Abstract
Three succinate coenzyme A (succinate-CoA) ligases (SucCD) from Escherichia coli, Advenella mimigardefordensis DPN7T, and Alcanivorax borkumensis SK2 were characterized regarding their substrate specificity concerning succinate analogues. Previous studies had suggested that SucCD enzymes might be promiscuous toward succinate analogues, such as itaconate and 3-sulfinopropionate (3SP). The latter is an intermediate of the degradation pathway of 3,3′-dithiodipropionate (DTDP), a precursor for the biotechnical production of polythioesters (PTEs) in bacteria. The sucCD genes were expressed in E. coli BL21(DE3)/pLysS. The SucCD enzymes of E. coli and A. mimigardefordensis DPN7T were purified in the native state using stepwise purification protocols, while SucCD from A. borkumensis SK2 was equipped with a C-terminal hexahistidine tag at the SucD subunit. Besides the preference for the physiological substrates succinate, itaconate, ATP, and CoA, high enzyme activity was additionally determined for both enantiomeric forms of malate, amounting to 10 to 21% of the activity with succinate. Km values ranged from 2.5 to 3.6 mM for l-malate and from 3.6 to 4.2 mM for d-malate for the SucCD enzymes investigated in this study. As l-malate-CoA ligase is present in the serine cycle for assimilation of C1 compounds in methylotrophs, structural comparison of these two enzymes as members of the same subsubclass suggested a strong resemblance of SucCD to l-malate-CoA ligase and gave rise to the speculation that malate-CoA ligases and succinate-CoA ligases have the same evolutionary origin. Although enzyme activities were very low for the additional substrates investigated, liquid chromatography/electrospray ionization-mass spectrometry analyses proved the ability of SucCD enzymes to form CoA-thioesters of adipate, glutarate, and fumarate. Since all SucCD enzymes were able to activate 3SP to 3SP-CoA, we consequently demonstrated that the activation of 3SP is not a unique characteristic of the SucCD from A. mimigardefordensis DPN7T. The essential role of sucCD in the activation of 3SP in vivo was proved by genetic complementation.
INTRODUCTION
Succinyl coenzyme A (succinyl-CoA) synthetases (succinate-CoA ligase; SucCD; EC 6.2.1.4 and 6.2.1.5) catalyze the reversible conversion of succinyl-CoA to succinate under the concomitant formation of a nucleoside triphosphate (NTP) in the citric acid cycle (1, 2). The enzyme consists of two different subunits forming a heterodimer or a heterotetramer structure (3, 4). The α subunit (SucD) and the β subunit (SucC) have molecular masses of 29 to 34 kDa and 41 to 45 kDa, respectively (5). The β subunit is responsible for the binding of the NTP, whereas the α subunit binds CoA (6). So far, the binding site for the substrate succinate has not been located; however, it is assumed that it occurs at the dimer interface (6–9). A conserved histidine residue of the α subunit is phosphorylated during catalysis. The phosphate moiety is conferred to a nucleoside diphosphate to yield an NTP. Replacement of the histidine residue with any other amino acid residue results in an inactive enzyme (3, 7). The reverse reaction leading from succinate to succinyl-CoA is important in the reductive citric acid cycle in many bacteria, as well as part of heme biosynthesis and ketone body activation in higher organisms (10).
Although much interest was devoted to the structure (6, 11, 12), function, regulation (13), and nucleotide specificity (14, 15), only little is known about the substrate range concerning carbon acids. Among the more than 30 members of the subsubclass acid CoA ligases (EC 6.2.1), miscellaneous descriptions about extended ranges concerning the acid substrates for the enzymes have been published. The greatest flexibility was shown for long-chain fatty acid CoA ligases (EC 6.2.1.3) (16), but acetate-CoA ligases (17, 18), a propionate-CoA ligase (19), and a butyrate-CoA ligase (20) were also shown to react with more than one carbon acid substrate. In 2007, the succinate-CoA ligase from the hyperthermophilic archaeon Thermococcus kodakaraensis, which structurally resembles the acetate-CoA ligase from Pyrococcus furiosus, was described (21). This enzyme exhibits a subunit domain distribution which is different from the αβ heterodimer/αβ tetramer structure typical for SucCD enzymes. This enzyme showed an extended substrate range and was also active with isovalerate, 3-methyl thiopropionate, glutarate, adipate, and butyrate. However, for succinate-CoA ligases with a classical domain structure, relevant investigations are still missing. In 1957, the formation of itaconyl-CoA from itaconate, a structural analogue to succinate, in mammalian liver mitochondria catalyzed by SucCD was reported (22). Later, this reaction was proved for the SucCD from Micrococcus sp. and Pseudomonas fluorescens (22–24).
Later, other investigators showed the participation of the SucCD of Advenella mimigardefordensis DPN7T in the degradation pathway of 3,3′-dithiodipropionic acid (DTDP), a precursor for the production of polythioesters in bacteria (25–28). In this strain, DTDP is metabolized via the intermediate product 3-sulfinopropionate (3SP) (29–31). This xenobiotic structural analogue to succinate carries a sulfino group instead of the carboxyl group in succinate, and it is converted to 3SP-CoA in vivo (26). The authors also showed that the mutant strain A. mimigardefordensis DPN7T ΔsucCD was no longer able to grow on DTDP and 3SP. Furthermore, Schürmann et al. demonstrated the conversion of itaconate to itaconyl-CoA by SucCD from A. mimigardefordensis DPN7T (SucCDAm) in vitro (26). In addition to that, the authors observed the formation of 3SP-CoA by a crude extract of the expression strain Escherichia coli BL21(DE3)/pLysS not harboring genes for SucCDAm (26). These findings suggested that the formation of 3SP-CoA from 3SP is not a unique characteristic of the A. mimigardefordensis DPN7T SucCD, and it raised the question of whether SucCD enzymes in general have an extended substrate range like other members of enzyme subsubclass 6.2.1 and whether other SucCD enzymes are also able to form itaconyl-CoA and 3SP-CoA.
In this study, we purified three homologous SucCD enzymes and characterized these enzymes with regard to their ability to convert different carbon acid substrates as analogues of succinate to their corresponding CoA-thioesters in vitro. These included the SucCD enzyme from Escherichia coli BL21 (SucCDBL21), SucCDAm, and the SucCD enzyme from Alcanivorax borkumensis SK2 (SucCDAboHis). SucCDBL21 is identical to the E. coli K-12 (ATCC 47076) enzyme (32) and has been of scientific interest for the last 60 years. The enzyme is able to use ATP as well as GTP as a cosubstrate (15). Several crystallographic structures proved the locations of the binding domains for the nucleotide and the CoA involved in catalysis (11, 33). A. mimigardefordensis is a bacterium that can grow on the sulfur-containing precursor DTDP and is a suitable host for polythioester production (28). As mentioned above, the activation of itaconate, as well as that of 3SP, was shown for SucCDAm (26). A. borkumensis SK2 is a marine gammaproteobacterium whose genome was published in 2006 (34). SucCDAbo has not been investigated yet. Alignments of the primary structures show high sequence similarity among the SucCD enzymes investigated in this study.
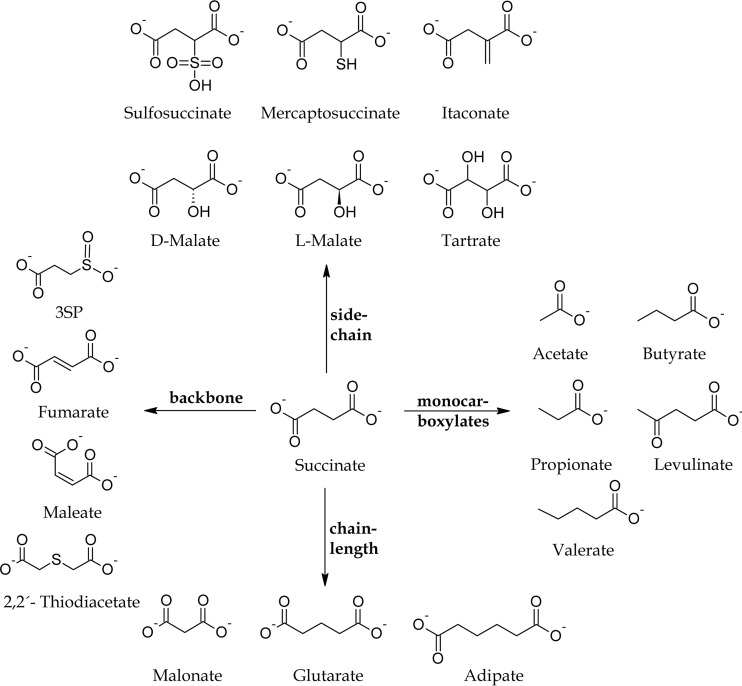
As there are no data on the extended substrate ranges of SucCD except from itaconate for mammalian SucCD enzymes and itaconate and 3SP for SucCDAm, we aimed at using substrate compounds that differ from succinate with respect to their carbon acid backbone, their chain length, or their side chain; in addition, we analyzed monocarboxylate compounds (Fig. 1). We also investigated the SucCD-dependent activation of the side chain variant malate to malyl-CoA, and we analyzed the relation between our findings for malate-CoA ligase activity exhibited by succinate-CoA ligases in this study and further report on the participation of a l-malate-CoA ligase in the serine cycle in some methylotrophic bacteria.
FIG 1.
Succinate and analogous compounds investigated as potential substrates for SucCD enzymes.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
All strains used in this study are listed in Table 1. Cells of A. mimigardefordensis DPN7T and A. mimigardefordensis DPN7T ΔsucCD were cultivated at 30°C in mineral salt medium (MSM) (35) containing 20 mM gluconate, 50 mM DTDP, or 20 mM 3SP as the sole source of carbon and energy. Carbon sources were added to MSM from filter-sterilized stock solutions as indicated in the text. For maintenance of plasmids, solutions of antibiotics were prepared according to the method of Sambrook et al. (36) at the following concentrations: 100 μg/ml ampicillin [for cultivation of strains harboring the pET-23a(+) vector system], 150 μg/ml [for cultivation of strains harboring the pBluescriptSK(−) vector system], and 34 μg/ml chloramphenicol.
TABLE 1.
Strains, plasmids and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Description or sequence (5′–3′)a | Source or reference |
|---|---|---|
| Strains | ||
| A. mimigardefordensis DPN7 | Wild type, DTDP-degrading bacterium | 30 (DSM 17166T, LMG 22922T) |
| A. mimigardefordensis DPN7 ΔsucCD | sucCD deletion mutant of strain DPN7 | 26 |
| E. coli Top10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 nupG deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL(Strr) endA1 λ− | Invitrogen, Carlsbad, CA |
| E. coli S17-1 | thi-1 proA hsdR17(rK− mK+) recA1; the tra gene of plasmid RP4 is integrated into the genome | 42 |
| E. coli BL21(DE3) pLysS | F− ompT hsdSB(rB− mB−) gal dcm (DE3)/pLysS (Cmr) | Novagen, Madison, WI |
| Alcanivorax borkumensis SK2 | Wild type | DSM 11573 |
| Plasmids | ||
| pBluescriptSK(−) | Apr, lacPOZ′ | Stratagene, San Diego, CA |
| pET-23a(+) | pBR322 ori f1 ori His6 Apr T7lac | Novagen, Madison, WI |
| pBBR1MCS5 | Broad-host-range expression vector, Gmr | 45 |
| pBluescriptSK(-)::sucCDAm | Apr | 26 |
| pBluescriptSK(-)::sucCDBL21 | Apr | This study |
| pET-23a(+)::sucCDAbo | Apr | This study |
| pET-23a(+)::sucCDAboHis | Apr | This study |
| pBBR1MCS5::sucCDAm | Gmr | This study |
| Oligonucleotides | ||
| sucCDforward_PstI | CTGCAGCAGTCTCAATTCGTGTGCTCGC | 26 |
| sucCDreverse_XhoI_stop | CTCGAGTTACAGTACTGATTTGAGCAGTTTG | 26 |
| sucCDBL21_forward_EcoRI | AAAAGAATTCTCCGACAAGCGATGCCTGATG | |
| suCDBL21_reverse_HindIII | AAAAAAGCTTTTATTTCAGAACAGTTTTCAGTGCTTCACC | |
| sucCDAbo_forward_NdeI | AAAACATATGAATCTCCATGAATATCAGTCAAAACAGC | |
| sucCDAbo_reverse_SalI | AAAAGTCGACTTACCAGCCAGTCGCTTCTTTCAC | |
| P_Abo_rev_mutagenesis | CCAGCCAGTCGCTTCTTTCAC (5′ phosphorylation) | |
| P_forward_XhoI_Histag_Abo | CTCGAGCACCACCACCACC (5′ phosphorylation) | |
| SKF2 (Abo seq) | TCTGAGCGCAGTGCTGG | |
| T7 terminator pET-23a | GCTAGTTATTGCTCAGCGG | |
| T7 promoter pET-23a | TAATACGACTCACTATAGGG | |
| For3 (BL21 seq) | ACTTTTTGCCCGCTATGG | |
| For2 (BL21 seq) | GTATCAAACAGATGCCAATGG | |
| Rev2 (BL21 seq) | GTAGTGCCGCCTTTACCTG | |
| SucCDAm_Prom_fw_XhoI | CTCGAGTTGCTGGTCACGCAGGAAGG |
For the abbreviations used in the E. coli genotypes, see reference 58.
Strains of E. coli were cultivated in lysogeny broth (LB) medium (36) or ZYP-5052 complex medium for autoinduction according to the method of Studier (37). The latter was used for homo- and heterologous expression of genes in E. coli under the control of the lac promoter and T7 promoter, respectively. For expression of sucCD, a single colony of the expression strain harboring the expression plasmid was used to inoculate a preculture of 50 ml LB medium in a baffled flask containing antibiotics, which was then incubated at 105 rpm and 30°C. After 6 to 9 h, the main culture containing 500 ml ZYP-5052 autoinduction medium in a 2.5-liter baffled flask with antibiotics was inoculated with 2% (vol/vol) of the preculture and the culture was incubated for 36 to 48 h at 105 rpm and 30°C. For complementation experiments with A. mimigardefordensis DPN7T strains, precultures grown in the presence of 20 mM gluconate were used to inoculate the main cultures (50 mM DTDP), resulting in an optical density (600 nm) of 0.1.
Chemicals.
d-Malic acid of high-purity grade (Chemical Abstracts Service [CAS] registry number 636-61-3) was purchased from Alfa Aeser (Karlsruhe, Germany), and l-malic acid of high-purity grade (CAS registry number 97-67-6) was purchased from Applichem (Darmstadt, Germany). DTDP of high-purity grade was purchased from Sigma-Aldrich (Steinheim, Germany). 3SP was synthesized as described by Jollès-Bergeret (38); the procedure was modified by one repetition of the step for alkaline cleavage of the intermediate bis-(2-carboxyethyl)sulfone, as described earlier (29). The synthesis and purity of the substance were confirmed by gas chromatography (GC) and GC/mass spectrometry (MS). According to GC/MS, the purity of the 3SP used was 98.7% for qualitative and quantitative enzyme assay and at least 95% when it was used as the carbon and energy source in MSM.
Analysis of 3SP by GC or GC/MS.
For purity analysis, the synthesized 3SP was analyzed by GC or GC/MS as described previously (26). For this, 3SP was subjected to methylation in the presence of 1 ml chloroform, 0.850 ml methanol, and 0.150 ml sulfuric acid for 2 h at 100°C. Upon methylation, 2 ml H2O was added and the samples were vigorously shaken for 30 s. After phase separation, the organic layer containing the resulting methyl esters of the organic acids was analyzed in an HP6850 gas chromatograph equipped with a BP21 capillary column (50 m by 0.22 mm; film thickness, 250 nm; SGE, Darmstadt, Germany) and a flame ionization detector. Helium was used as the carrier gas at a flow rate of 0.6 ml/min. The temperatures of the injector and detector were 250 and 240°C, respectively. Identification of peaks was performed by using AMDIS software in combination with the NIST database (39).
Analysis of CoA-thioester formation by LC/ESI-MS.
CoA-thioesters, which were formed during enzyme assays, were monitored by high-pressure liquid chromatography (HPLC) in combination with MS (liquid chromatography [LC]/electrospray ionization [ESI]-MS]), as described previously (26). LC/ESI-MS analysis was carried out with an UltiMate 3000 HPLC apparatus (Dionex GmbH, Idstein, Germany) connected directly to a LXQ Finnigan mass spectrometer (Thermo Scientific, Dreieich, Germany). An Acclaim 120 C18 reversed-phase LC column (4.6 by 250 mm, 5 μm, with 120 Å pores) was used to separate the CoA-thioesters at 30°C. The eluents used were an ammonium acetate buffer (50 mM, pH 5.0) adjusted with acetic acid (eluent A) and 100% (vol/vol) methanol (eluent B). Elution occurred at a flow rate of 0.3 ml/min. Ramping was performed as follows: equilibration with 90% eluent A for 2 min before injection and 90 to 45% eluent A for 20 min, followed by holding for 2 min and then a return to 90% eluent A within 5 min after injection. Detection of CoA-thioesters occurred at 259 nm with a photodiode array detector. A solution of 0.4 mM CoA was used to tune the instrument by direct infusion at a flow rate of 10 μl/min into the ion source of the mass spectrometer to optimize the ESI-MS system for maximum generation of protonated molecular ions (parents) of CoA derivatives. The following tuning parameters were retained for optimum detection of CoA-thioesters: capillary temperature, 300°C; sheath gas flow, 12 liters/h; auxiliary gas flow, 6 liters/h; and sweep gas flow, 1 liter/h. The mass range was set to m/z equal to 50 to 1,000 Da when the system was run in the scan mode. The collision energy in the MS mode was set to 30 V and yielded fragmentation patterns that were in good accordance with those found in other publications (26, 40).
Isolation and manipulation of DNA.
Chromosomal DNA of A. mimigardefordensis strain DPN7T was isolated according to the method of Marmur (41). Plasmid DNA was isolated from E. coli using a peqGOLD plasmid miniprep kit I from Peqlab Biotechnologie GmbH (Erlangen, Germany) according to the manufacturer's manual. DNA was digested with restriction endonucleases (Fermentas GmbH, St. Leon-Rot, Germany) under the conditions described by the manufacturer. PCRs were carried out in an Omnigene HBTR3CM DNA thermal cycler (Hybaid, Heidelberg, Germany) or a PeqSTAR 2× gradient thermal cycler (Peqlab Biotechnologie GmbH, Erlangen, Germany) using Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA) and Phusion high-fidelity DNA polymerase (Fermentas GmbH, St. Leon-Rot, Germany). T4 DNA ligase was purchased from Fermentas (Fermentas GmbH, St. Leon-Rot, Germany). Primers were synthesized by MWG-Biotech AG (Ebersberg, Germany) and are listed in Table 1.
Transfer of DNA.
Competent cells of E. coli strains were prepared and transformed by the CaCl2 procedure (36). Plasmids were transferred to A. mimigardefordensis DPN7T cells by conjugation (42).
DNA sequencing and sequence data analysis.
Sequence analysis was performed by Seqlab (Göttingen, Germany). Sequences were analyzed using the program BLAST (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/BLAST/) (43).
Cloning of sucCD genes for expression in E. coli BL21(DE3)/pLysS.
The corresponding sucCD genes were amplified from total genomic DNA of A. mimigardefordensis strain DPN7T, E. coli BL21, and A. borkumensis SK2 by PCR using Phusion high-fidelity DNA polymerase (New England BioLabs GmbH, Frankfurt am Main, Germany) or Biomix containing Taq DNA polymerase (Bioline GmbH, Luckenwalde, Germany). Information on genomic sequences was obtained from the Integrated Microbial Genomes nameplate (https://img.jgi.doe.gov/cgi-bin/er/main.cgi) (44). Oligonucleotides are listed in Table 1. Amplification of sucCDAm from genomic DNA of A. mimigardefordensis DPN7T was performed previously (26) using oligonucleotides sucCDforward_PstI and sucCDreverse_XhoIstopp. Amplification of sucCDBL21 (GenBank accession no. P0A836 and P0AGE9) from the genomic DNA of E. coli BL21 was performed by use of oligonucleotides sucCDBL21_forward_EcoRI and sucCDBL21_reverse_HindIII and yielded a fragment of 2,171 bp. Amplification of the sucCD genes for the native form of SucCD from A. borkumensis SK2 (locus tags ABO_1493 and ABO_1492) was performed by using oligonucleotides sucCDAbo_forward_NdeI and sucCDAbo_reverse_SalI and gave a fragment of 2,041 bp.
PCR products were isolated from agarose gels using a peqGOLD gel extraction kit (Peqlab Biotechnologie GmbH, Erlangen, Germany), digested with the appropriate restriction enzymes provided in the primer name, and ligated with digested pET-23a(+) (Novagen, Madison, WI) or pBluescriptSK(−) (Stratagene, San Diego, CA), yielding pBluescriptSK(−)::sucCDAm, pBluescriptSK(−)::sucCDBL21, and pET-23a(+)::sucCDAbo.
Ligation products were used for transformation of CaCl2-competent cells of E. coli Top10, and transformants were selected on LB agar plates containing ampicillin. After that, the hybrid plasmids were isolated, analyzed by sequencing, and used for transformation of CaCl2-competent cells of E. coli BL21(DE3)/pLysS (New England BioLabs Inc., Ipswich, MA). Plasmid pET-23a(+)::sucCDAboHis was generated by PCR-based mutagenesis using 5′-phosphorylated oligonucleotides P_forward_XhoI_Histag_Abo and P_Abo_rev_mutagenesis and pET-23a(+)::sucCDAbo as the template. This PCR led to the deletion of the terminal stop codon of the sucD gene. After amplification, a ligation reaction was performed in the buffer used for PCR, and the sample was used for transformation of CaCl2-competent cells of E. coli Top10.
Construction of plasmid for complementation experiments.
For complementation studies in the broad-host-range vector pBBR1MCS-5 (45), sucCDAm and a 478-bp upstream region were amplified by PCR using Phusion high-fidelity DNA polymerase (New England BioLabs GmbH, Frankfurt am Main, Germany) and applying oligonucleotides sucCDAm_Prom_fw_XhoI and sucCDreverse_XhoI_stop (Table 1) (26). The PCR product was ligated into the pJet1.2 blunt vector. After digestion using XhoI, the gene fragment of 2,541 bp was extracted from the agarose gel using the peqGOLD gel extraction kit (Peqlab Biotechnologie GmbH, Erlangen, Germany) and ligated with pBBR1MCS-5, which had previously been linearized with the XhoI restriction enzyme. The ligation products were transferred to CaCl2-competent cells of E. coli S17-1 and E. coli Top10. The hybrid plasmid pBBR1MCS-5::sucCDAm was then transferred into A. mimigardefordensis DPN7T ΔsucCD by conjugation (42).
Preparation of crude extracts.
Cells from 50- to 500-ml cultures were harvested by centrifugation (20 min, 4°C, 4,000 × g) and stored at −20°C until use. Cells were resuspended in 50 mM Tris-HCl buffer (pH 7.4) for purification of native SucCD or in 50 mM Tris-HCl, 500 mM NaCl, and 20 mM imidazole (pH 7.4) for purification of the hexahistidine-tagged variant of SucCD. Cells were subsequently disrupted by applying a 3-fold passage through a cooled French press (100 × 106 Pa; Aminco, Silver Spring, MD) (46) or a Sonoplus GM200 sonication apparatus (Bandelin, Berlin, Germany) equipped with an SH 213G booster horn and MS 72 or MS 73 microtip probes. The amplitude was 16 μm (1 min/ml), while cooling was performed in an NaCl-ice bath. Soluble protein fractions of crude extracts were obtained in the supernatants after 1.5 h of centrifugation at 100,000 × g and 4°C and were used for enzyme purifications.
Coupling of succinic acid anhydride to EAH-Sepharose 4B matrix.
In order to functionalize the EAH (epoxy-coupled 1,6-diaminohexane spacer group)-Sepharose 4B matrix with succinate as the ligand, EAH-Sepharose 4B (GE Healthcare, Munich, Germany) was dissolved in water and incubated under slight shaking at 4°C. Solid succinic acid anhydride was added until insolubility was reached. The pH was kept at 6 by addition of HCl during the reaction. Succinic acid anhydride was added stepwise. The functionalization was performed over a period of 3 days. This method represents a modification of the carbodiimide method, according to the instructions in the manufacturer's manual.
Purification of homo- and heterologously expressed sucCD genes in native state.
After expression of sucCD in E. coli BL21(DE3)/pLysS, the cells were harvested and stored at −20°C until use. Cells were disrupted, and the soluble fraction was generated as described above. All purification steps were performed at 4°C. In the case of SucCDAm and SucCDBL21, the soluble fraction was applied to Q-Sepharose fast-flow chromatography (32 ml; GE Healthcare, Munich, Germany) as described by Schürmann et al. (26). The matrix was equilibrated with 50 mM Tris-HCl (pH 7.4) and 0 mM NaCl at a flow rate of 4 ml/min. The proteins were eluted by a step gradient with increasing sodium chloride concentrations at a flow rate of 4 ml/min, as follows: 0 to 20 min, 0 mM NaCl; 20 to 65 min, 50 mM NaCl; 65 to 110 min, 75 mM NaCl; 110 to 155 min, 100 mM NaCl; and 155 to 210 min, 150 mM NaCl. The majority of SucCDAm and SucCDBL21 eluted at 150 mM NaCl. In the case of SucCDBL21 and SucCDAm, the eluted fractions were concentrated and buffered to the binding conditions using ultrafiltration (stirred cell model 8200; Amicon; Millipore Corporation, Billerica, MA). An Ultracel regenerated cellulose membrane with a nominal molecular mass limit of 10 kDa was used for protein concentration and buffer exchange to the binding conditions for chromatography.
Purification of SucCDAm.
A protein solution containing enriched SucCDAm after Q-Sepharose chromatography (see the previous paragraph) was then applied to a DEAE-Sepharose column (27 ml; GE Healthcare, Munich, Germany) equilibrated to the binding conditions (50 mM Tris-HCl [pH 7.4], 0 mM NaCl at a flow rate of 1 ml/min). Proteins were eluted by a linear gradient with increasing NaCl concentrations at a flow rate of 1 ml/min, as follows: 0 to 40 min, 0 mM NaCl; 40 to 110 min, a linear gradient of 0 to 1 M NaCl (change in concentration [Δ], 14.3 mmol/min); and 110 to 160 min, 1 M NaCl. SucCDAm eluted at a concentration between 60 and 350 mM NaCl. The eluted protein was concentrated, buffered to the binding conditions, and applied to a modified EAH-Sepharose 4B column (25 ml; GE Healthcare, Munich, Germany) carrying a succinate functionalization. Equilibration to the binding conditions was performed with 50 mM Tris-HCl (pH 7.4) and 0 mM NaCl at a flow rate of 1 ml/min. Proteins were eluted with the same protocol used for DEAE-Sepharose chromatography. Impurities were eluted from the matrix, and pure SucCDAm was located in the flowthrough. The latter was concentrated, buffered to the storage conditions as described by Gibson et al. (47) (100 mM Tris-HCl, 150 mM NaCl, pH 7.5, in 50% [vol/vol] glycerol), and used for enzyme assays. The SucCD concentration was determined using the specific molar extinction coefficient at 280 nm calculated for the αβ dimer by the software tool Protparam (48).
Purification of SucCDBL21.
A protein solution enriched SucCDBL21 after Q-Sepharose chromatography (see “Purification of homo- and heterologously expressed sucCD genes in native state” above) was applied to a Cibacron Blue F3GA column (50 ml; GE Healthcare, Munich, Germany) equilibrated to the binding conditions (50 mM Tris-HCl [pH 7.4], 0 mM NaCl) at a flow rate of 3 ml/min. The proteins were eluted by a linear gradient of increasing sodium chloride concentrations at a flow rate of 3 ml/min, as follows: 0 to 40 min, 0 mM NaCl, and 40 to 240 min, 0 to 1 M NaCl (Δ, 5 mmol/min). SucCDBl21 eluted after 101 to 141 min according to the purification protocol. Pooled samples were concentrated, buffered to the storage conditions (100 mM Tris-HCl, 150 mM NaCl, pH 7.5 in 50% [vol/vol] glycerol), and used for enzyme assays. The SucCD concentration was determined using the specific molar extinction coefficient at 280 nm calculated for the αβ dimer by the software tool Protparam (48).
Purification of SucCDAboHis.
In an initial attempt to purify SucCDAbo, it was expressed from the vector pET-23a(+)::sucCDAbo (Table 1). Both subunits were expressed in equimolar amounts, as judged by SDS-PAGE, but were repeatedly separated during purification with Q-Sepharose. Hence, the vector pET-23a(+)::sucCDAboHis, encoding a C-terminal hexahistidine tag on the SucD subunit, was generated (Table 1). After expression of sucCDAboHis in E. coli BL21(DE3)/pLysS, cell harvest, and cell disruption, the soluble fraction was applied to an Ni-nitrilotriacetic acid (NTA) Sepharose column (1 ml; HisTrap HP; GE Healthcare, Munich, Germany). The SucD subunit was provided with a hexahistidine tag at the C terminus. The SucC subunit coeluted from the column matrix. Binding conditions were 50 mM Tris-HCl, 500 mM NaCl, and 20 mM imidazole (pH 7.4). Equilibration was performed following the manufacturer's instructions. The flow rate was 1 ml/min. For elution purposes, a gradient of imidazole was applied. The elution program was as follows: 0 to 5 min, 20 mM imidazole; 5 to 10 min, 40 mM imidazole; and 10 to 50 min, 40 to 500 mM imidazole (Δ, 11.5 mmol/min). SucCD elution started at a concentration of 50 ± 5 mM imidazole. The SucCD concentration was determined as described previously by Bradford (49).
Enzyme assays.
The standard in vitro activity of succinate-CoA ligase in the direction of ADP formation was assayed by a continuous spectrophotometric assay according to the method of Cha and Parks (50). Measurements of succinate, itaconate, malate, and 3SP were carried out at 30°C in the presence of 50 mM Tris-HCl (pH 7.4), 0.4 mM ATP, 0.1 mM CoA, 7 mM MgCl2, 2 mM phosphoenolpyruvate, and 0.1 mM NADH, together with 6 U of pyruvate kinase and 6 U of lactate dehydrogenase from rabbit muscle (Sigma) as auxiliary enzymes. The concentrations of the organic acids were assayed over the range of 0.1 to 10 mM (succinate), 0.3 to 10 mM (itaconate), 0.625 to 15 mM (malate), and 0.625 to 25 mM (3SP). ATP and CoA measurements were carried out as described above in the presence of 10 mM succinate. The formation of ADP and the concomitant equimolar formation of the CoA-thioester were measured as a decrease in NADH absorption at 340 nm. The auxiliary enzymes were tested to ensure that they were not rate limiting. The formation of the expected CoA-thioesters was verified by liquid chromatography/electrospray ionization-mass spectrometry (LC/ESI-MS). For this analysis, the reactions were stopped by the addition of 30 μl of 15% (wt/vol) trifluoroacetic acid. The samples were subsequently analyzed as described above.
The following compounds (each at 10 mM) were investigated: succinate, sulfosuccinate, mercaptosuccinate, itaconate, d-malate, l-malate, tartrate, acetate, butyrate, propionate, levulinate, valerate, malonate, glutarate, adipate, 3SP, fumarate, maleate, and 2,2′-thiodiacetate. Stock solutions (500 mM) of the respective compounds were prepared in 50 mM Tris-HCl and were neutralized prior to application.
RESULTS
In silico analysis of SucCD enzymes.
In this study, we characterized three different bacterial SucCD enzymes with regard to their substrate ranges concerning structural analogues to succinate. An amino acid sequence alignment showed 100% sequence identity of the SucCDBL21 and the E. coli K-12 enzymes. A multiple-sequence alignment of SucC subunits revealed the following: SucCBL21/SucCAm, 53% identical (72% similar) amino acid residues; SucCBL21/SucCAbo, 74% identical (87% similar) amino acid residues; and SucCAm/SucCAbo, 52% identical (71% similar) amino acid residues. For SucD subunits, the following sequence similarities were determined: SucDBL21/SucDAm, 55% identical (71% similar) amino acid residues; SucDBL21/SucDAbo, 84% identical (90% similar) amino acid residues; and SucDAm/SucDAbo, 54% identical (73% similar) amino acid residues. The theoretical molecular masses for SucCDBL21 are 41.4 kDa for the SucC subunit and 29.7 kDa for the SucD subunit. The calculated molecular masses of the A. mimigardefordensis DPN7T enzyme are 41.3 kDa for the SucC subunit and 30.9 kDa for the SucD subunit. The A. borkumensis SK2 SucCD molecular masses were calculated to be 41.4 kDa (SucC) and 29.9 kDa (SucD).
Purification of SucCD enzymes.
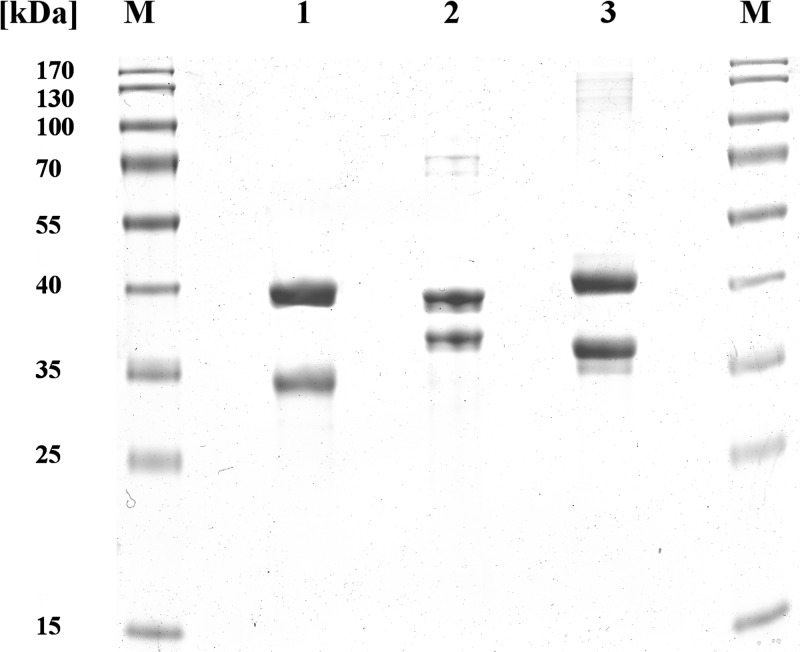
The sucCD genes from E. coli BL21 and A. borkumensis SK2 were amplified from genomic DNA and cloned into expression vectors, yielding pBluescriptSK(−)::sucCDBL21 and pET-23a(+)::sucCDAbo(His), respectively, as described in the Material and Methods section. Optimal expression of all sucCD enzyme in this study was achieved using expression host E. coli BL21(DE3)/pLysS in ZYP-5052 autoinduction medium (37). Despite a structural relation of at least 50% sequence identity, the SucCD enzymes showed a quite distinct binding behavior on chromatography matrices, resulting in three different purification protocols (Table 2). Provision of a terminal hexahistidine tag for a more efficient purification protocol for SucCDAm led to the formation of inclusion bodies. The amplified fragment of sucCDAm, performed by Schürmann et al. (26), contained 68 bp of the sucC upstream region. The fragment amplified from E. coli BL21 genomic DNA contained the bicistronic operon for sucC plus sucD as well as the 135-bp region upstream of the sucC initiation codon. In the case of sucCDAboHis, the relevant fragment contained only the gene loci for sucC and sucD. The sucCD genes investigated in this study were expressed independently in E. coli BL21(DE3)/pLysS as soluble proteins. SucC and SucD were synthesized in almost equimolar amounts, according to SDS-PAGE analysis by application of crude extracts and the soluble protein fraction (not shown). SucCDAm and SucCDBL21 were purified as native proteins using Q-Sepharose anion-exchange chromatography as a capture step. SucCDBL21 was further purified to electrophoretic homogeneity by Cibacron Blue F3GA-Sepharose affinity chromatography. Enriched SucCDAm in the Q-Sepharose eluate was further purified to electrophoretic homogeneity by DEAE-Sepharose anion-exchange chromatography and by a final polishing step using modified EAH-Sepharose 4B chromatography. SucCDAboHis, unlike SucCDBL21 and SucCDAm, carried a hexahistidine tag at the C terminus of SucD. The SucC subunit coeluted with SucD from the Ni-NTA matrix. The purity of the proteins was confirmed by applying 10 μg of protein to SDS-polyacrylamide gels (Fig. 2).
TABLE 2.
Purification of SucCD enzymes from E. coli BL21, A. mimigardefordensis DPN7T, and A. borkumensis SK2
| SucCD origin | Purification step | Total amt of protein (mg) | Total enzyme activitya (U) | Sp act (U/mg) | Yield (%) | Level (fold) |
|---|---|---|---|---|---|---|
| E. coli BL21 | Soluble protein fraction | 2,961 | 37,365 | 12.6 | 100.0 | 1.00 |
| Q-Sepharose | 1,050 | 12,573 | 12.0 | 33.6 | 0.95 | |
| Cibacron Blue F3GA-Sepharose | 147 | 2,734 | 18.6 | 7.3 | 1.47 | |
| A. mimigardefordensis DPN7T | Soluble protein fraction | 2,047 | 22,829 | 11.2 | 100.0 | 1.00 |
| Q-Sepharose | 277 | 3,194 | 11.5 | 14.0 | 1.03 | |
| DEAE-Sepharose | 111 | 1,437 | 12.9 | 6.3 | 1.16 | |
| EAH-Sepharose 4B | 57 | 682 | 12.0 | 3.0 | 1.07 | |
| A. borkumensis SK2 | Soluble protein fraction | 279 | 744 | 2.67 | 100.0 | 1.00 |
| Ni-NTA-Sepharose | 42 | 173 | 4.14 | 23.3 | 1.55 |
Enzyme activity was determined at 30°C in the direction of succinyl-CoA/ADP formation, as described in the Materials and Methods section. One unit corresponds to the formation of 1 μmol ADP per minute.
FIG 2.
Purification of SucCD enzymes from E. coli, A. mimigardefordensis DPN7T, and A. borkumensis SK2 revealed by SDS-PAGE (11.5%, wt/vol, acrylamide). Lane 1, 10 μg purified SucCD from E. coli; lane 2, 10 μg purified SucCD from A. mimigardefordensis DPN7T; lane 3, 10 μg of purified SucCD from A. borkumensis SK2; lanes M, molecular mass standard (PageRuler prestained protein ladder; Thermo Fisher Scientific, Rockford, IL). The gel was stained with Coomassie brilliant blue R.
Carbon acid specificity of SucCDBL21, SucCDAm, and SucCDAboHis.
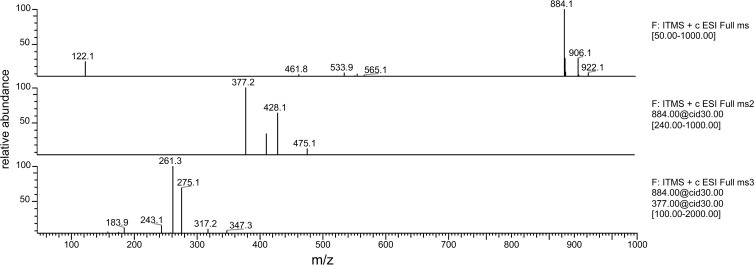
After expression and purification of SucCD enzymes, the enzyme activity was determined by using a continuous spectrophotometric assay, as described in the Material and Methods section. Several long-term storage conditions were investigated for each SucCD. Optimal stability was observed with 100 mM Tris, 150 mM NaCl and storage at 4°C or after the addition of 50% (vol/vol) glycerol and storage at −20°C. For verification of the formation of the expected CoA-thioesters, the samples obtained by in vitro catalysis were subjected to LC/ESI-MS. Dalluge et al. established an LC/ESI-MS-based method for detection and verification of CoA-thioesters (40). By use of electrospray ionization, the authors showed that CoA-thioesters from various organic acids showed a specific parental ion mass spectrum. The mass of the parental ion from various CoA-thioesters was obtained by the following equation (40): 768 Da (mass of free CoA) + x Da (mass of the various organic acids) − 18 Da (mass of H2O). Cleavage of this parental ion results in two distinct daughter ions with m/z equal to 428 Da and m/z equal to 261 Da + x Da (mass of the organic acid) − 18 Da (mass of H2O).
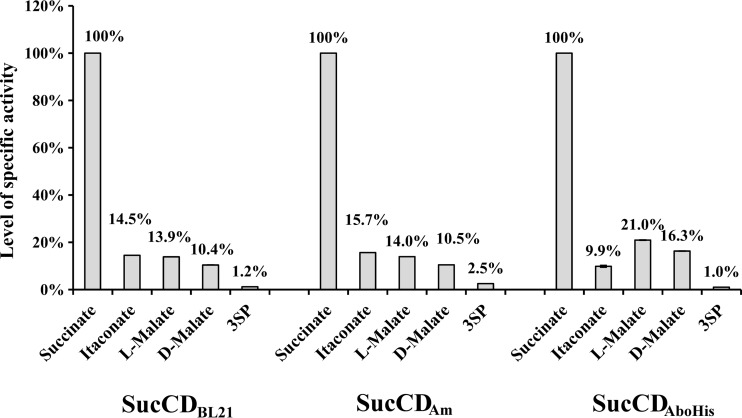
For the identification of CoA-thioesters it was therefore essential to detect the masses of the parental ion and of the organic acid covalently bound to 4-phosphopantetheine. The SucCD enzymes investigated in this study showed almost identical properties regarding the activation of substrate analogues as well as regarding kinetic properties with substrates that showed the highest activity (Table 3). The SucCD enzymes employed in this study were able to activate succinate, itaconate, 3SP, l-malate, d-malate, glutarate, and fumarate to their corresponding CoA-thioester. Typical fragmentation of malyl-CoA (Fig. 3), as observed by ESI/MS, is shown as an example in Fig. 4. SucCDBL21 was also able to activate adipate to adipyl-CoA; however, in corresponding samples containing SucCDAm and SucCDAboHis, only typical parent ions were detected. No clear evidence for the formation of tartryl-CoA from tartrate was obtained. Either only parental ions (in samples containing SucCDAm or SucCDAboHis) or typical fragments from daughter ions (428 Da and 393 Da; when SucCDBL21 was applied in enzyme assay) were observed. Nonetheless, the amounts detected were very low. No formation of CoA-thioesters could be demonstrated for monocarboxylic acid variants, such as propionate, butyrate, acetate, levulinate, and valerate. Kinetic data suggest that all enzymes strongly preferred the physiological substrates CoA, ATP, and succinate. Comparably high enzyme activity was determined for itaconate, l-malate, and d-malate (Fig. 5; Table 3). A common characteristic was the reduced SucCD activity with 3SP to only 1.0 to 2.5% or even less of the activity with succinate. The Km values calculated for itaconate were 2- to 10-fold higher than the Km values calculated for succinate. l-Malate showed 18- to 20-fold higher Km values and d-malate showed 20- to 27-fold higher Km values than those for succinate. The SucCD enzymes investigated here also exhibited comparably low affinities for 3SP (10- to 16-fold higher Km values than the value for succinate).
TABLE 3.
Kinetic parameters determined for SucCDBL21, SucCDAm, and SucCDAboHisa
| Enzyme | Substrate | Vmax (U/mg) | Km (mM) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|---|---|
| SucCDBL21 | Succinate | 12.06 ± 0.03 | 0.141 ± 0.003 | 14.3 | 101.4 |
| Itaconate | 1.30 ± 0.01 | 0.475 ± 0.019 | 1.5 | 3.3 | |
| l-Malate | 1.51 ± 0.04 | 2.558 ± 0.106 | 1.8 | 0.7 | |
| d-Malate | 0.98 ± 0.02 | 3.635 ± 0.223 | 1.2 | 0.3 | |
| 3SP | 0.15 ± 0.00 | 1.520 ± 0.081 | 0.2 | 0.1 | |
| CoA | 22.48 ± 0.92 | 0.058 ± 0.005 | 26.7 | 461.3 | |
| ATP | 16.49 ± 0.10 | 0.055 ± 0.002 | 19.6 | 354.4 | |
| SucCDAm | Succinate | 25.86 ± 0.06 | 0.182 ± 0.003 | 31.1 | 171.1 |
| Itaconate | 4.42 ± 0.02 | 0.351 ± 0.003 | 5.3 | 15.1 | |
| l-Malate | 2.15 ± 0.09 | 3.095 ± 0.354 | 2.6 | 0.8 | |
| d-Malate | 1.77 ± 0.02 | 3.588 ± 0.111 | 2.1 | 0.6 | |
| 3SP | 0.14 ± 0.01 | 2.964 ± 0.275 | 0.2 | 0.1 | |
| CoA | 33.47 ± 0.23 | 0.037 ± 0.001 | 40.3 | 1,099.6 | |
| ATP | 48.89 ± 0.41 | 0.201 ± 0.002 | 58.8 | 292.5 | |
| SucCDAboHis | Succinate | 2.23 ± 0.01 | 0.157 ± 0.003 | 2.7 | 17.2 |
| Itaconate | 0.39 ± 0.00 | 1.509 ± 0.048 | 0.5 | 3.1 | |
| l-Malate | 0.88 ± 0.00 | 3.270 ± 0.017 | 1.1 | 0.3 | |
| d-Malate | 1.38 ± 0.02 | 4.243 ± 0.089 | 1.7 | 0.4 | |
| 3SP | ND | ND | ND | ND | |
| CoA | 1.33 ± 0.03 | 0.004 ± 0.001 | 1.6 | 372.3 | |
| ATP | 2.96 ± 0.02 | 0.241 ± 0.050 | 3.6 | 14.8 |
Enzyme activities for Vmax values were determined in the direction of CoA-thioester/ADP formation. One unit corresponds to the formation of 1 μmol ADP per minute. kcat values are given for the number of active sites (αβ dimer). ND, not determined.
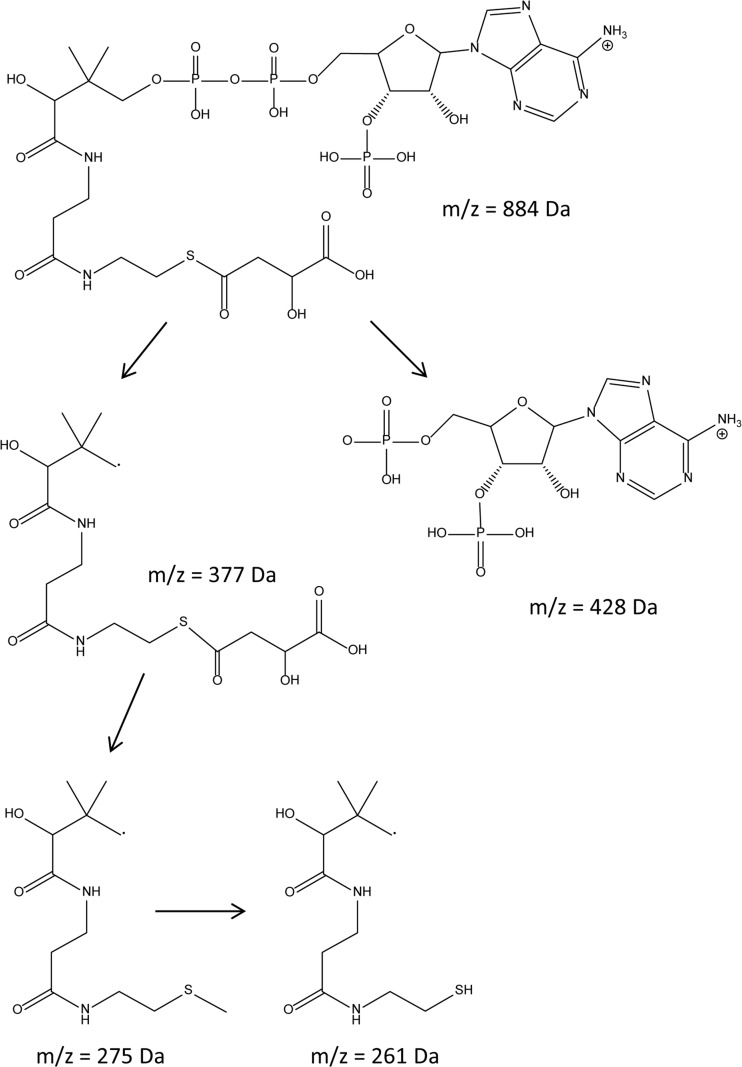
FIG 3.
Structural formula of malyl-CoA and pattern of fragmentation of the parent ion into several daughter ions in ESI-MS.
FIG 4.
Analyses of malyl-CoA. (Top) ESI spectrum of malyl-CoA in the positive mode; (middle) MS spectrum of the parent ion (m/z = 884 Da); two main fragments (m/z = 428 Da and m/z = 377 Da) were obtained; (bottom) further fragmentation of the parent ion with an m/z of 377 Da yielded the daughter ions with m/z equal to 275 kDa and 261 kDa. ITMS + c ESI Full ms, ion trap mass spectrometry, positive mode with electrospray ionization (full MS spectrum recorded); cid, collision-induced dissociation (typically with an energy of 30.00 eV [electron volts]).
FIG 5.
Levels of acyl-CoA-forming activity (from ADP formation) of SucCDBL21, SucCDAm, and SucCDAboHis. Activity values were normalized to the activity with succinate. Each of the various organic acids was applied to the assay mixture at 10 mM. Activity was determined in duplicate experiments. Black error bars, standard deviations.
The kcat values calculated for the substrates l-malate and d-malate were similar to the corresponding values for itaconate (Table 3). According to Shikata et al., activity levels for different substrates were obtained and normalized to the activity with the substrate succinate at a final concentration of 10 mM (21) (Fig. 5). Vmax values for both enantiomeric forms of malate were of the same order of magnitude and comparable to the Vmax of the physiological substrate itaconate (Fig. 5; Table 3).
Complementation studies applying pBBR1MCS-5::sucCDAm.
In an attempt to complement the A. mimigardefordensis ΔsucCD mutant, which exhibited a negative phenotype on MSM agar plates containing either DTDP or 3SP as the sole carbon and energy source in comparison to that of the wild type, the hybrid plasmid pBBR1MCS-5::sucCDAm was transferred to the deletion mutant by conjugation. Transconjugants were selected on MSM containing 0.5% (wt/vol) gluconate and gentamicin. As expected, growth of the complemented mutant was observed in liquid MSM containing 50 mM DTDP (and gentamicin for plasmid stability). While the wild type grew normally, the deletion mutant showed no growth at all. Growth of the complemented mutant was delayed in comparison to that of the wild type but reached at least 56% of the wild type's cell density over the given time range (see Fig. S1 in the supplemental material).
DISCUSSION
In this study, we purified and analyzed three different SucCD enzymes with respect to their substrate range. Additionally, growth of the mutant strain A. mimigardefordensis DPN7T ΔsucCD on DTDP was restored by pBBR1MCS-5::sucCDAm. The plasmid was constructed with a 478-bp upstream region to apply the endogenous promoter region for expression of sucCD. Xia et al. showed that with this plasmid, an efficient gene expression is achieved with the endogenous promoter independent of the orientation of the gene (28). These results complete the findings of Schürmann et al., who described the essential role of SucCD in A. mimigardefordensis DPN7T in the degradation of DTDP (26). Expression of the relevant genes sucC and sucD in about equimolar amounts in a successful purification protocol was observed to require 68 or 135 bp of the corresponding sucC upstream regions in the expression vector pBluescriptSK(−) for sucCDAm and sucCDBL21, respectively. During the experiments, the best expression was obtained when the sucCD genes of A. mimigardefordensis DPN7T and E. coli BL21 were each applied in one bicistronic operon that included the strain-specific Shine-Dalgarno sequence upstream of sucC. The genes encoding SucCDAboHis were also expressed in one bicistronic operon. In this case, the vector-specific Shine-Dalgarno sequence was used. Efficient purification of SucCDAboHis was achieved with a hexahistidine-tagged variant of SucD. Both subunits coeluted from the column matrix. All SucCD enzymes were isolated in an active state. SucCDAboHis showed a reduced specific activity in comparison to that of the other SucCD enzymes investigated in this study. The lysate obtained from cells of E. coli BL21(DE3)/pLysS expressing either sucCDAbo or sucCDAboHis led to similar results concerning the level of expression of sucCD as well as the specific activity of SucCD, and the results indicate that SucCD from A. borkumensis SK2 might, in general, have comparably low activity.
Kinetic parameters for all three SucCD enzymes indicate a preference for the physiological substrates CoA, ATP, and succinate; therefore, this clearly allocated this enzyme to the citric acid cycle. Low levels of activity were determined for SucCDBL21 and SucCDAm with 3SP as a substrate. Due to the generally reduced activity of SucCDAboHis, kinetic parameters for 3SP were not determined, as use of a strong excess of enzyme within the photometric assay would have been necessary. SucCDAm showed an approximately 3.6-fold higher Km value for 3SP than that indicated previously (26). This could be explained by use of a different method with altered concentrations of Mg2+ and ATP which had been adapted for the determination of enzyme activity with SucCDBL21 in this study. Nonetheless, the other kinetic parameters in this study were in good accordance with published data for SucCDAm (26). Activity was also proven for itaconate, which has been described to be the substrate for A. mimigardefordensis DPN7T and for mammalian SucCD (22, 26). The ability to convert both enantiomers of malate, with a slight preference for l-malate, was a general feature of the SucCD enzymes investigated in this study. The Km values for these substrates were in the same range as those for 3SP obtained in this study.
d-Malate has no relevance in vivo, as no metabolic pathways involving this compound are known yet. The conversion of the stereoisomer l-malate to l-malyl-CoA is a crucial catalytic step in the serine cycle in one group of the methylotrophic bacteria. This pathway serves for the efficient assimilation of C1 compounds, such as methanol or methylamine (51, 52). The genes responsible for the catalytic step from l-malate to l-malyl-CoA in Methylobacterium extorquens strain AM1 have been identified to be mtkA and mtkB, encoding the malate-CoA ligases, also known as malate thiokinase (51). The malate-CoA ligase of Aminobacter aminovorans, which was formerly known as Pseudomonas sp. strain MA (ATCC 23819) and which is also a member of the group of microorganisms able to assimilate C1 compounds, was biochemically characterized in the past (53–56). Surprisingly, Hersh determined a 5% higher activity with succinate compared to the activity with l-malate in vitro (55). Besides these kinetic data obtained by Hersh (55), malate-CoA ligases show similarities to SucCD enzymes concerning the amino acid sequence (51), subunit distribution, and molecular weight (53). Although M. extorquens AM1 possesses both the mtkAB and sucCD genes in its genome (44), it was shown that mtkAB is essential for growth on C1 and C2 compounds, because an insertion mutant lacking intact mtkA did not grow on these compounds. Growth was restored by applying a rescue vector for mtkAB in complementation experiments (51). All these data suggest that these two enzyme subsubclasses, succinate-CoA ligase (EC 6.2.1.4 and 6.2.1.5) and malate-CoA ligase (EC 6.2.1.9), share the same evolutionary origin. Since SucCD from M. extorquens AM1 is not able to compensate for the MtkAB deficiency in the mutant strain (51), an in vitro characterization of corresponding SucCD and MtkAB might elucidate the mechanistic and kinetic differences relative to the SucCD enzymes found in this study.
In addition to the compounds succinate, itaconate, 3SP, l-malate, and d-malate, CoA-thioester formation with fumarate, glutarate, and adipate was verified with the same LC/ESI-MS method established by Dalluge et al. (40). However, the SucCD activity obtained with the last three compounds was below 1% of that obtained with succinate; therefore, it is assumed that these activities do not have any relevance in vivo. Nonetheless, the ability to form CoA-thioesters of these dicarboxylic acids might be used for the analysis of substrate specificities in further enzyme characterization experiments (57).
No activation to CoA-thioesters was observed with monocarboxylic acids, such as acetate, propionate, butyrate, valerate, and levulinate. Thus, a second carboxyl group appears to be mandatory for proper binding within the active site of SucCD. Although maleate carries a second carboxyl group, maleyl-CoA was not detected during LC/ESI-MS analyses. Hence, the cis double bond of maleate (in contrast to a trans double bond in fumarate) might also impair proper binding to the active site. With regard to chain length, succinate (C4) was found to be the best substrate, whereas CoA-thioesters of glutarate (C5) and adipate (C6) were formed in only trace amounts; malonate (C3) was not activated at all. While d-malate and l-malate were activated to the corresponding CoA-thioester, the structural analogue mercaptosuccinate (Fig. 1) could not be utilized by any of the investigated SucCD enzymes. This might be due to the higher acidity of sulfhydryl groups than hydroxyl groups, which consequently results in an additional negative charge at the sulfur atom. The lack of activation of sulfosuccinate might be explained by the steric hindrance caused by the sulfonic acid group; in comparison, malate has a hydroxyl group, which is smaller and which does not cause steric hindrance.
This study proved the ability of different SucCD enzymes to form CoA-thioesters with succinate analogues as well, such as malate and 3SP. Concomitantly, this study showed that activation of the latter is not a unique characteristic of SucCDAm in the degradation of DTDP.
Supplementary Material
ACKNOWLEDGMENT
The LC/ESI-MS device used in this study was provided by funds of the DFG (Deutsche Forschungsgemeinschaft, grant no. INST 211/415-1 FUGG), which we gratefully acknowledge.
Footnotes
Published ahead of print 18 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03075-13.
REFERENCES
- 1.Bridger WA. 1974. Succinyl-CoA synthetase, p 581–606 In Boyer PD. (ed), The enzymes. Academic Press, Inc., New York, NY [Google Scholar]
- 2.Kaufman S. 1955. Studies on the mechanism of the reaction catalyzed by the phosphorylating enzyme. J. Biol. Chem. 216:153–164 [PubMed] [Google Scholar]
- 3.Bridger WA. 1971. Evidence for two types of subunits in succinyl coenzyme A synthetase. Biochem. Biophys. Res. Commun. 42:948–954. 10.1016/0006-291X(71)90522-5 [DOI] [PubMed] [Google Scholar]
- 4.Wolodko WT, Kay CM, Bridger WA. 1986. Active enzyme sedimentation, sedimentation velocity, and sedimentation equilibrium studies of succinyl-CoA synthetases of porcine heart and Escherichia coli. Biochemistry 25:5420–5425. 10.1021/bi00367a012 [DOI] [PubMed] [Google Scholar]
- 5.Studart-Guimaraes C, Gibon Y, Frankel N, Wood CC, Zanor MI, Fernie AR, Carrari F. 2005. Identification and characterisation of the alpha and beta subunits of succinyl CoA ligase of tomato. Plant Mol. Biol. 59:781–791. 10.1007/s11103-005-1004-1 [DOI] [PubMed] [Google Scholar]
- 6.Joyce MA, Fraser ME, Brownie ER, James MN, Bridger WA, Wolodko WT. 1999. Probing the nucleotide-binding site of Escherichia coli succinyl-CoA synthetase. Biochemistry 38:7273–7283. 10.1021/bi990527s [DOI] [PubMed] [Google Scholar]
- 7.Majumdar R, Guest JR, Bridger WA. 1991. Functional consequences of substitution of the active site (phospho)histidine residue of Escherichia coli succinyl-CoA synthetase. Biochim. Biophys. Acta 1076:86–90. 10.1016/0167-4838(91)90223-M [DOI] [PubMed] [Google Scholar]
- 8.Pearson PH, Bridger WA. 1975. Catalysis of a step of the overall reaction by the alpha subunit of Escherichia coli succinyl-coenzyme A synthetase. J. Biol. Chem. 250:8524–8529 [PubMed] [Google Scholar]
- 9.Wolodko WT, Fraser ME, James MN, Bridger WA. 1994. The crystal structure of succinyl-CoA synthetase from Escherichia coli at 2.5-Å resolution. J. Biol. Chem. 269:10883–10890 [DOI] [PubMed] [Google Scholar]
- 10.Jenkins TM, Weitzman PD. 1986. Distinct physiological roles of animal succinate thiokinases. Association of guanine nucleotide-linked succinate thiokinase with ketone body utilization. FEBS Lett. 205:215–218 [DOI] [PubMed] [Google Scholar]
- 11.Fraser ME, James MN, Bridger WA, Wolodko JWT. 1999. A detailed structural description of Escherichia coli succinyl-CoA synthetase. J. Mol. Biol. 285:1633–1653. 10.1006/jmbi.1998.2324 [DOI] [PubMed] [Google Scholar]
- 12.Wolodko WT, James MN, Bridger WA. 1984. Crystallization of succinyl-CoA synthetase from Escherichia coli. J. Biol. Chem. 259:5316–5320 [PubMed] [Google Scholar]
- 13.Birney M, Um HD, Klein C. 1996. Novel mechanisms of Escherichia coli succinyl-coenzyme A synthetase regulation. J. Bacteriol. 178:2883–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapatral V, Bina X, Chakrabarty AM. 2000. Succinyl coenzyme A synthetase of Pseudomonas aeruginosa with a broad specificity for nucleoside triphosphate (NTP) synthesis modulates specificity for NTP synthesis by the 12-kilodalton form of nucleoside diphosphate kinase. J. Bacteriol. 182:1333–1339. 10.1128/JB.182.5.1333-1339.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami K, Mitchell T, Nishimura JS. 1972. Nucleotide specificity of Escherichia coli succinic thiokinase. Succinyl coenzyme A-stimulated nucleoside diphosphate kinase activity of the enzyme. J. Biol. Chem. 247:6247–6252 [PubMed] [Google Scholar]
- 16.Noy N, Zakim D. 1985. Fatty acids bound to unilamellar lipid vesicles as substrates for microsomal acyl-CoA ligase. Biochemistry 24:3521–3525. 10.1021/bi00335a020 [DOI] [PubMed] [Google Scholar]
- 17.Bräsen C, Schönheit P. 2005. AMP-forming acetyl-CoA synthetase from the extremely halophilic archaeon Haloarcula marismortui: purification, identification and expression of the encoding gene, and phylogenetic affiliation. Extremophiles 9:355–365. 10.1007/s00792-005-0449-0 [DOI] [PubMed] [Google Scholar]
- 18.Mai X, Adams MW. 1996. Purification and characterization of two reversible and ADP-dependent acetyl coenzyme A synthetases from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 178:5897–5903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajashekhara E, Watanabe K. 2004. Propionyl-coenzyme A synthetases of Ralstonia solanacearum and Salmonella choleraesuis display atypical kinetics. FEBS Lett. 556:143–147. 10.1016/S0014-5793(03)01394-2 [DOI] [PubMed] [Google Scholar]
- 20.Shimizu S, Inoue K, Tani Y, Yamada H. 1981. Butyryl-CoA synthetase of Pseudomonas aeruginosa—purification and characterization. Biochem. Biophys. Res. Commun. 103:1231–1237. 10.1016/0006-291X(81)90254-0 [DOI] [PubMed] [Google Scholar]
- 21.Shikata K, Fukui T, Atomi H, Imanaka T. 2007. A novel ADP-forming succinyl-CoA synthetase in Thermococcus kodakaraensis structurally related to the archaeal nucleoside diphosphate-forming acetyl-CoA synthetases. J. Biol. Chem. 282:26963–26970. 10.1074/jbc.M702694200 [DOI] [PubMed] [Google Scholar]
- 22.Adler J, Wang SF, Lardy HA. 1957. The metabolism of itaconic acid by liver mitochondria. J. Biol. Chem. 229:865–879 [PubMed] [Google Scholar]
- 23.Cooper RA, Itiaba K, Kornberg HL. 1965. The utilization of aconate and itaconate by Micrococcus sp. Biochem. J. 94:25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagai J. 1963. Studies on itaconate metabolism, I. Itaconyl-CoA synthesizing reaction in cell-free extract. J. Biochem. 53:181–187 [DOI] [PubMed] [Google Scholar]
- 25.Lütke-Eversloh T, Steinbüchel A. 2003. Novel precursor substrates for polythioesters (PTE) and limits of PTE biosynthesis in Ralstonia eutropha. FEMS Microbiol. Lett. 221:191–196. 10.1016/S0378-1097(03)00185-X [DOI] [PubMed] [Google Scholar]
- 26.Schürmann M, Wübbeler JH, Grote J, Steinbüchel A. 2011. Novel reaction of succinyl coenzyme A (succinyl-CoA) synthetase: activation of 3-sulfinopropionate to 3-sulfinopropionyl-CoA in Advenella mimigardefordensis strain DPN7T during degradation of 3,3′-dithiodipropionic acid. J. Bacteriol. 193:3078–3089. 10.1128/JB.00049-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wübbeler JH, Bruland N, Wozniczka M, Steinbüchel A. 2010. Biodegradation of the xenobiotic organic disulphide 4,4′-dithiodibutyric acid by Rhodococcus erythropolis strain MI2 and comparison with the microbial utilization of 3,3′-dithiodipropionic acid and 3,3′-thiodipropionic acid. Microbiology 156:1221–1233. 10.1099/mic.0.036178-0 [DOI] [PubMed] [Google Scholar]
- 28.Xia Y, Wübbeler JH, Qi Q, Steinbüchel A. 2012. Employing a recombinant strain of Advenella mimigardefordensis for biotechnical production of homopolythioesters from 3,3′-dithiodipropionic acid. Appl. Environ. Microbiol. 78:3286–3297. 10.1128/AEM.00007-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wübbeler JH, Bruland N, Kretschmer K, Steinbüchel A. 2008. Novel pathway for catabolism of the organic sulfur compound 3,3′-dithiodipropionic acid via 3-mercaptopropionic acid and 3-sulfinoproppionic acid to propionyl-coenzyme A by the aerobic bacterium Tetrathiobacter mimigardefordensis strain DPN7. Appl. Environ. Microbiol. 74:4028–4035. 10.1128/AEM.00422-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wübbeler JH, Lütke-Eversloh T, Van Trappen S, Vandamme P, Steinbüchel A. 2006. Tetrathiobacter mimigardefordensis sp. nov., isolated from compost, a betaproteobacterium capable of utilizing the organic disulfide 3,3′-dithiodipropionic acid. Int. J. Syst. Evol. Microbiol. 56:1305–1310. 10.1099/ijs.0.64126-0 [DOI] [PubMed] [Google Scholar]
- 31.Wübbeler JH, Raberg M, Brandt U, Steinbüchel A. 2010. Dihydrolipoamide dehydrogenases of Advenella mimigardefordensis and Ralstonia eutropha catalyze cleavage of 3,3′-dithiodipropionic acid into 3-mercaptopropionic acid. Appl. Environ. Microbiol. 76:7023–7028. 10.1128/AEM.01706-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buck D, Spencer ME, Guest JR. 1985. Primary structure of the succinyl-CoA synthetase of Escherichia coli. Biochemistry 24:6245–6252. 10.1021/bi00343a031 [DOI] [PubMed] [Google Scholar]
- 33.Joyce MA, Fraser ME, James MN, Bridger WA, Wolodko WT. 2000. ADP-binding site of Escherichia coli succinyl-CoA synthetase revealed by X-ray crystallography. Biochemistry 39:17–25. 10.1021/bi991696f [DOI] [PubMed] [Google Scholar]
- 34.Schneiker S, Martins dos Santos VA, Bartels D, Bekel T, Brecht M, Buhrmester J, Chernikova TN, Denaro R, Ferrer M, Gertler C, Goesmann A, Golyshina OV, Kaminski F, Khachane AN, Lang S, Linke B, McHardy AC, Meyer F, Nechitaylo T, Puhler A, Regenhardt D, Rupp O, Sabirova JS, Selbitschka W, Yakimov MM, Timmis KN, Vorholter FJ, Weidner S, Kaiser O, Golyshin PN. 2006. Genome sequence of the ubiquitous hydrocarbon-degrading marine bacterium Alcanivorax borkumensis. Nat. Biotechnol. 24:997–1004. 10.1038/nbt1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlegel HG, Kaltwasser H, Gottschalk G. 1961. A submersion method for culture of hydrogen-oxidizing bacteria: growth physiological studies. Arch. Mikrobiol. 38:209–222. 10.1007/BF00422356 [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207–234. 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 38.Jollès-Bergeret B. 1974. Enzymatic and chemical synthesis of 3-sulfinopropionic acid, an analog of succinic acid. Eur. J. Biochem. 42:349–353. 10.1111/j.1432-1033.1974.tb03346.x [DOI] [PubMed] [Google Scholar]
- 39.Stein S, Levitsky A, Fateev O, Mallard G. 1998. The NIST mass spectral search program, Windows software version 1.6d. National Institute of Standards and Technology, Gaithersburg, MD [Google Scholar]
- 40.Dalluge JJ, Gort S, Hobson R, Selifonova O, Amore F, Gokarn R. 2002. Separation and identification of organic acid-coenzyme A thioesters using liquid chromatography/electrospray ionization-mass spectrometry. Anal. Bioanal. Chem. 374:835–840. 10.1007/s00216-002-1554-x [DOI] [PubMed] [Google Scholar]
- 41.Marmur J. 1961. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3:208–218. 10.1016/S0022-2836(61)80047-8 [DOI] [Google Scholar]
- 42.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology 1:784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- 43.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 40:D115–D122. 10.1093/nar/gkr1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. 10.1016/0378-1119(95)00584-1 [DOI] [PubMed] [Google Scholar]
- 46.Milner HW, Lawrence NS, French CS. 1950. Colloidal dispersion of chloroplast material. Science 111:633–634. 10.1126/science.111.2893.633 [DOI] [PubMed] [Google Scholar]
- 47.Gibson J, Upper CD, Gunsalus IC. 1967. Succinyl coenzyme A synthetase from Escherichia coli. I. Purification and properties. J. Biol. Chem. 242:2474–2477 [PubMed] [Google Scholar]
- 48.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. 1999. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112:531–552 [DOI] [PubMed] [Google Scholar]
- 49.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 50.Cha S, Parks RE. 1964. Succinic thiokinase. II. Kinetic studies: initial velocity, product inhibition, and effect of arsenate. J. Biol. Chem. 239:1968–1977 [PubMed] [Google Scholar]
- 51.Chistoserdova LV, Lidstrom ME. 1994. Genetics of the serine cycle in Methylobacterium extorquens AM1: identification, sequence, and mutation of three new genes involved in C1 assimilation, orf4, mtkA, and mtkB. J. Bacteriol. 176:7398–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones JG, Bellion E. 1991. In vivo 13C and 15N NMR studies of methylamine metabolism in Pseudomonas species MA. J. Biol. Chem. 266:11705–11713 [PubMed] [Google Scholar]
- 53.Elwell M, Hersh LB. 1979. Substrate-dependent dissociation of malate thiokinase. J. Biol. Chem. 254:2434–2438 [PubMed] [Google Scholar]
- 54.Hersh LB. 1974. Malate thiokinase. The reaction mechanism as determined by initial rate studies. J. Biol. Chem. 249:6264–6271 [PubMed] [Google Scholar]
- 55.Hersh LB. 1973. Malate adenosine triphosphate lyase. Separation of the reaction into a malate thiokinase and malyl coenzyme A lyase. J. Biol. Chem. 248:7295–7303 [PubMed] [Google Scholar]
- 56.Hersh LB, Peet M. 1981. Half-of-the-sites reactivity in the malate thiokinase reaction. J. Biol. Chem. 256:1732–1737 [PubMed] [Google Scholar]
- 57.Schürmann M, Deters A, Wübbeler JH, Steinbüchel A. 2013. A novel 3-sulfinopropionyl coenzyme A (3SP-CoA) desulfinase from Advenella mimigardefordensis strain DPN7T acting as a key enzyme during catabolism of 3,3′-dithiodipropionic acid is a member of the acyl-CoA dehydrogenase superfamily. J. Bacteriol. 195:1538–1551. 10.1128/JB.02105-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bachmann BJ. 1990. Linkage map of Escherichia coli K-12, edition 8. Microbiol. Rev. 54:130–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.