantis LMG 11588 | + (up to xylotetraose) | + (limited degradation) |
| IV | B. longum subsp. longum LMG 11047 | + (up to xylotetraose) | + (broad degradation) |
| V | B. catenulatum LMG 11043T | + (up to xylohexaose) | + (broad degradation) |
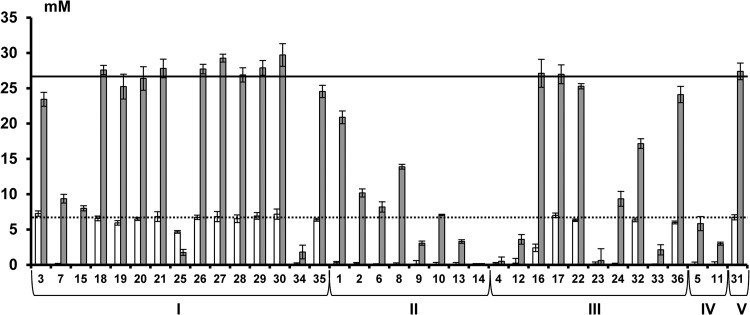
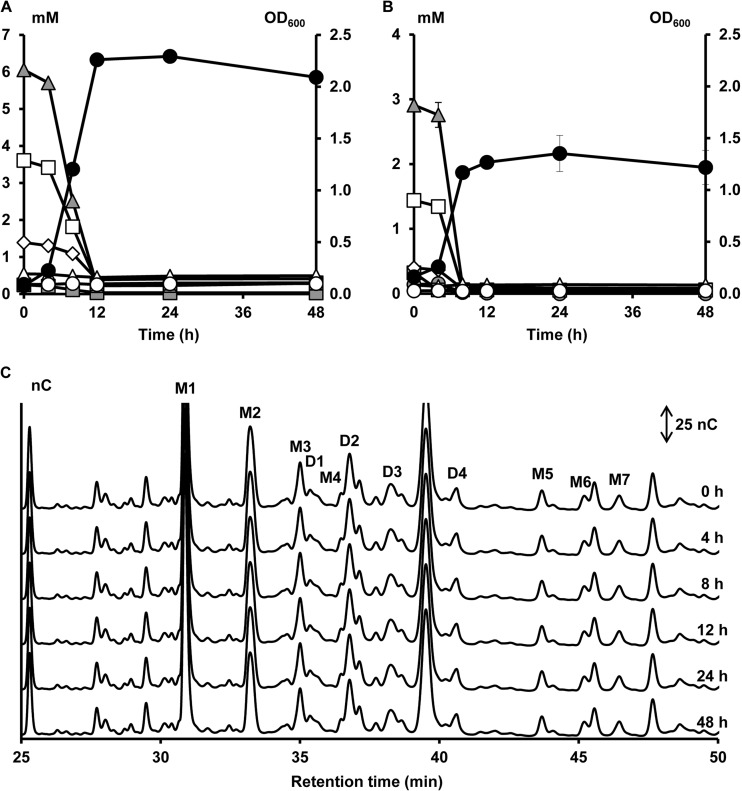
The first and largest cluster (cluster I, cluster center [PC1, −2.50; PC2, −2.18; PC3, −0.29], meaning a low score on all PCs [Fig. 1]) was composed of 15 bifidobacterial strains belonging to seven different species (B. adolescentis, B. angulatum, B. bifidum, B. breve, B. dentium, B. longum, and B. thermophilum). Strains belonging to this cluster could not use the xylose backbones or the AXOS substituents, except for B. adolescentis LMG 10734 (with a high score on PC2) that could use xylobiose, xylotriose, and xylotetraose of XOS but not of (A)XOS (see Fig. S1N in the supplemental material). A representative of this cluster is B. bifidum LMG 11583 (Fig. 2). Within this cluster, strains displaying the lowest scores for PC1 (Fig. 1 and see Table S3 in the supplemental material) could neither utilize free arabinose nor free xylose during fermentation in MCB with A+X (Fig. 3). In contrast, strains displaying less negative scores for PC1 could use both arabinose and xylose but to a different extent (Fig. 3). Strains belonging to the B. longum species and B. angulatum LMG 11568 consumed more arabinose than xylose, while B. dentium LMG 10507 metabolized more xylose than arabinose (Fig. 3). Graphical representations of the other strains within this cluster are provided as supplemental material (see Fig. S1A to N in the supplemental material).
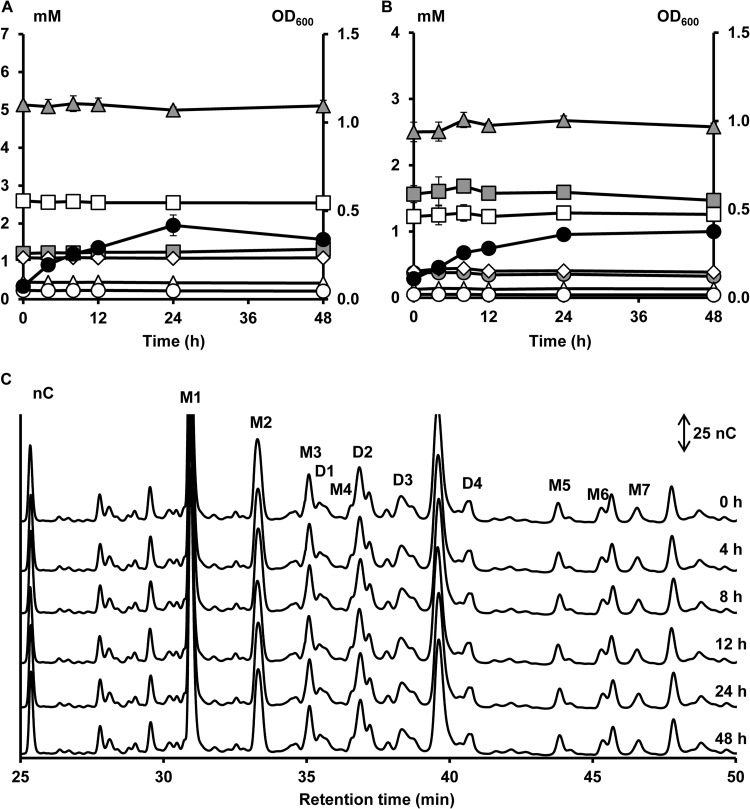
FIG 2.
Growth of and carbohydrate degradation by B. bifidum LMG 11583, a strain representing cluster I, in a medium for colon bacteria with 5 g of XOS liter−1 (A) or 5 g of (A)XOS liter−1 (B and C) as a function of time (samples taken at 0, 4, 8, 12, 24, and 48 h). Growth is represented as OD600 measurements (●). Monosaccharide, XOS, and XOS(A)XOS consumption are represented as residual concentrations of arabinose ( ), xylose (
), xylose ( ), xylobiose (
), xylobiose ( ), xylotriose (□), xylotetraose (◊), xylopentaose (△), and xylohexaose (○) in mM. (C) Enlargement of the AXOS HPAEC-PAD chromatograms between 25.0 and 50.0 min of elution. D1 to D4, AXOS peaks containing at least one doubly substituted Xylp residue; M1 to M7, AXOS peaks containing at least one monosubstituted Xylp residue.
), xylotriose (□), xylotetraose (◊), xylopentaose (△), and xylohexaose (○) in mM. (C) Enlargement of the AXOS HPAEC-PAD chromatograms between 25.0 and 50.0 min of elution. D1 to D4, AXOS peaks containing at least one doubly substituted Xylp residue; M1 to M7, AXOS peaks containing at least one monosubstituted Xylp residue.
FIG 3.
Residual concentrations (in mM) of arabinose (□) and xylose ( ) after 24 h of fermentation in a medium for colon bacteria supplemented with 1 g liter−1 (6.7 mM) of arabinose plus 4 g liter−1 (26.6 mM) of xylose. Horizontal lines represent the initial arabinose concentration (dotted line) and the initial xylose concentration (solid line). Numbers correspond to the strains listed in Table 1.
) after 24 h of fermentation in a medium for colon bacteria supplemented with 1 g liter−1 (6.7 mM) of arabinose plus 4 g liter−1 (26.6 mM) of xylose. Horizontal lines represent the initial arabinose concentration (dotted line) and the initial xylose concentration (solid line). Numbers correspond to the strains listed in Table 1.
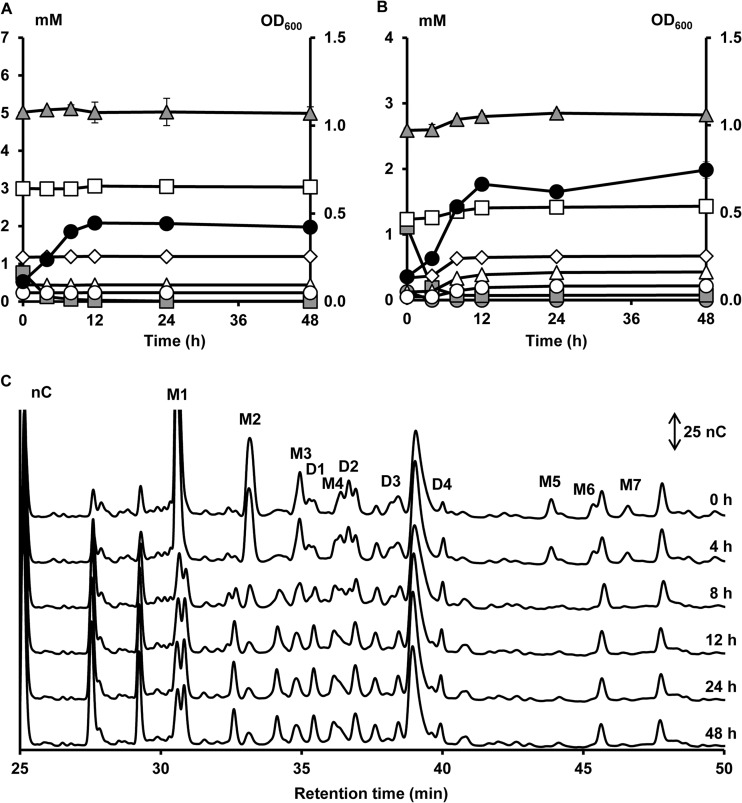
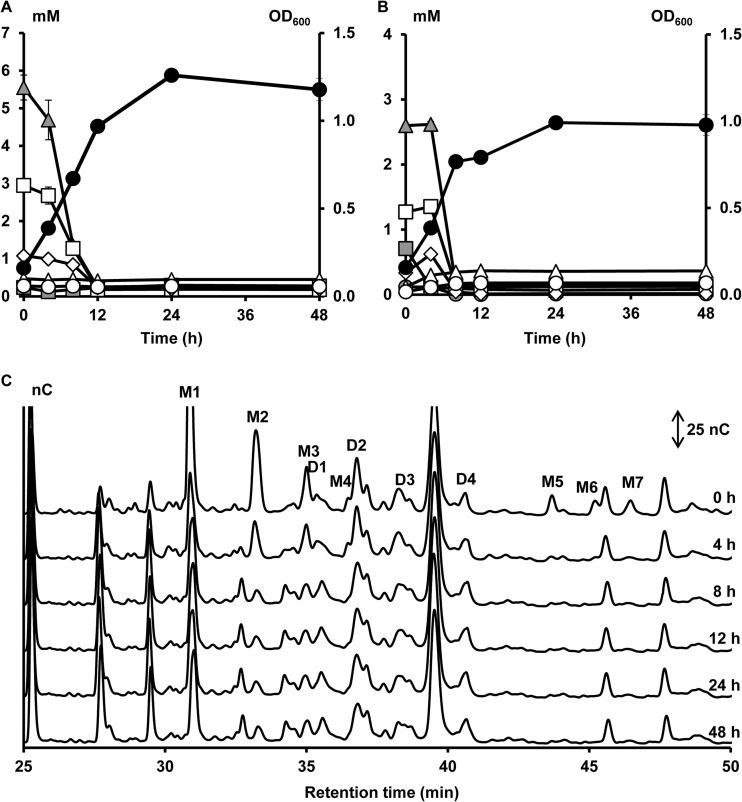
A second cluster (cluster II, cluster center [PC1, 4.80; PC2, −1.90; PC3, −0.03], meaning a high score on PC1 [Fig. 1]), consisting of eight B. longum strains, was characterized by the inability to consume the xylose backbones and the ability to consume the arabinose substituents of AXOS, both mono- and disubstituted, which resulted in the release of xylobiose, xylotriose, xylotetraose, xylopentaose, xylohexaose, and possibly also longer xylose backbones in the medium. A representative of cluster II is B. longum subsp. longum NCC2705 (Fig. 4). Within this cluster, all strains could use free arabinose and free xylose during fermentation in MCB with A+X but consumed more arabinose than xylose, except for B. longum subsp. longum CUETM 171, which could use arabinose and xylose equally well (Fig. 3). The latter strain was an exception with respect to its inability to use XOS, since it could use xylobiose and xylotriose of the (A)XOS substrate, whereas no consumption of these compounds was found when grown on XOS (see Fig. S2G in the supplemental material). In the case of B. longum subsp. longum CUETM 290 during fermentation in MCB with (A)XOS, an increase of the arabinose concentration in the medium, after an initial decrease, was found (see Fig. S2F in the supplemental material). Graphical representations of the other strains within this cluster are provided as supplemental material (see Fig. S2A to G in the supplemental material).
FIG 4.
Growth of and carbohydrate degradation by B. longum subsp. longum NCC2705, a strain representing cluster II, in a medium for colon bacteria with 5 g of XOS liter−1 (A) or 5 g of (A)XOS liter−1 (B and C) as a function of time (samples taken at 0, 4, 8, 12, 24, and 48 h). Peaks and symbols are named as in Fig. 2.
The third cluster [cluster III, cluster center (PC1, −1.11; PC2, 3.92; PC3, −0.77), meaning a high score on PC2 (Fig. 1)] included 10 strains belonging to six different species (B. adolescentis, B. angulatum, B. animalis, B. gallicum, B. longum, and B. pseudolongum). A representative of this cluster is B. longum subsp. infantis LMG 11588 (Fig. 5). Cluster III was characterized by the consumption of the xylose backbones, albeit only up to xylotetraose, and no or only limited degradation of AXOS substituents, which was not accompanied by an increase in xylose backbone concentrations in the medium, except for B. longum subsp. longum CUETM 239 (see Fig. S3A in the supplemental material). With the latter strain, an increase in xylose backbone concentrations was found after arabinose substituent degradation. Besides an increase in xylose concentration, an increase in xylobiose concentration was found during XOS metabolism by B. pseudolongum subsp. pseudolongum LMG 11594 and B. angulatum LMG 11039T (see Fig. S3B and H in the supplemental material). Degradation of xylobiose, xylotriose, and xylotetraose by the members of this cluster started simultaneously. However, for most strains of cluster III, except for the two B. longum strains, xylobiose was hydrolyzed less preferentially than xylotriose and xylotetraose (see Fig. S3A to I in the supplemental material). A variation occurred in the distribution of the strains in this cluster (Fig. 1), indicating differences in xylose backbone degradation between the XOS and (A)XOS substrates; this degradation took place with or without an increase of the xylose concentration in the medium. Strains displaying the highest scores for PC2 (Fig. 1 and see Table S3 in the supplemental material) consumed xylobiose, xylotriose, and xylotetraose of both the XOS and (A)XOS substrates used, with an increase of the xylose concentration in the medium (see Fig. S3 in the supplemental material). Although the other strains, displaying lower scores for PC2, showed differences in xylose backbone metabolism between the two substrates and/or their backbone degradation behavior, B. longum subsp. infantis LMG 11588, for instance, consumed the xylose backbones up to xylotetraose without an increase in xylose concentration in the medium (Fig. 5). Alternatively, for B. adolescentis LMG 10502T, differences in XOS degradation between the XOS and (A)XOS substrates were found (see Fig. S3I in the supplemental material). Also, differences in monosaccharide consumption occurred within this cluster during fermentation in MCB with A+X (Fig. 3). Bifidobacterial strains displaying more positive PC1 scores (> −0.96; see Table S3 in the supplemental material) used both arabinose and xylose, while the other strains were not able to use any of the monosaccharides or consumed only one of the two monosaccharides (Fig. 3). During fermentation of (A)XOS with B. longum subsp. longum CUETM 239, arabinose concentrations did not decrease in the extracellular medium (see Fig. S3A in the supplemental material). After arabinose was completely consumed, an increase of the arabinose concentration was found during the utilization of AXOS.
FIG 5.
Growth of and carbohydrate degradation by B. longum subsp. infantis LMG 11588, a strain representing cluster III, in a medium for colon bacteria with 5 g of XOS liter−1 (A) or 5 g of (A)XOS liter−1 (B and C) as a function of time (samples taken at 0, 4, 8, 12, 24, and 48 h). Peaks and symbols are named as in Fig. 2.
Cluster four (cluster IV, cluster center [PC1, 5.19; PC2, 3.31; PC3, −0.32], meaning high scores for both PC1 and PC2 [Fig. 1]) included B. longum subsp. longum LMG 11047 (representative of cluster IV, Fig. 6) and B. longum subsp. longum CUETM 193, which shared the ability to use the complete AXOS molecules by first cleaving the arabinose substituents, both mono- and disubstituted, followed by using the released xylose backbones. Xylobiose, xylotriose, and xylotetraose were simultaneously and equally consumed by both strains, while the longer xylose backbones were left unaffected. The degradation of these oligosaccharides was accompanied by a slight increase in xylose concentration in the medium, except during the (A)XOS fermentation with B. longum subsp. longum CUETM 193 (see Fig. S4A in the supplemental material). For none of the strains in this cluster, an increase of the arabinose concentration in the medium was found, which was in accordance with a greater consumption of arabinose than xylose during fermentation in MCB with A+X (Fig. 3).
FIG 6.
Growth of and carbohydrate degradation by B. longum subsp. longum LMG 11047, a strain representing cluster IV, in a medium for colon bacteria with 5 g of XOS liter−1 (A) or 5 g of (A)XOS liter−1 (B and C) as a function of time (samples taken at 0, 4, 8, 12, 24, and 48 h). Peaks and symbols are named as in Fig. 2.
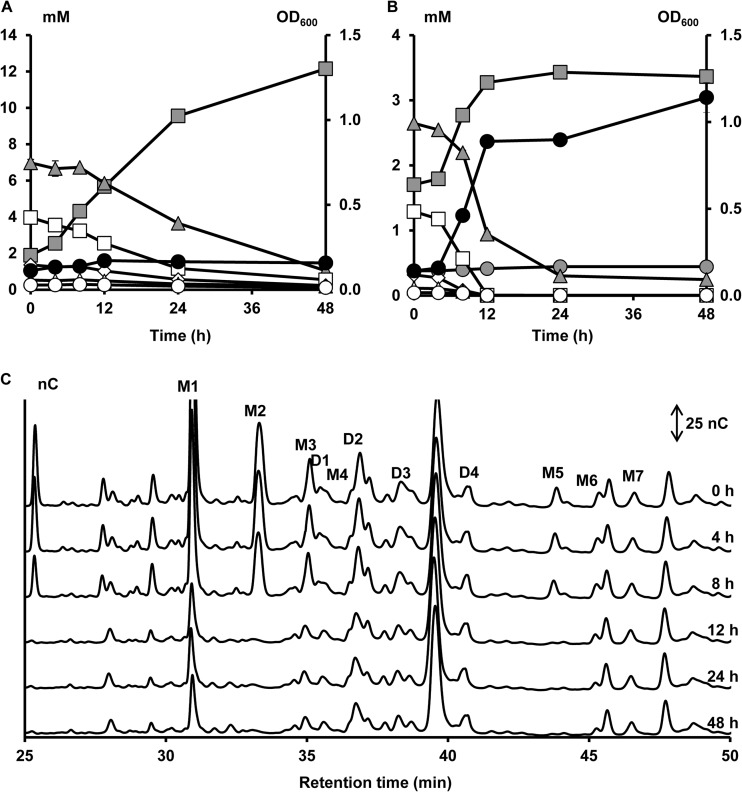
Cluster five (cluster V, cluster center [PC1, −0.14; PC2, 2.16; PC3, 12.94], meaning high scores for PC2 and PC3 [Fig. 1]) consisted only of B. catenulatum LMG 11043T (Fig. 7). Regarding the degradation of arabinose substituents, this strain showed a profile comparable to that of the bifidobacterial strains belonging to clusters II and IV (Fig. 7C). However, B. catenulatum LMG 11043T was the only strain that could use all XOS fractions in a nonpreferential way, up to xylohexaose, and possibly also longer xylose backbones for which no commercial standards were available. Degradation of the xylose backbones during fermentation of XOS and (A)XOS was characterized by a high increase in xylose concentrations in the medium. Also, arabinose concentrations slightly increased in the medium. These increases in monosaccharide concentrations are in accordance with the inability of this strain to consume arabinose and xylose during fermentation in MCB with A+X (Fig. 3).
FIG 7.
Growth of and carbohydrate degradation by Bifidobacterium catenulatum LMG 11043T, a strain representing cluster V, in a medium for colon bacteria with 5 g of XOS liter−1 (A) or 5 g of (A)XOS liter−1 (B and C) as a function of time (samples taken at 0, 4, 8, 12, 24, and 48 h). Peaks and symbols are named as in Fig. 2.
Genetic AX(OS)-degrading potential of bifidobacterial strains.
The genetic screening indicates that genes encoding AX(OS)-degrading enzymes, as those present in B. longum subsp. longum NCC2705, are widespread in B. longum strains (Table 4). A total of 16 of the 36 bifidobacterial strains possessed a putative α-arabinofuranosidase type I gene (BL0181), and 21 strains possessed at least one α-arabinofuranosidase type A gene (BL0544/BL1611, BL1138, or BL1166). An abundant presence of an α-arabinofuranosidase type I gene was found in the B. longum strains (60%), whereas only 32% of the other bifidobacterial strains tested contained this gene. The same was found for the α-arabinofuranosidase type A genes (BL0544/BL1611), present in 80% of the B. longum strains, in contrast to only 10% of the other bifidobacterial strains; the α-arabinofuranosidase type A gene (BL1138), present in 53% versus 29% of the strains; and the α-arabinofuranosidase type A gene (BL1166), present in 87% versus 0% of the strains. Twelve B. longum strains contained a gene encoding a putative xylan esterase (BL0682). Fourteen of the 15 B. longum strains possessed a putative extracellular β-endoxylanase gene (BL1543). All putative extracellular β-xylosidase genes (BL1544), present in 13 B. longum strains, showed the same order of multiple domains (in casu SignP-GH43-CBM22-TmD; SignP = signal peptide, GH43 = glycoside hydrolase family 43, CBM = carbohydrate-binding module, TmD = membrane anchor) as that of B. longum subsp. longum NCC2705. However, a nonsynonymous sequence variation (c.791C>T) was found in B. longum subsp. longum BB536, B. longum subsp. longum LMG 13197T, B. longum subsp. longum 46, and B. longum subsp. longum LMG 13196 that resulted in an altered protein sequence (p.Thr264Met) in the GH43 domain. Also, an additional nonsynonymous sequence variation (c.2111T>G) was found outside the predicted protein domains in B. longum subsp. longum 46 that resulted in an altered protein sequence (p.Val704Gly). Further, three synonymous sequence variations (c.543T>C in B. longum subsp. longum PRO-16-10 and c.2058C>T in B. longum subsp. longum BB536 and B. longum subsp. longum 46) were found. In addition, an insertion (c.2686_2687ins TGGTGATCAGACCAGTGA) in the extracellular β-xylosidase gene of B. longum subsp. longum 46 was found in a noncoding domain. Bifidobacterium breve Yakult was the only non-B. longum strain that possessed an extracellular β-xylosidase gene.
TABLE 4.
Genetic screening for putative AX(OS)-degrading enzymes in 36 bifidobacterial strainsa
| Bifidobacterium strain | Presence (+) or absence (−) of genes |
||||||
|---|---|---|---|---|---|---|---|
| BL1543 | BL1544 | BL0682 | BL0181 | BL0544/BL1611 | BL1138 | BL1166 | |
| B. longum subsp. longum BB536 | + | + | + | + | + | + | + |
| B. longum subsp. longum LMG 13197T | + | + | + | – | + | – | + |
| B. longum subsp. infantis LMG 11570 | + | – | – | – | – | – | – |
| B. longum subsp. infantis LMG 11588 | – | – | – | – | – | – | – |
| B. longum subsp. longum LMG 11047 | + | + | + | – | + | – | + |
| B. longum subsp. longum 46 | + | + | + | – | + | – | + |
| B. longum subsp. longum LMG 13196 | + | + | + | + | + | + | + |
| B. longum subsp. longum NCC2705 | + | + | + | + | + | + | + |
| B. longum subsp. longum CUETM 172 | + | + | + | + | + | + | + |
| B. longum subsp. longum PRO-16-10 | + | + | + | + | + | + | + |
| B. longum subsp. longum CUETM 193 | + | + | + | + | + | + | + |
| B. longum subsp. longum CUETM 239 | + | + | + | + | + | + | + |
| B. longum subsp. longum CUETM 290 | + | + | + | + | + | + | + |
| B. longum subsp. longum CUETM 171 | + | + | + | + | – | – | + |
| B. longum subsp. longum ATCC 51870 | + | + | – | – | + | – | + |
| B. pseudolongum subsp. pseudolongum LMG 11594 | – | – | – | – | – | – | – |
| B. pseudolongum subsp. globosum LMG 11614 | – | – | – | – | – | – | – |
| B. bifidum DSM 20082 | – | – | – | – | – | – | – |
| B. bifidum LMG 11583 | – | – | – | + | – | – | – |
| B. bifidum LMG 11582 | – | – | – | – | – | – | – |
| B. bifidum LMG 13195 | – | – | – | – | – | – | – |
| B. animalis subsp. lactis DN-173 010 | – | – | – | – | + | – | – |
| B. animalis subsp. lactis R-17143 | – | – | – | – | – | – | – |
| B. animalis subsp. lactis BB-12 | – | – | – | – | + | – | – |
| B. dentium LMG 10507 | – | – | – | + | – | + | – |
| B. breve LMG 13194 | – | – | – | – | – | – | – |
| B. breve LMG 13208 | – | – | – | – | – | – | – |
| B. breve LMG 11040 | – | – | – | – | – | – | – |
| B. breve Yakult | – | + | – | – | – | – | – |
| B. thermophilum LMG 11574 | – | – | – | – | – | – | – |
| B. catenulatum LMG 11043T | – | – | – | + | – | + | – |
| B. gallicum LMG 11596T | – | – | – | – | – | – | – |
| B. angulatum LMG 11039T | – | – | – | + | – | + | – |
| B. angulatum LMG 11568 | – | – | – | + | – | + | – |
| B. adolescentis LMG 10734 | – | – | – | + | – | + | – |
| B. adolescentis LMG 10502T | – | – | – | + | – | + | – |
The genes listed represent the original reference sequences of B. longum subsp. longum NCC2705 (BL1543, extracellular β-endoxylanase; BL1544, extracellular β-xylosidase; BL0181, α-arabinofuranosidase type I; BL0544/BL1611, BL1138, and BL1166, α-arabinofuranosidase type A; and BL0682, xylan esterase).
DISCUSSION
The present study of monoculture fermentations with 36 bifidobacterial strains belonging to 11 different species highlighted the complexity with which bifidobacteria break down AXOS and underlined that this breakdown is a strain-specific characteristic. Some strains prefer arabinose and/or xylose, while some others consume derived oligosaccharides (XOS and AXOS). Two mechanisms have been proposed for the degradation of fructo-oligosaccharides by bifidobacteria, namely, transport of small oligosaccharides into the cell and subsequent intracellular degradation, as well as external degradation by membrane-bound and/or extracellular enzymes, followed by uptake of the resulting monosaccharides (19, 21, 36). The bifidobacterial strains tested during the present study probably either used one of the two mechanisms or a combination of the two mechanisms to degrade (A)XOS. No clear-cut correlations were found between AXOS degradation mechanisms and the presence of genes encoding AX(OS)-degrading enzymes.
Arabinose substituent consumption patterns.
In the present study, cleavage of arabinose substituents of AXOS was found for strains belonging to clusters II, III, IV, and V. The increase of the concentrations of the AXOS-derived xylose backbones in the medium for strains belonging to clusters II and IV indicates an extracellular arabinose substituent-oriented AXOS metabolism. Since these strains consumed the free arabinose, it is suggested that the extracellular cleavage of these substituents is followed by their immediate transport into the cell. However, for some strains, an increase in arabinose concentration in the medium was found, suggesting that the extracellularly released arabinose was consumed to a lesser extent compared to other bifidobacterial strains. This is in contrast with the finding that these strains contained several intracellular α-arabinofuranosidase genes, which indicates transport of AXOS into the cell and subsequent intracellular degradation. Extracellular arabinose substituent cleavage has been reported for B. longum VTT E-96664 and B. longum VTT E-96702 when grown on AX (16) and for B. adolescentis ATCC 15703 and B. longum ATCC 15707 when grown on AXOS (17), albeit based on endpoint determinations or making use of purified short-chain AXOS standards, respectively. Although the extracellular cleavage of arabinose substituents by cluster II and IV strains suggests the presence of extracellular α-arabinofuranosidases, no such enzymes have been characterized in B. longum until now (11, 37). Similarly, in B. adolescentis ATCC 15703, only α-arabinofuranosidases have been characterized that are located intracellularly (38). This suggests that other genes must be responsible for the extracellular cleavage of arabinose by the strains tested. Indeed, the gene BL1544, predicted to encode an extracellular β-xylosidase in B. longum subsp. longum NCC2705, represents an enzyme with a high amino acid similarity with an AX α-arabinofuranosidase of B. adolescentis DSM 20083 that hydrolyzes Araf residues from doubly substituted Xylp residues at position C-(O)-3 (39). Also, B. catenulatum LMG 11043T of cluster V consumed almost all arabinose substituents of AXOS, but this was not accompanied by an increase of the concentrations of the AXOS-derived xylose backbones in the medium. This indicates a more preferential degradation of the xylose backbones through extracellular β-xylosidase activity (40) or import of AXOS before hydrolysis. In contrast, strains of cluster III displayed only a limited degradation of the AXOS substituents without an increase of the concentrations of the xylose backbones in the medium. This might be explained by the existence of α-arabinofuranosidases with different specificities. Indeed, several α-arabinofuranosidases have been characterized that are specific for Araf residues from monosubstituted Xylp, Araf residues from both mono- and disubstituted Xylp, and Araf residues from doubly substituted Xylp residues at position C-(O)-3 (38, 39, 41, 42). Also, the degree of substitution (not determined in the present study) may be an important factor in the degradation of AXOS (less degradation with increasing A/X) and hence explain preferences for certain AXOS molecules (5, 43). In addition, arabinose residues can be esterified with ferulic acid and could hinder the formation of α-arabinofuranosidase-substrate complexes (44). In the present study, no correlation could be found between degradation of AXOS substituents and the presence of a xylan esterase.
Xylose backbone consumption patterns.
In the present study, we detected the consumption of decorticated xylose backbones for strains belonging to clusters III, IV, and V. Most strains showed a preferential simultaneous metabolism of the short XOS fractions (xylobiose, xylotriose, and xylotetraose), while the longer xylose backbones were left unaffected. This is in accordance with the fact that particularly AXOS with a low average DP, including the AXOS-5-0.27 and AXOS-3-0.26 substrates, have significant bifidogenic properties in rats (6). Preferential metabolism of short oligosaccharide fractions by bifidobacterial strains has already been demonstrated in the case of fructo-oligosaccharides (19, 21, 45, 46), galacto-oligosaccharides (46, 47), and XOS (48). The preferential metabolism of short xylose backbones suggests the presence of specific transport systems for translocation of these XOS into the cell followed by intracellular hydrolysis (49–51). Indeed, ca. 5% of the total bifidobacterial gene content is dedicated to carbohydrate internalization, either through ATP-binding cassette (ABC) transporters, permeases, or proton symporters (52). For example, genome analysis of and microarray-based gene expression analysis in B. longum subsp. longum NCC2705 revealed 15 transport systems that could be involved in the transport of oligosaccharides (11, 53). The use of free xylose by B. longum subsp. longum NCC2705 (cluster II) and its inability to ferment XOS, as shown in the present study, reflects the presence of ABC transporters for xylose uptake in this strain, whereas no transporters for XOS are present (11, 53, 54). Finally, the XOS breakdown profile of B. catenulatum LMG 11043T suggests extracellular degradation and simultaneous uptake of the different fractions released (4). However, this might be less competitive than internal or cell wall-bound degradation, since it may lead to substrate losses through diffusion or consumption by competitors (4, 19, 21, 45).
The preference for xylotriose and xylotetraose by some cluster III strains may be ascribed to specific enzymes. Indeed, a reducing-end xylose-releasing exo-oligoxylanase (rexA) and an outer membrane β-xylosidase (xylB), both enzymes lacking a signal peptide, that possess a weak activity on xylobiose and a preferred activity on XOS with a DP of >2, have been characterized in B. adolescentis LMG 10502T (14, 49).
The increase of the xylose concentration during XOS breakdown for some of the xylose backbone-degrading strains indicates extracellular β-xylosidase activity. Increases in extracellular xylose concentrations during XOS metabolism have been shown for B. infantis 420 (55), B. longum 2 (55), B. animalis subsp. lactis BB-12 (50, 55), B. adolescentis ATCC 15703 (17), and B. adolescentis DSMZ 18350 (51), albeit based on endpoint determinations (except for the latter study). Xylose accumulation in the medium in the case of B. longum subsp. longum LMG 11047, B. longum subsp. longum CUETM 193, and B. longum subsp. longum CUETM 239 could be linked to the presence of a putative extracellular β-xylosidase gene (BL1544), which is an extracellular multidomain (SignP-GH43-CBM22-TmD) glycoside hydrolase. However, in the case of some other B. longum strains without β-xylosidase activity, including B. longum subsp. longum NCC2705, the BL1544 gene was present. Since there was no correlation between the presence of a nonsynonymous sequence variation in the GH43 domain and β-xylosidase activity, it is suggested that different regulation mechanisms may be responsible for this observation or, more likely, that the putative BL1544 gene indeed encodes an extracellular α-arabinofuranosidase (supported by the experiments of the present study) instead of an extracellular β-xylosidase (as annotated in reference 11). Furthermore, several microbial β-xylosidases have been characterized that exhibit both β-xylosidase and α-arabinofuranosidase activities, as Xylp and Araf are structurally similar (42, 56). Similarly, the presence of the putative β-endoxylanase gene (BL1543) could not be linked with an increase of the xylose concentration in the medium. Although a significant increase in the expression of the BL1543 gene has been found in B. longum subsp. longum NCC2705 when grown on AX (57), the production of a β-endoxylanase has not been reported on protein level nor has the backbone degradation of xylans by bifidobacteria been demonstrated until now. Alternatively, the absence of xylose in the supernatant could indicate efficient consumption of the extracellularly released xylose.
Arabinose and xylose consumption patterns.
The B. bifidum strains of cluster I did not use XOS, AXOS, or the corresponding free monosaccharides nor oligofructose and inulin (19). This suggests that these bifidobacterial strains probably evolved to use energy sources other than from plant origin. Indeed, genome analysis has shown that B. bifidum PRL2010 contains a low number of carbohydrate transporters compared to other Bifidobacterium spp. and that this species is particularly specialized in the consumption of human milk oligosaccharides and mucin (58, 59). However, other strains in this cluster that consumed one or both monosaccharides could possibly benefit from extracellularly released arabinose and xylose during XOS and (A)XOS consumption by bifidobacterial strains belonging to other clusters. A more preferential metabolism of arabinose compared to xylose by cluster II strains is in agreement with their use of the arabinose substituents but not the xylose backbones. In cluster III, more diverse monosaccharide preferences occurred among the bifidobacterial strains. Some strains displayed xylose backbone degradation, while xylose could not be consumed, indicating the presence of transport systems for XOS and not for xylose. Similar degradation preferences have been found for other bifidobacterial strains (16, 40, 60). This could be an advantage for these strains, since in the human colon only low concentrations of monosaccharides occur, while oligo- and polysaccharides are the main carbohydrates. The finding that B. longum subsp. longum LMG 11047 and B. longum subsp. longum CUETM 193, belonging to cluster IV, were the only strains in the present study that could use all substrates, encompassing free arabinose, free xylose, XOS, and AXOS, suggests that they may play a pivotal role in the fermentation of AXOS. Further, this could partly explain the dominance of B. longum subsp. longum among the bifidobacterial species present in the human colon (61).
In conclusion, AXOS degradation by bifidobacterial strains is a complex and strain-dependent process that involves consumption of arabinose substituents, whether or not followed by the consumption of the xylose backbones, either up to xylotetraose or longer XOS and either intracellularly or extracellularly. Strain-dependent differences in (A)XOS degradation could avoid competition among different bifidobacterial strains and favor coexistence whether or not through cooperation. Indeed, the release of partial carbohydrate breakdown products (arabinose, xylose, xylose backbones, or AXOS) in the medium by bifidobacterial strains belonging to one cluster may open cross-feeding opportunities with other bifidobacteria belonging to other clusters. This possible cross-feeding hypothesis has been shown by Pastell et al. (17) in a triculture of B. longum ATCC 15707, B. adolescentis ATCC 15703, and B. breve ATCC 15700. Finally, the intra- and interspecies differences in (A)XOS degradation potential within the bifidobacterial genus could have in vivo implications on the specificity of the stimulatory effect of AXOS and could help in the rational design of tailor-made prebiotics with a maximal bifidogenic effect in the colon.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the financial support of the Research Council of the Vrije Universiteit Brussel (SRP, IRP, and IOF projects), the Hercules Foundation, and the Research Foundation-Flanders (FWO-Vlaanderen). A.R. is the recipient of a Ph.D. fellowship of the FWO-Vlaanderen. We thank Christophe Pierlot for his contribution to the experimental work. We thank Christophe Courtin (KU Leuven) for providing the AXOS. We thank Shandong Longlive Bio-Technology for providing the XOS. We also thank Filip Timmermans (Thermo Scientific) for kindly providing the CarboPac PA200 column. We acknowledge Denis Roy (Université Laval, Québec, Canada) for kindly providing several bifidobacterial strains.
Footnotes
Published ahead of print 18 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02853-13.
REFERENCES
- 1.Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan shapes the human gut microbiota. Nat. Rev. Microbiol. 10:323–335. 10.1038/nrmicro2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flint HJ. 2004. Polysaccharide breakdown by anaerobic microorganisms inhabiting the mammalian gut. Adv. Appl. Microbiol. 56:89–120. 10.1016/S0065-2164(04)56003-3 [DOI] [PubMed] [Google Scholar]
- 3.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3:289–306. 10.4161/gmic.19897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Vuyst L, Leroy F. 2011. Cross-feeding between bifidobacteria and butyrate-producing colon bacteria explains bifidobacterial competitiveness, butyrate production, and gas production. Int. J. Food Microbiol. 149:73–80. 10.1016/j.ijfoodmicro.2011.03.003 [DOI] [PubMed] [Google Scholar]
- 5.Grootaert C, Delcour JA, Courtin CM, Broekaert WF, Verstraete W, Van de Wiele T. 2007. Microbial metabolism and prebiotic potency of arabinoxylan oligosaccharides in the human intestine. Trends Food Sci. Technol. 18:64–71. 10.1016/j.tifs.2006.08.004 [DOI] [Google Scholar]
- 6.Van Craeyveld V, Swennen K, Dornez E, Van de Wiele T, Marzorati M, Verstraete W, Delaedt Y, Onagbesan O, Decuypere E, Buyse J, De Ketelaere B, Broekaert WF, Delcour JA, Courtin CM. 2008. Structurally different wheat-derived arabinoxylo-oligosaccharides have different prebiotic and fermentation properties in rats. J. Nutr. 138:2348–2355. 10.3945/jn.108.094367 [DOI] [PubMed] [Google Scholar]
- 7.Cloetens L, Broekaert WF, Delaedt Y, Ollevier F, Courtin CM, Delcour JA, Rutgeerts P, Verbeke K. 2010. Tolerance of arabinoxylan-oligosaccharides and their prebiotic activity in healthy subjects: a randomised, placebo-controlled cross-over study. Br. J. Nutr. 103:703–713. 10.1017/S0007114509992248 [DOI] [PubMed] [Google Scholar]
- 8.Broekaert WF, Courtin CM, Verbeke K, Van de Wiele T, Verstraete W, Delcour JA. 2011. Prebiotic and other health-related effects of cereal-derived arabinoxylans, arabinoxylan-oligosaccharides, and xylooligosaccharides. Crit. Rev. Food Sci. Nutr. 51:178–194. 10.1080/10408390903044768 [DOI] [PubMed] [Google Scholar]
- 9.Neyrinck AM, Possemiers S, Druart C, van de Wiele T, De Backer F, Cani PD, Larondelle Y, Delzenne NM. 2011. Prebiotic effects of wheat arabinoxylan related to the increase in bifidobacteria, roseburia and bacteroides/prevotella in diet-induced obese mice. PLoS One 6:1–12. 10.1371/journal.pone.0020944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van den Abbeele P, Gerard P, Rabot S, Bruneau A, El Aidy S, Derrien M, Kleerebezem M, Zoetendal EG, Smidt H, Verstraete W, Van de Wiele T, Possemiers S. 2011. Arabinoxylans and inulin differentially modulate the mucosal and luminal gut microbiota and mucin-degradation in humanized rats. Environ. Microbiol. 13:2667–2680. 10.1111/j.1462-2920.2011.02533.x [DOI] [PubMed] [Google Scholar]
- 11.Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, Pridmore RD, Arigoni F. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U. S. A. 99:14422–14427. 10.1073/pnas.212527599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damen B, Verspreet J, Pollet A, Broekaert WF, Delcour JA, Courtin CM. 2011. Prebiotic effects and intestinal fermentation of cereal arabinoxylans and arabinoxylan oligosaccharides in rats depend strongly on their structural properties and joint presence. Mol. Nutr. Food Res. 55:1862–1874. 10.1002/mnfr.201100377 [DOI] [PubMed] [Google Scholar]
- 13.Izydorczyk MS, Biliaderis CG. 1995. Cereal arabinoxylans: advances in structure and physicochemical properties. Carbohydr. Polym. 28:33–48. 10.1016/0144-8617(95)00077-1 [DOI] [Google Scholar]
- 14.Lagaert S, Van Campenhout S, Pollet A, Bourgois TM, Delcour JA, Courtin CM, Volckaert G. 2007. Recombinant expression and characterization of a reducing-end xylose-releasing exo-oligoxylanase from Bifidobacterium adolescentis. Appl. Environ. Microbiol. 73:5374–5377. 10.1128/AEM.00722-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Laere KMJ, Hartemink R, Bosveld M, Schols HA, Voragen AGJ. 2000. Fermentation of plant cell wall derived polysaccharides and their corresponding oligosaccharides by intestinal bacteria. J. Agric. Food Chem. 48:1644–1652. 10.1021/jf990519i [DOI] [PubMed] [Google Scholar]
- 16.Crittenden R, Karppinen S, Ojanen S, Tenkanen M, Fagerstrom R, Matto J, Saarela M, Mattila-Sandholm T, Poutanen K. 2002. In vitro fermentation of cereal dietary fibre carbohydrates by probiotic and intestinal bacteria. J. Sci. Food Agric. 82:1–9. 10.1002/jsfa.1053 [DOI] [Google Scholar]
- 17.Pastell H, Westermann P, Meyer AS, Tuomainen P, Tenkanen M. 2009. In vitro fermentation of arabinoxylan-derived carbohydrates by bifidobacteria and mixed faecal microbiota. J. Agric. Food Chem. 57:8598–8606. 10.1021/jf901397b [DOI] [PubMed] [Google Scholar]
- 18.Macfarlane S, Macfarlane GT, Cummings JH. 2006. Prebiotics in the gastrointestinal tract. Aliment. Pharmacol. Ther. 24:701–714. 10.1111/j.1365-2036.2006.03042.x [DOI] [PubMed] [Google Scholar]
- 19.Falony G, Lazidou K, Verschaeren A, Weckx S, Maes D, De Vuyst L. 2009. In vitro kinetic analysis of fermentation of prebiotic inulin-type fructans by Bifidobacterium species reveals four different phenotypes. Appl. Environ. Microbiol. 75:454–461. 10.1128/AEM.01488-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kok RG, De Waal A, Schut F, Welling GW, Weenk G, Hellingwerf KJ. 1996. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl. Environ. Microbiol. 62:3668–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van der Meulen R, Makras L, Verbrugghe K, Adriany T, De Vuyst L. 2006. In vitro kinetic analysis of oligofructose consumption by Bacteroides and Bifidobacterium spp. indicates different degradation mechanisms. Appl. Environ. Microbiol. 72:1006–1012. 10.1128/AEM.72.2.1006-1012.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rivière A, Eeltink S, Pierlot C, Balzarini T, Moens F, Selak M, De Vuyst L. 2013. A high-resolution ion-exchange chromatography method for monitoring of prebiotic arabinoxylan-oligosaccharide degradation in a complex fermentation medium. Anal. Chem. 85:4982–4990. 10.1021/ac400187f [DOI] [PubMed] [Google Scholar]
- 23.Vrancken G, Rimaux T, De Vuyst L, Leroy F. 2008. Kinetic analysis of growth and sugar consumption by Lactobacillus fermentum IMDO 130101 reveals adaptation to the acidic sourdough ecosystem. Int. J. Food Microbiol. 128:58–66. 10.1016/j.ijfoodmicro.2008.08.001 [DOI] [PubMed] [Google Scholar]
- 24.Braem G, De Vliegher S, Supré K, Haesebrouck F, Leroy F, De Vuyst L. 2011. (GTG)5-PCR fingerprinting for the classification and identification of coagulase-negative Staphylococcus species from bovine milk and teat apices: a comparison of type strains and field isolates. Vet. Microbiol. 147:67–74. 10.1016/j.vetmic.2010.05.044 [DOI] [PubMed] [Google Scholar]
- 25.Resource Coordinators NCBI 2013. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 41:8–20. 10.1093/nar/gks1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 27.Rozen S, Skaletsky HJ. 2000. Primer3 on the www for general users and for biologist programmers, p 365–386 In Krawetz S, Misener S. (ed), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 28.Weckx S, De Rijk P, Van Broeckhoven C, Del-Favero J. 2004. SNPbox: web-based high-throughput primer design from gene to genome. Nucleic Acids Res. 32:170–172. 10.1093/nar/gkh369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. (Oxford) 41:95–98 [Google Scholar]
- 30.Ewing B, Green P. 1998. Basecalling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186–194 [PubMed] [Google Scholar]
- 31.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. CLUSTAL W and CLUSTAL X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 32.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 33.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 34.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769–781. 10.1042/BJ20040892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH. 2013. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41:348–352. 10.1093/nar/gks1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amaretti A, Tamburini E, Bernardi T, Pompei A, Zanoni S, Vaccari G, Matteuzzi D, Rossi M. 2006. Substrate preference of Bifidobacterium adolescentis MB 239: compared growth on single and mixed carbohydrates. Appl. Microbiol. Biotechnol. 73:654–662. 10.1007/s00253-006-0500-9 [DOI] [PubMed] [Google Scholar]
- 37.van den Broek LAM, Hinz SWA, Beldman G, Vincken JP, Voragen AGJ. 2008. Bifidobacterium carbohydrases: their role in breakdown and synthesis of (potential) prebiotics. Mol. Nutr. Food Res. 52:146–163. 10.1002/mnfr.200700121 [DOI] [PubMed] [Google Scholar]
- 38.Lagaert S, Pollet A, Delcour JA, Lavigne R, Courtin CM, Volckaert G. 2010. Substrate specificity of three recombinant α-l-arabinofuranosidases from Bifidobacterium adolescentis and their divergent action on arabinoxylan and arabinoxylan oligosaccharides. Biochem. Biophys. Res. Commun. 402:644–650. 10.1016/j.bbrc.2010.10.075 [DOI] [PubMed] [Google Scholar]
- 39.van den Broek LAM, Lloyd RM, Beldman G, Verdoes JC, McCleary BV, Voragen AGJ. 2005. Cloning and characterization of arabinoxylan arabinofuranohydrolase-D3 (AXHd3) from Bifidobacterium adolescentis DSM20083. Appl. Microbiol. Biotechnol. 67:641–647. 10.1007/s00253-004-1850-9 [DOI] [PubMed] [Google Scholar]
- 40.Palframan RJ, Gibson GR, Rastall RA. 2003. Carbohydrate preferences of Bifidobacterium species isolated from the human gut. Curr. Issues Intest. Microbiol. 4:71–75 [PubMed] [Google Scholar]
- 41.Van Laere KMJ, Voragen CHL, Kroef T, van den Broek LAM, Beldman G, Voragen AGJ. 1991. Purification and mode of action of two different arabinoxylan arabinofuranohydrolases from Bifidobacterium adolescentis DSM 20083. J. Microbiol. Biotechnol. 51:606–613 [Google Scholar]
- 42.Numan MD, Bhosle NB. 2006. a-l-Arabinofuranosidases: the potential applications in biotechnology. J. Ind. Microbiol. Biotechnol. 33:247–260. 10.1007/s10295-005-0072-1 [DOI] [PubMed] [Google Scholar]
- 43.Pollet A, Van Craeyveld V, Van de Wiele T, Verstraete W, Delcour JA, Courtin CM. 2012. In vitro fermentation of arabinoxylan oligosaccharides and low molecular mass arabinoxylans with different structural properties from wheat (Triticum aestivum L.) bran and psyllium (Plantago ovata Forsk) seed husk. J. Agric. Food Chem. 60:946–954. 10.1021/jf203820j [DOI] [PubMed] [Google Scholar]
- 44.Saha BC. 2000. α-l-Arabinofuranosidases: biochemistry, molecular biology and application in biotechnology. Biotechnol. Adv. 18:403–423. 10.1016/S0734-9750(00)00044-6 [DOI] [PubMed] [Google Scholar]
- 45.Van der Meulen R, Avonts L, De Vuyst L. 2004. Short fractions of oligofructose are preferentially metabolized by Bifidobacterium animalis DN-173 010. Appl. Environ. Microbiol. 70:1923–1930. 10.1128/AEM.70.4.1923-1930.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watson D, O'Connell Motherway M, Schoterman MHC, Joost van Neerven RJ, Nauta A, van Sinderen D. 2013. Selective carbohydrate utilization by lactobacilli and bifidobacteria. J. Appl. Microbiol. 114:1132–1146. 10.1111/jam.12105 [DOI] [PubMed] [Google Scholar]
- 47.Amaretti A, Bernardi T, Tamburini E, Zanoni S, Lomma M, Matteuzzi D, Rossi M. 2007. Kinetics and metabolism of Bifidobacterium adolescentis MB 239 growing on glucose, galactose, lactose, and galactooligosaccharides. Appl. Environ. Microbiol. 73:3637–3644. 10.1128/AEM.02914-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moura P, Barata R, Carvalheiro F, Girio F, Loureiro-Dias MC, Esteves MP. 2007. In vitro fermentation of xylo-oligosaccharides from corn cobs autohydrolysis by Bifidobacterium and Lactobacillus strains. Food Sci. Technol. 40:963–972. 10.1016/j.lwt.2006.07.013 [DOI] [Google Scholar]
- 49.Lagaert S, Pollet A, Delcour JA, Lavigne R, Courtin CM, Volckaert G. 2011. Characterization of two β-xylosidases from Bifidobacterium adolescentis and their contribution to the hydrolysis of prebiotic xylooligosaccharides. Appl. Microbiol. Biotechnol. 92:1179–1185. 10.1007/s00253-011-3396-y [DOI] [PubMed] [Google Scholar]
- 50.Gilad O, Jacobsen S, Stuer-Lauridsen B, Pedersen MB, Garrigues C, Svensson B. 2010. Combined transcriptome and proteome analysis of Bifidobacterium animalis subsp. lactis BB-12 grown on xylo-oligosaccharides and a model of their utilization. Appl. Environ. Microbiol. 76:7285–7291. 10.1128/AEM.00738-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amaretti A, Bernardi T, Leonardi A, Raimondi S, Zanoni S, Rossi M. 2013. Fermentation of xylo-oligosaccharides by Bifidobacterium adolescentis DSMZ 18350: kinetics, metabolism, and β-xylosidase activities. Appl. Microbiol. Biotechnol. 97:3109–3117. 10.1007/s00253-012-4509-y [DOI] [PubMed] [Google Scholar]
- 52.Ventura M, O'Flaherty S, Claesson MJ, Turroni F, Klaenhammer TR, van Sinderen D, O'Toole PW. 2009. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat. Rev. Microbiol. 7:61–71. 10.1038/nrmicro2047 [DOI] [PubMed] [Google Scholar]
- 53.Parche S, Amon J, Jankovic I, Rezzonico E, Beleut M, Barutcu H, Schendel I, Eddy MP, Burkovski A, Arigoni F, Titgemeyer F. 2007. Sugar transport systems of Bifidobacterium longum NCC2705. J. Mol. Microbiol. Biotechnol. 12:9–19. 10.1159/000096455 [DOI] [PubMed] [Google Scholar]
- 54.Liu DW, Wang SA, Xu B, Guo YH, Zhao JL, Liu W, Sun ZK, Shao CL, Wei X, Jiang Z, Wang XS, Liu F, Wang J, Huang LY, Hu DR, He X, Riedel CU, Yuan J. 2011. Proteomics analysis of Bifidobacterium longum NCC2705 growing on glucose, fructose, mannose, xylose, ribose, and galactose. Proteomics 11:2628–2638. 10.1002/pmic.201100035 [DOI] [PubMed] [Google Scholar]
- 55.Jaskari J, Kontula P, Siitonen A, Jousimies-Somer H, Mattila-Sandholm T, Poutanen K. 1998. Oat β-glucan and xylan hydrolysates as selective substrates for Bifidobacterium and Lactobacillus strains. Appl. Microbiol. Biotechnol. 49:175–181. 10.1007/s002530051155 [DOI] [PubMed] [Google Scholar]
- 56.Wagschal K, Heng C, Lee CC, Wong DWS. 2009. Biochemical characterization of a novel dual-function arabinofuranosidase/xylosidase isolated from a compost starter mixture. Appl. Microbiol. Biotechnol. 81:855–863. 10.1007/s00253-008-1662-4 [DOI] [PubMed] [Google Scholar]
- 57.Savard P, Roy D. 2008. Determination of differentially expressed genes involved in arabinoxylan degradation by Bifidobacterium longum NCC2705 using real-time RT-PCR. Probiotics Antimicrob. Proteins 1:121–129. 10.1007/s12602-009-9015-x [DOI] [PubMed] [Google Scholar]
- 58.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sánchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald GF, Mills D, Margolles A, Kelly F, van Sinderen D, Ventura M. 2010. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. U. S. A. 107:19514–19519. 10.1073/pnas.1011100107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turroni F, Strati F, Foroni E, Serafini F, Duranti S, van Sinderen D, Ventura M. 2012. Analysis of predicted carbohydrate transport systems encoded by Bifidobacterium bifidum PRL2010. Appl. Environ. Microbiol. 78:5002–5012. 10.1128/AEM.00629-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, Sun BG, Cao YP, Wang CT. 2010. In vitro fermentation of xylooligosaccharides from wheat bran insoluble dietary fibre by bifidobacteria. Carbohydr. Polym. 82:419–423. 10.1016/j.carbpol.2010.04.082 [DOI] [Google Scholar]
- 61.Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, Ugarte E, Muñoz-Tamayo R, Paslier DLE, Nalin R, Dore J, Leclerc M. 2009. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 11:2574–2584. 10.1111/j.1462-2920.2009.01982.x [DOI] [PubMed] [Google Scholar]
- 62.Bahaka D, Neut C, Khattabi A, Monget D, Gavini F. 1993. Phenotypic and genomic analyses of human strains belonging or related to Bifidobacterium longum, Bifidobacterium infantis, and Bifidobacterium breve. Int. J. Syst. Bacteriol. 43:565–573. 10.1099/00207713-43-3-565 [DOI] [PubMed] [Google Scholar]
- 63.Matamoros S, Savard P, Roy D. 2011. Genotyping of Bifidobacterium longum subsp. longum strains by multilocus variable number of tandem repeat analysis. J. Microbiol. Methods 87:378–380. 10.1016/j.mimet.2011.10.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.