Abstract
The diversity of deep-sea high-pressure-adapted (piezophilic) microbes in isolated monoculture remains low. In this study, a novel obligately psychropiezophilic bacterium was isolated from seawater collected from the Puerto Rico Trench at a depth of ∼6,000 m. This isolate, designated YC-1, grew best in a nutrient-rich marine medium, with an optimal growth hydrostatic pressure of 50 MPa (range, 20 to 70 MPa) at 8°C. Under these conditions, the maximum growth rate was extremely slow, 0.017 h−1, and the maximum yield was 3.51 × 107 cells ml−1. Cell size and shape changed with pressure, shifting from 4.0 to 5.0 μm in length and 0.5 to 0.8 μm in width at 60 MPa to 0.8- to 1.0-μm diameter coccoid cells under 20 MPa, the minimal pressure required for growth. YC-1 is a Gram-negative, facultatively anaerobic heterotroph. Its predominant cellular fatty acids are the monounsaturated fatty acids (MUFAs) C16:1 and C18:1. Unlike many other psychropiezophiles, YC-1 does not synthesize any polyunsaturated fatty acids (PUFAs). Phylogenetic analysis placed YC-1 within the family of Oceanospirillaceae, closely related to the uncultured symbiont of the deep-sea whale bone-eating worms of the genus Osedax. In common with some other members of the Oceanospirillales, including those enriched during the Deepwater Horizon oil spill, YC-1 is capable of hydrocarbon utilization. On the basis of its characteristics, YC-1 appears to represent both a new genus and a new species, which we name Profundimonas piezophila gen. nov., sp. nov.
INTRODUCTION
Psychropiezophilic (low-temperature and high-pressure adapted) and obligately psychropiezophilic (requiring high pressure for growth) bacteria have been isolated from a variety of deep-sea environments below 2,000 m and 5,000 m, respectively, as reviewed by Eloe et al. (1). To date, all of these bacteria fall into the genera Colwellia, Moritella, Psychromonas, and Shewanella within the Alteromonadaceae family within the Gammaproteobacteria, the genus Photobacterium within the Vibrionaceae family within the Gammaproteobacteria (2, 3), and (for one recent isolate) within the Roseobacter clade of the Rhodobacterales order within the Alphaproteobacteria (1). A common characteristic shared by all psychropiezophiles examined is the production of large proportions of their membrane phospholipid-associated fatty acids as unsaturated fatty acids, present as monounsaturated fatty acids (MUFAs), long-chain omega-3 polyunsaturated fatty acids (PUFAs), or both (4). Unsaturated fatty acids are needed to counteract high-pressure increases in the packing density of fatty-acyl chains, which reduce membrane fluidity, permeability, and elasticity, as well as membrane protein function (5, 6). In the psychrotolerant piezophilic bacterium Photobacterium profundum strain SS9, the monounsaturated fatty acid cis-vaccenic acid (C18:1) is required for high-pressure adaptation, but surprisingly, eicosapentaenoic acid (EPA; C20:5), a PUFA produced in greater abundance at high pressure than at low pressure, is not required for high-pressure growth (7). In contrast, EPA is required for high-pressure growth, a late step in cell division, and the maintenance of proper membrane fluidity in the psychropiezophile Shewanella violacea strain DSS12 (8, 9). Thus, while unsaturated fatty acids appear to be a universal requirement for psychropiezophiles, the relative importance of MUFAs and PUFAs may depend on the species and physiological state. Additional adaptations for life at low temperature and high pressure are also needed. For example, conditionally pressure sensitive mutants of P. profundum strain SS9 indicate the occurrence of high-pressure-specific modifications in chromosome structure and function and in ribosome assembly and function (10).
Oceanospirillales are heterotrophic bacteria both associated with oil spills, including the Deepwater Horizon deep-sea oil plume (11, 12), and involved in symbiotic interactions with various marine invertebrates, including sea urchins, corals, mussels, and bone-eating worms (13). The connection between members of the Oceanospirillales and bone-eating worms is particularly relevant to this study. Bone-eating worms of the genus Osedax have been discovered in whale carcasses from both shallow-water and deep-sea benthoses (14). Female worms possess a posterior root-like structure that infiltrates whale bones, apparently as a result of acid secretion (15). Although Osedax females lack a mouth and a gut, they harbor heterotrophic symbiotic bacteria in bacteriocytes within their roots. It is believed that the root epithelium absorbs bone collagen and/or lipids, which are utilized by the bacterial symbionts, and that the symbionts, in turn, serve as a food source for their hosts (16, 17). Phylogenetic analyses together with fluorescent in situ hybridization (FISH) experiments have shown that Osedax species harbor intracellular microbes belonging to the order Oceanospirillales (13, 16). The free-living hydrocarbon-degrading bacterium Neptunomonas naphthovorans, isolated from Puget Sound sediment at a depth of 15.5 m, and Neptunomonas japonica strains isolated from sediment adjacent to a sperm whale carcass off Kagoshima, Japan, at a depth of 226 to 246 m are the cultured relatives most closely related to the shallow-water Osedax symbionts (13, 18, 19). However, no close relatives of the clades of Osedax symbionts that include those present at a deep-sea whale fall have yet been cultivated under laboratory conditions (13).
In this study, we report on the isolation of the first psychropiezophilic member of the order Oceanospirillales of the Gammaproteobacteria. In order to understand more about Oceanospirillales bacteria in the deep ocean, we characterized the general growth, morphology, and phylogenetic and physiological properties of strain YC-1. This piezophilic bacterium is identified as the first cultured deep-sea bacterium closely related to the clade Osedax symbionts containing deep-sea whale fall representatives. The purpose of this study was to describe the isolation of strain YC-1 and to place it in a meaningful taxonomic framework. Strain YC-1 represents a new genus and species that we have designated Profundimonas piezophila gen. nov., sp. nov.
MATERIALS AND METHODS
Sample collection and high-pressure cultivation conditions.
Strain YC-1 was isolated from deep-sea seawater collected from the Puerto Rico Trench at a depth of 6,000 m (19.667°N, 65.966°W), by the R/V Atlantic Explorer (Bermuda Atlantic Time Series [BATS]) with a conductivity-temperature-depth (CTD) rosette (with 24 12-liter bottles) in October 2008. The water sample was carried to the sea surface and was then placed in sterilized bags (Kapak, USA). The plastic bags were sealed and were subsequently pressurized to 60 MPa in pressure vessels that were maintained at 2°C. High-pressure cultivation utilized a liquid hydraulic system. For enrichment, the water sample was cultivated in marine broth 2216 (Difco, USA) at 2°C and 60 MPa for about 6 weeks. Strain YC-1 was isolated using the silica gel method by following the general procedures described by Dietz and Yayanos (20). The piezophilic strain was maintained in pressurizable polyethylene transfer pipette bulbs containing marine broth 2216, and it grew at 8°C and 60 MPa. Optimal growth pressure and temperature were determined by total-cell counts with epifluorescence microscopy. The culture samples were fixed with formaldehyde and were stained with 4′,6-diamidino-2-phenylindole (DAPI).
Morphological analysis.
Cell morphologies at low pressure (20 MPa) and high pressure (60 MPa) were determined by scanning electron microscopy as described previously (21). Strain YC-1 was cultivated in marine broth 2216 at 8°C for each pressure. Mid-exponential-phase cultures were fixed with glutaraldehyde (final concentration, 2%) and were filtered through 0.2-μm polycarbonate filters (Nuclepore, USA). Cells were dehydrated by transferring the filters through a series of increasing concentrations of ethanol (10, 30, 50, 70, and 95%, and three times at 100%, for 10 min each time), followed by critical-point drying with CO2 and sputter-coating with gold. Samples were examined with an environmental scanning electron microscope (Quanta 600; FEI, USA).
Characterization of growth under high-pressure conditions.
All high-pressure physiological tests were repeated three times with uninoculated blank controls. Acid production from sugars (l-arabinose, d-cellobiose, d-fructose, d-galactose, d-glucose, lactose, d-mannose, maltose, d-raffinose, l-rhamnose, d-ribose, sucrose, starch, d-trehalose, and d-xylose) was assessed in modified O/F medium (22) containing 0.5× artificial seawater (1.5% NaCl, 0.27% MgSO4·7H2O, 0.035% KCl, 0.05% CaCl2·2H2O, 0.54% MgCl2·6H2O), 0.05% NH4H2PO4, 0.005% yeast extract, 0.1% Na2CO3, 1% sugar, and 0.003% bromothymol blue (the pH was adjusted to 7.1 on ice). The cultures were incubated at 60 MPa and 8°C for 4 weeks. Tests for the production of hydrogen sulfide from thiosulfate, indole production, and cell motility were performed using SIM (sulfide indole motility) medium (23) prepared with 0.5× artificial seawater instead of water. Enzymatic activities were tested as described previously (24). Cells were cultivated in marine broth 2216. After centrifugation at 14,000 rpm for 20 min, the cell pellet was spread on oxidase test paper (25) for the oxidase test. Catalase activity was tested by putting 3% H2O2 on the cell pellet. The DNase test was performed by inoculating stab cultures in DNase medium (Difco, USA) containing 0.5× artificial seawater and 0.01% methyl green. To test for gelatinase activity, cells were inoculated by stab cultures in marine broth 2216 medium containing 2% gelatin. After incubation, the stab culture was decompressed and was placed in a 30% trichloroacetic acid solution for clear-zone examination. Tests for growth with sole carbon sources and sole electron acceptors were performed according to the methods described by DeLong et al. (26). For instance, tests for lactate as the sole carbon source and trimethylamine oxide (TMAO) as the sole electron acceptor were performed as follows: four parts of inoculated mineral medium (27) were mixed with no addition, 15 mM lactate, 25 mM TMAO, and 15 mM lactate plus 25 mM TMAO, respectively, and after incubation, the cultures were decompressed and checked for growth.
The cellular fatty acid content of strain YC-1 were analyzed at Microbial Identification, Inc. (MIDI), using the MIDI system (28). Chromosomal DNA was extracted and was purified according to the standard method (29). The DNA G+C content was analyzed using reversed-phase high-performance liquid chromatography (HPLC) (30).
16S rRNA sequencing and phylogenetic analysis.
For phylogenetic analysis, the 16S rRNA gene was PCR amplified with primers 27F and 1492R (31). The PCR conditions were as follows: initial denaturation at 94°C for 10 min; 35 cycles of denaturation (94°C for 30 s), annealing (52°C for 2 min), and extension (72°C for 2 min); and a final extension of 72°C for 7 min. The purified PCR product was sequenced by SeqXcel Inc. using BigDye Terminator chemistry (Applied Biosystems, USA) with the 3730 DNA analyzer (Applied Biosystems, USA). The 16S rRNA gene sequence of strain YC-1 was first compared to other gene sequences in the NCBI GenBank database using a BLAST search (32). The sequence was later aligned using the ARB software package with the ARB_EDIT4 tool (33). Aligned sequences were exported to PHYLIP (34) and FastTree (35) for neighbor-joining (NJ) and maximum-likelihood (ML) analysis with bootstrap sampling, respectively. Profundimonas piezophilia strain YC-1 is available through the American Type Culture Collection (catalog no. BP-BAA-2591).
Nucleotide sequence accession number.
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain YC-1 is HQ230045.
RESULTS AND DISCUSSION
YC-1 is an obligate psychropiezophile with morphology varying as a function of pressure.
Strain YC-1 grew well in pressure vessels incubated at temperatures of 4 to 14°C at 60 MPa, with a temperature optimum of 8°C. No growth was observed at temperatures higher than 16°C. Growth occurred under hydrostatic pressures of 20 to 70 MPa at 8°C. The optimal pressure for growth was determined to be 50 MPa at 8°C, and no growth occurred at pressures either below 10 MPa or above 80 MPa (Fig. 1). The minimal doubling time was about 41 h, and the optimal growth yield was 3.5 × 107 cells ml−1. The maximal growth rate of YC-1 (0.017 h−1) was much lower than those of other cultured psychropiezophilic isolates within the Gammaproteobacteria (0.07 to 0.5 h−1). The growth rate of YC-1 is comparable to that of the recently isolated psychropiezophilic Alphaproteobacterium strain PRT1, which was isolated from hadal seawater (collected from 8,350 m) in the Puerto Rico Trench and has a minimal doubling time of 36 h (1). This suggests that the low growth rates of YC-1 and PRT1 could reflect adaptation to their oligotrophic environments.
FIG 1.
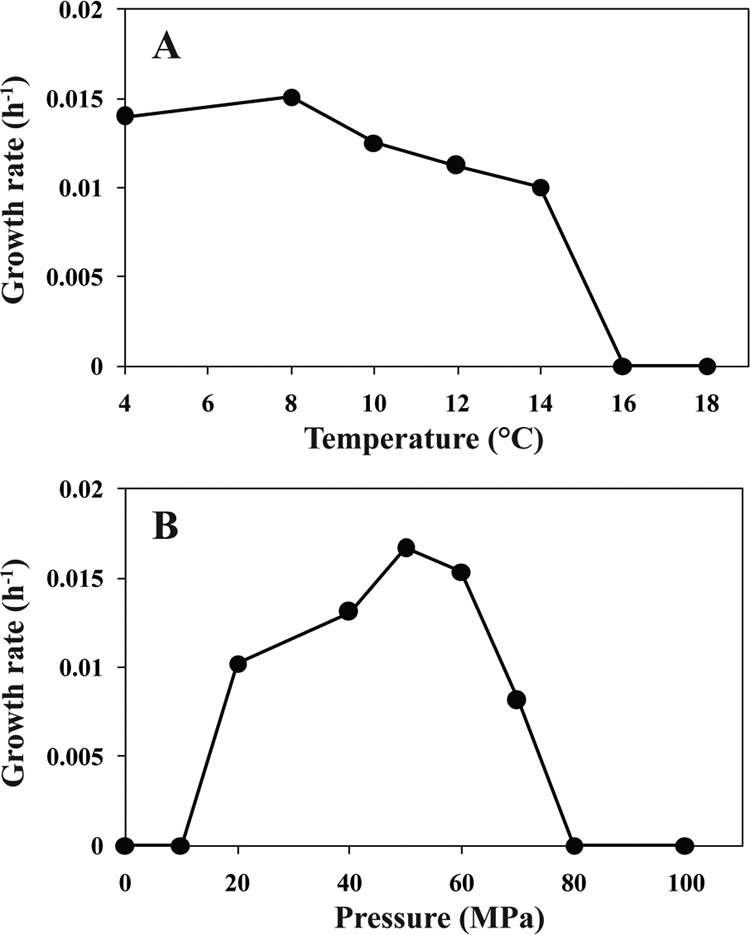
Growth of strain YC-1. (A) Effect of temperature on the growth rate of strain YC-1 under 60 MPa, equivalent to the pressure at a depth of 6,000 m. (B) Growth rate of strain YC-1 under different pressures at 8°C.
Cells of strain YC-1 were Gram-negative rods, 4.0 to 5.0 μm long and 0.5 to 0.8 μm wide under 60 MPa, which is equivalent to the hydrostatic pressure at its depth of isolation, 6,000 m. Coccoid bodies (diameter, 0.8 to 1.0 μm) were observed at a pressure of 20 MPa, which is the minimal pressure permitting growth of strain YC-1 (Fig. 2). These results indicate that strain YC-1 changes morphology under different pressures, shifting toward more filamentous forms at higher pressures, a result similar to those obtained previously for pressure-sensitive bacteria (36).
FIG 2.
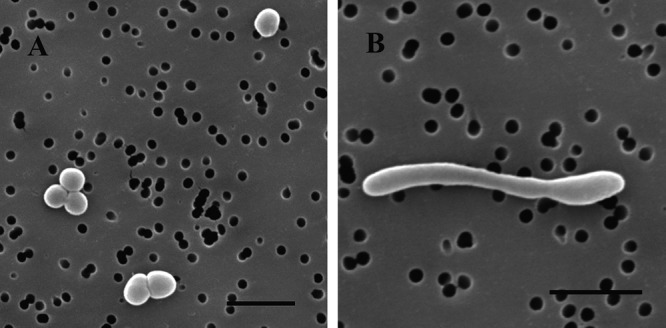
Scanning electron micrographs of cells of strain YC-1 cultivated in marine broth 2216 at 8°C under pressures of 20 MPa (A) and 60 MPa (B). Bars, 2 μm.
MUFAs are the predominant cellular fatty acids of YC-1.
The whole-cell fatty acid compositions of the novel strain YC-1 and related species are shown in Table 1. The fatty acid profile is similar to those of members of the genus Neptunomonas (37). The predominant nonpolar cellular fatty acids of YC-1 were C16:1 (palmitoleic acid) and C18:1 (oleic acid and cis-vaccenic acid). YC-1 produced more C16:1 (44.9%) than C18:1 (30.7%) at low pressure (20 MPa). At high pressure (60 MPa), the proportions of C18:1 fatty acids (45.6%), especially cis-vaccenic acid, of YC-1 increased dramatically, while that of the C16:1 fatty acid (35.8%) was reduced. The major hydroxy fatty acid was C10:0 3-OH at both low and high pressures. Unlike most other piezophilic bacteria within the Gammaproteobacteria, strain YC-1, like Colwellia piezophila Y223G, did not produce either EPA or DHA (38). The ratio of unsaturated fatty acids to saturated fatty acids in YC-1 changed from 6.8 to 8.8 when the pressure was increased from 20 MPa to 60 MPa.
TABLE 1.
Whole-cell fatty acid compositions of piezophilic strain YC-1, related Neptunomonas strains, and piezophilic species of related genera
| Fatty acid | Whole-cell fatty acid composition (%)a of: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain YC-1 grown at: |
N. concharum LHW37T | N. japonica JAMM 0745T | N. antarctica S3-22T | N. naphthovorans NAG-2N-126T | Colwellia piezophila Y223GT | Moritella japonica JCM 10249T | Photobacterium profundum JCM 10084T | Psychromonas kaikoae JCM 11054T | Shewanella benthica ATCC 43992T | ||
| 20 MPa | 60 MPa | ||||||||||
| C10:0 | Tr | Tr | 2.9 | 2.9 | TR | — | — | — | — | — | — |
| C12:0 | Tr | Tr | 1.3 | 1.7 | TR | 3.7 | 1–2 | — | 2 | 1 | 2 |
| C14:0 | Tr | Tr | — | — | — | — | 9 | 18 | 3 | 6 | 14 |
| C15:0 | Tr | Tr | — | — | — | — | 3 | 1 | 1 | 1 | — |
| C16:0 | 7.7 | 7.0 | 8.7 | 7.0 | 14.7 | 17.1 | 31–33 | 21 | 9 | 15 | 14 |
| C13:0 iso | — | — | — | — | — | — | — | — | 2 | — | 5 |
| C14:0 iso | — | — | — | — | — | — | — | — | 4 | — | — |
| C15:0 iso | — | — | — | — | — | — | — | — | 2 | — | 11 |
| C16:0 iso | Tr | — | — | — | — | — | — | — | 15 | — | — |
| C14:1 | — | — | — | — | — | — | 2 | 2 | 3 | 10 | — |
| C15:1 | Tr | Tr | — | — | — | — | 3 | — | — | — | — |
| C16:1 | 44.9 | 35.8 | 38.0 | 45.2 | 64.0 | 37.9 | 48–50 | 50 | 31 | 54 | 31 |
| C18:1 | 30.7 | 45.6 | 39.9 | 32.5 | 9.9 | 34.3 | — | 2 | 9 | 2 | 2 |
| C20:5 (EPA) | — | — | — | — | — | — | — | — | — | 2 | 16 |
| C22:6 (DHA) | — | — | — | — | — | — | — | 6 | 13 | 2 | — |
| C10:0 3-OH | 11.0 | 7.3 | 5.1 | 4.8 | 4.5 | 6.1 | — | — | — | — | — |
| C12:0 3-OH | — | — | — | — | — | — | 1 | — | 5 | 2 | 1 |
| C14:0 3-OH | — | — | — | — | — | — | — | — | — | 4 | — |
| C13:0 iso 3-OH | — | — | — | — | — | — | — | — | — | — | 5 |
YC-1 is a novel member of the Oceanospirillales.
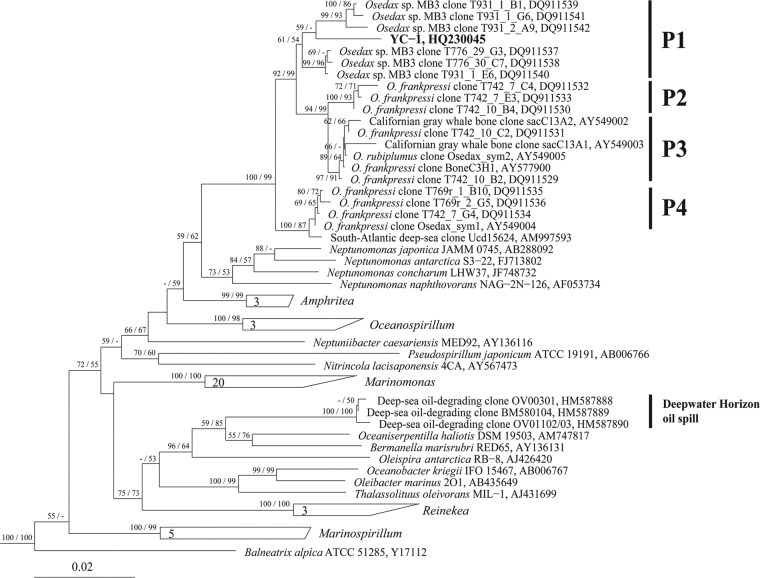
Phylogenetic analyses based on the 16S rRNA gene sequence place YC-1 within the Oceanospirillales order of the Gammaproteobacteria (Fig. 3). More specifically, YC-1 is localized within the P1 clade, a grouping that, until now, was associated exclusively with uncultured symbionts of bone-eating worms of the genus Osedax (13, 14). All of the sequences assigned to this clade are from the first and the deepest whale fall yet characterized for Osedax worms, a gray whale carcass present at 2,891 m within Monterey Canyon, CA (16). Strain YC-1 is closely related to the symbiont bacterial clone Osedax_sym1 (97.2% 16S rRNA sequence identity). Curiously, the worm tissues from which Osedax_sym1 nucleic acid were derived were also found to be rich in vaccenic acid (C18:1) and the long-chain polyunsaturated fatty acid EPA, features shared with many psychropiezophilic bacteria. The cultivated organisms most closely related to strain YC-1 belong to the genus Neptunomonas and include Neptunomonas antarctica S3-22T, Neptunomonas japonica JAMM 0745T, Neptunomonas naphthovorans NAG-2N-126T, and Neptunomonas concharum LHW37T, which share 93.2, 93.1, 92.7, and 92.6% 16S rRNA sequence identity with YC-1, respectively. Neptunomonas japonica also is connected to whale falls. The one member of this species was isolated from sediment adjacent to a sperm whale carcass off Kagoshima, Japan (19), and is closely related to symbionts of Osedax present at shallow-water minke, pilot, and sperm whale falls in the North Atlantic (13). Phylogenetically, strain YC-1 falls next to the genus Neptunomonas and within clades P1 to P4 of uncultured California Osedax symbionts and uncultured sea ice and seawater microbes (14). The G+C content of the DNA of strain YC-1 is 44.6 mol%.
FIG 3.
Phylogenetic tree depicting the relationship, based on 16S rRNA gene sequences, of strain YC-1 (shown in boldface) from the Puerto Rico Trench and related strains within the family Oceanospirillaceae. Bootstrap support for nodes is indicated (PHYLIP NJ method/FastTree ML method; only values above 50% are shown). The outgroup used to calculate phylogeny was Escherichia coli ATCC 11775T (GenBank accession number X80725). The bar represents 0.02 substitution per nucleotide position. Clusters P1 to P4, previously identified by Goffredi et al. (14), and Deepwater Horizon oil spill-associated clones identified by Hazen et al. (11) are indicated.
Previous FISH investigations of Osedax symbionts revealed that the symbionts are present in the ovisac and proliferative “root” tissues of mature females (13, 39) and are absent from Osedax eggs, sperm, and presettlement larvae (13, 40). It is believed that Osedax worms acquire the symbionts as free-living bacteria from the environment before settlement. Given that the symbionts of deep-sea Osedax hosts are distinct from those of their shallow-water counterparts, horizontal transfer of symbionts could be controlled by both host and nonhost environmental factors, including those associated with increasing water depth (13, 14). Indeed, a recent study of temporal variation with Osedax symbiont diversity provided evidence that the Oceanospirillales symbionts associated with Osedax are dynamic and differ according to environmental conditions. The acquisition of symbionts by Osedax was affected by the composition of free-living microbes in the local environment (41). Species related to the psychropiezophilic strain YC-1 could be selectively favored by Osedax larvae during symbiont acquisition in deeper waters. However, the relationship between free-living strain YC-1 and bone-eating worm symbionts still needs further study.
YC-1 can degrade hydrocarbons and is metabolically versatile.
The characteristics of the isolated piezophilic strain YC-1 and reference Neptunomonas strains are shown in Table 2. YC-1 is a facultatively anaerobic heterotroph. The results of oxidase and catalase tests were positive, and the tests of DNase and gelatinase activities were negative. Hydrogen sulfide and indole were not produced, and motility was not detected. Acid was not produced from sugar. Strain YC-1 oxidized Tween 80, Triton X-100, Triton X-114, hexadecane, citrate, formate, lactate, l-alanine, l-arginine, l-aspartic acid, l-lysine, l-methionine, l-phenylalanine, l-serine, and l-threonine as sole carbon sources. An electron acceptor was required for anaerobic growth, and YC-1 was able to use nitrate, sulfate, and TMAO as terminal electron acceptors.
TABLE 2.
Phenotypic characteristics of strain YC-1 and related species of the genus Neptunomonasa
| Characteristic | Strain YC-1 | N. japonica JAMM 0745T | N. naphthovorans NAG-2N-126T | N. antarctica S3-22T | N. concharum LHW37T |
|---|---|---|---|---|---|
| Cell shape | Rod/cocci | Rod | Rod | Rod | Rod |
| Motility | − | + | + | + | + |
| Growth temp (°C) | |||||
| Range | 2–14 | 5–25 | 4–30 | 4–25 | 10–45 |
| Optimum | 8 | 20 | ND | 15 | 37 |
| Optimum pressure (MPa) | 50 | 0.1 | 0.1 | 0.1 | 0.1 |
| Growth at atmospheric pressure | − | + | + | + | + |
| Oxidase | + | + | + | + | + |
| Catalase | + | + | + | + | + |
| DNase | − | + | − | − | + |
| Gelatinase | − | + | − | − | + |
| Indole production | − | − | + | − | − |
| Thiosulfate reduction | − | − | − | − | − |
| Nitrate reduction | + | + | − | + | + |
| Acid from sugars | − | − | + | + | + |
| DNA G+C content (mol%) | 44.6 | 43.6 | 46.3 | 45.6 | 48.2 |
The utilization of hexadecane by YC-1 as its sole carbon source indicated an ability to degrade aliphatic hydrocarbon. Studies of the 2010 Deepwater Horizon oil spill in the Gulf of Mexico revealed that microbial processes were the key factor accounting for the degradation of the 1,100- to 1,200-m-deep plume of light crude oil and chemical dispersants (11, 12, 42). Microbial community analyses revealed that a single operational taxonomic unit (OTU) within the Oceanospirillales accounted for more than 90% of the 16S rRNA gene sequences recovered from plume samples (11). This OTU shares 90.0% similarity with YC-1. Metagenomics, metatranscriptomics, and single-cell sequencing of the Deepwater Horizon plume-associated microbes has identified the entire pathways for degradation of alkanes in these uncultured Oceanospirillales species (12). However, the lack of a cultivated isolate within the Oceanospirillales from the deep sea has prevented in-depth physiological studies of hydrocarbon degradation within this order. The isolation of the psychropiezophilic Oceanospirillales isolate YC-1 provides an opportunity for such studies.
Consistent with the phylogenetic analysis, strain YC-1 shares a number of physiological and biochemical properties with members of the genus Neptunomonas, including positive oxidase and catalase activities, C16:1 and C18:1 fatty acids as dominant components of MUFAs, and DNA G+C contents of 43.6 to 46.3 mol%. However, there were also some major differences between strain YC-1 and members of the genus Neptunomonas. YC-1 is a deep-sea psychropiezophilic bacterium that cannot survive at atmospheric pressure. It also exhibits no motility. The low 16S rRNA gene sequence similarity and the significant phenotypic differences between strain YC-1 and the representatives of related genera within the family Oceanospirillaceae suggest that strain YC-1 represents a novel deep-sea species within a new genus, which we designate Profundimonas piezophila gen. nov., sp. nov.
Description of Profundimonas gen. nov.
Profundimonas (Pro.fun.di.mo′nas. L. neut. n. profundum, the depths of the sea; L. fem. n. monas, a unit, monad; N.L. fem. n. Profundimonas, a unit [bacterium] from the depths of the sea). Cells are nonmotile, facultatively anaerobic, Gram-negative rods at optimal high hydrostatic pressures. Coccoid bodies formed under low pressures. Oxidase and catalase positive, and predominant fatty acids are C16:1 and C18:1. Psychropiezophilic growth, with pressure range from 20 to 70 MPa, temperature range from <4 to 14°C. 16S rRNA gene sequence analysis places the genus close to the genus Neptunomonas within the family Oceanospirillaceae. The type species is Profundimonas piezophila.
Description of Profundimonas piezophila sp. nov.
Profundimonas piezophila (pi.e.zo.phi′la. Gr. v. piezo, to press; N.L. adj. philus, -a, -um [from Gr. adj. philos, -ê, -on], friend, loving; N.L. fem. adj. piezophila, loving pressure). The main characteristics of the species are the same as those reported for the genus. The cell size is 4.0 to 5.0 by 0.5 to 0.8 μm as a rod under 60 MPa, 0.8 to 1.0 μm in diameter as a coccoid under 20 MPa. Optimal growth occurs at 50 MPa and 8°C when cells are cultivated in marine broth 2216. The main nonpolar cellular fatty acids are C16:1 and C18:1.The major hydroxy fatty acid is C10:0 3-OH. Tests for oxidase and catalase activities are positive, and negative reactions are obtained for DNase and gelatinase. Negative for indole production, and H2S is not produced from thiosulfate. Acid is not produced from sugars. The following substrates are utilized as sole carbon sources for respiration: Tween 80, Triton X-100, Triton X-114, hexadecane, citrate, formate, lactate, l-alanine, l-arginine, l-aspartic acid, l-lysine, l-methionine, l-phenylalanine, l-serine, and l-threonine. The following compounds are used as terminal electron acceptors for anaerobic growth: nitrate, sulfate, and TMAO. The G+C content of the DNA is 44.6 mol%. The type strain, YC-1, was isolated from a deep-sea seawater sample collected in the Puerto Rico Trench. Profundimonas piezophilia strain YC-1 is available through the American Type Culture Collection (catalog no. BP-BAA-2591).
ACKNOWLEDGMENTS
We are grateful to the crew of the R/V Atlantic Explorer and to Christine Schulse for the collection of the deep-seawater sample.
This work was supported by NSF grants EF-0801973 and EF-0827051 to D.H.B. Y.C. was supported by a fellowship from the China Scholarship Council.
Footnotes
Published ahead of print 11 October 2013
REFERENCES
- 1.Eloe EA, Malfatti F, Gutierrez J, Hardy K, Schmidt WE, Pogliano K, Pogliano J, Azam F, Bartlett DH. 2011. Isolation and characterization of a psychropiezophilic alphaproteobacterium. Appl. Environ. Microbiol. 77:8145–8153. 10.1128/AEM.05204-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartlett DH. 1992. Microbial life at high-pressures. Sci. Prog. 76:479–496 [PubMed] [Google Scholar]
- 3.Lauro FM, Chastain RA, Blankenship LE, Yayanos AA, Bartlett DH. 2007. The unique 16S rRNA genes of piezophiles reflect both phylogeny and adaptation. Appl. Environ. Microbiol. 73:838–845. 10.1128/AEM.01726-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartlett DH. 2002. Pressure effects on in vivo microbial processes. Biochim. Biophys. Acta 1595:367–381. 10.1016/S0167-4838(01)00357-0 [DOI] [PubMed] [Google Scholar]
- 5.Abe F. 2007. Exploration of the effects of high hydrostatic pressure on microbial growth, physiology and survival: perspectives from piezophysiology. Biosci. Biotechnol. Biochem. 71:2347–2357. 10.1271/bbb.70015 [DOI] [PubMed] [Google Scholar]
- 6.Meersman F, Daniel I, Bartlett DH, Winter R, Hazael R, McMillan PF. 2013. High-pressure biochemistry and biophysics. Rev. Mineral. Geochem. 75:607–648. 10.2138/rmg.2013.75.19 [DOI] [Google Scholar]
- 7.Allen EE, Facciotti D, Bartlett DH. 1999. Monounsaturated but not polyunsaturated fatty acids are required for growth of the deep-sea bacterium Photobacterium profundum SS9 at high pressure and low temperature. Appl. Environ. Microbiol. 65:1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawamoto J, Sato T, Nakasone K, Kato C, Mihara H, Esaki N, Kurihara T. 2011. Favourable effects of eicosapentaenoic acid on the late step of the cell division in a piezophilic bacterium, Shewanella violacea DSS12, at high-hydrostatic pressures. Environ. Microbiol. 13:2293–2298. 10.1111/j.1462-2920.2011.02487.x [DOI] [PubMed] [Google Scholar]
- 9.Usui K, Hiraki T, Kawamoto J, Kurihara T, Nogi Y, Kato C, Abe F. 2012. Eicosapentaenoic acid plays a role in stabilizing dynamic membrane structure in the deep-sea piezophile Shewanella violacea: a study employing high-pressure time-resolved fluorescence anisotropy measurement. Biochim. Biophys. Acta 1818:574–583. 10.1016/j.bbamem.2011.10.010 [DOI] [PubMed] [Google Scholar]
- 10.Lauro FM, Tran K, Vezzi A, Vitulo N, Valle G, Bartlett DH. 2008. Large-scale transposon mutagenesis of Photobacterium profundum SS9 reveals new genetic loci important for growth at low temperature and high pressure. J. Bacteriol. 190:1699–1709. 10.1128/JB.01176-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, Jansson JK, Probst A, Borglin SE, Fortney JL, Stringfellow WT, Bill M, Conrad ME, Tom LM, Chavarria KL, Alusi TR, Lamendella R, Joyner DC, Spier C, Baelum J, Auer M, Zemla ML, Chakraborty R, Sonnenthal EL, D'haeseleer P, Holman HY, Osman S, Lu Z, Van Nostrand JD, Deng Y, Zhou J, Mason OU. 2010. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330:204–208. 10.1126/science.1195979 [DOI] [PubMed] [Google Scholar]
- 12.Mason OU, Hazen TC, Borglin S, Chain PS, Dubinsky EA, Fortney JL, Han J, Holman HY, Hultman J, Lamendella R, Mackelprang R, Malfatti S, Tom LM, Tringe SG, Woyke T, Zhou J, Rubin EM, Jansson JK. 2012. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J. 6:1715–1727. 10.1038/ismej.2012.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verna C, Ramette A, Wiklund H, Dahlgren TG, Glover AG, Gaill F, Dubilier N. 2010. High symbiont diversity in the bone-eating worm Osedax mucofloris from shallow whale-falls in the North Atlantic. Environ. Microbiol. 12:2355–2370. 10.1111/j.1462-2920.2010.02299.x [DOI] [PubMed] [Google Scholar]
- 14.Goffredi SK, Johnson SB, Vrijenhoek RC. 2007. Genetic diversity and potential function of microbial symbionts associated with newly discovered species of Osedax polychaete worms. Appl. Environ. Microbiol. 73:2314–2323. 10.1128/AEM.01986-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tresguerres M, Katz S, Rouse GW. 2013. How to get into bones: proton pump and carbonic anhydrase in Osedax boneworms. Proc. R. Soc. B 280:20130625. 10.1098/rspb.2013.0625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goffredi SK, Orphan VJ, Rouse GW, Jahnke L, Embaye T, Turk K, Lee R, Vrijenhoek RC. 2005. Evolutionary innovation: a bone-eating marine symbiosis. Environ. Microbiol. 7:1369–1378. 10.1111/j.1462-2920.2005.00824.x [DOI] [PubMed] [Google Scholar]
- 17.Rouse GW, Goffredi SK, Vrijenhoek RC. 2004. Osedax: bone-eating marine worms with dwarf males. Science 305:668–671. 10.1126/science.1098650 [DOI] [PubMed] [Google Scholar]
- 18.Hedlund BP, Geiselbrecht AD, Bair TJ, Staley JT. 1999. Polycyclic aromatic hydrocarbon degradation by a new marine bacterium, Neptunomonas naphthovorans gen. nov., sp. nov. Appl. Environ. Microbiol. 65:251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyazaki M, Nogi Y, Fujiwara Y, Kawato M, Kubokawa K, Horikoshi K. 2008. Neptunomonas japonica sp. nov., an Osedax japonicus symbiont-like bacterium isolated from sediment adjacent to sperm whale carcasses off Kagoshima, Japan. Int. J. Syst. Evol. Microbiol. 58:866–871. 10.1099/ijs.0.65509-0 [DOI] [PubMed] [Google Scholar]
- 20.Dietz AS, Yayanos AA. 1978. Silica gel media for isolating and studying bacteria under hydrostatic pressure. Appl. Environ. Microbiol. 36:966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yayanos AA, Dietz AS, Van Boxtel R. 1981. Obligately barophilic bacterium from the Mariana trench. Proc. Natl. Acad. Sci. U. S. A. 78:5212–5215. 10.1073/pnas.78.8.5212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hugh R, Leifson E. 1953. The taxonomic significance of fermentative versus oxidative metabolism of carbohydrates by various Gram-negative bacteria. J. Bacteriol. 66:24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards PR, Ewing WH. 1972. Identification of Enterobacteriaceae, 3rd ed. Burgess Publishing Company, Minneapolis, MN [Google Scholar]
- 24.Nogi Y, Kato C. 1999. Taxonomic studies of extremely barophilic bacteria isolated from the Mariana Trench and description of Moritella yayanosii sp. nov., a new barophilic bacterial isolate. Extremophiles 3:71–77. 10.1007/s007920050101 [DOI] [PubMed] [Google Scholar]
- 25.Baumann P, Baumann L. 1979. The marine gram-negative eubacteria: genera Photobacterium, Beneckea, Alteromonas, Pseudomonas and Alcaligenes, p 1302–1331 In Starr MP, Stolp H, Truper HG, Balows A, Schlegel HH. (ed), The prokaryotes. Springer-Verlag, New York, NY [Google Scholar]
- 26.DeLong EF, Franks DG, Yayanos AA. 1997. Evolutionary relationships of cultivated psychrophilic and barophilic deep-sea bacteria. Appl. Environ. Microbiol. 63:2105–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiChristina TJ, DeLong EF. 1994. Isolation of anaerobic respiratory mutants of Shewannella putrefaciens and genetic analysis of mutants deficient in anaerobic growth on Fe3+. J. Bacteriol. 176:1468–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sasser M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids, MIDI technical note 101. MIDI Inc., Newark, DE [Google Scholar]
- 29.Saito H, Miura K. 1963. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72:612–629 [PubMed] [Google Scholar]
- 30.Tamaoka J, Komagata K. 1984. Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol. Lett. 25:125–128. 10.1111/j.1574-6968.1984.tb01388.x [DOI] [Google Scholar]
- 31.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–148 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 33.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüssmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371. 10.1093/nar/gkh293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Felsenstein J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164–166 [Google Scholar]
- 35.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum-evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26:1641–1650. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zobell CE, Cobet AB. 1964. Filament formation by Escherichia coli at increased hydrostatic pressures. J. Bacteriol. 87:710–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee HW, Shin NR, Lee J, Roh SW, Whon TW, Bae JW. 2012. Neptunomonas concharum sp. nov., isolated from a dead ark clam, and emended description of the genus Neptunomonas. Int. J. Syst. Evol. Microbiol. 62:2657–2661. 10.1099/ijs.0.037473-0 [DOI] [PubMed] [Google Scholar]
- 38.Nogi Y, Hosoya S, Kato C, Horikoshi K. 2004. Colwellia piezophila sp. nov., a novel piezophilic species from deep-sea sediments of the Japan Trench. Int. J. Syst. Evol. Microbiol. 54:1627–1631. 10.1099/ijs.0.03049-0 [DOI] [PubMed] [Google Scholar]
- 39.Katz S, Klepal W, Bright M. 2011. The Osedax trophosome: organization and ultrastructure. Biol. Bull. 220:128–139 http://www.biolbull.org/content/220/2/128.long [DOI] [PubMed] [Google Scholar]
- 40.Rouse GW, Wilson NG, Goffredi SK, Johnson SB, Smart T, Widmer C, Young CM, Vrijenhoek RC. 2009. Spawning and development in Osedax boneworms (Siboglinidae, Annelida). Mar. Biol. 156:395–405. 10.1007/s00227-008-1091-z [DOI] [Google Scholar]
- 41.Salathé RM, Vrijenhoek RC. 2012. Temporal variation and lack of host specificity among bacterial endosymbionts of Osedax bone worms (Polychaeta: Siboglinidae). BMC Evol. Biol. 12:189. 10.1186/1471-2148-12-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baelum J, Borglin S, Chakraborty R, Fortney JL, Lamendella R, Mason OU, Auer M, Zemla M, Bill M, Conrad ME, Malfatti SA, Tringe SG, Holman HY, Hazen TC, Jansson JK. 2012. Deep-sea bacteria enriched by oil and dispersant from the Deepwater Horizon spill. Environ. Microbiol. 14:2405–2416. 10.1111/j.1462-2920.2012.02780.x [DOI] [PubMed] [Google Scholar]
- 43.Zhang XY, Zhang YJ, Yu Y, Li HJ, Gao ZM, Chen XL, Chen B, Zhang YZ. 2010. Neptunomonas antarctica sp. nov., isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 60:1958–1961. 10.1099/ijs.0.017756-0 [DOI] [PubMed] [Google Scholar]