Abstract
Ice nucleation-active (INA) bacteria may function as high-temperature ice-nucleating particles (INP) in clouds, but their effective contribution to atmospheric processes, i.e., their potential to trigger glaciation and precipitation, remains uncertain. We know little about their abundance on natural vegetation, factors that trigger their release, or persistence of their ice nucleation activity once airborne. To facilitate these investigations, we developed two quantitative PCR (qPCR) tests of the ina gene to directly count INA bacteria in environmental samples. Each of two primer pairs amplified most alleles of the ina gene and, taken together, they should amplify all known alleles. To aid primer design, we collected many new INA isolates. Alignment of their partial ina sequences revealed new and deeply branching clades, including sequences from Pseudomonas syringae pv. atropurpurea, Ps. viridiflava, Pantoea agglomerans, Xanthomonas campestris, and possibly Ps. putida, Ps. auricularis, and Ps. poae. qPCR of leaf washings recorded ∼108 ina genes g−1 fresh weight of foliage on cereals and 105 to 107 g−1 on broadleaf crops. Much lower populations were found on most naturally occurring vegetation. In fresh snow, ina genes from various INA bacteria were detected in about half the samples but at abundances that could have accounted for only a minor proportion of INP at −10°C (assuming one ina gene per INA bacterium). Despite this, an apparent biological source contributed an average of ∼85% of INP active at −10°C in snow samples. In contrast, a thunderstorm hail sample contained 0.3 INA bacteria per INP active at −10°C, suggesting a significant contribution to this sample.
INTRODUCTION
We have known for around 4 decades that soil, decaying vegetation, and plant surfaces harbor organic ice-nucleating particles (INP) and that their abundance and high temperature of activity suggest they are significant sources of atmospheric ice nuclei (1–8). The focus of more recent research has been to elucidate their identities, reservoirs, and emissions to and role, if any (9), in the troposphere (10–22).
DeMott and Prenni (23) identified key outstanding questions regarding the role of biogenic INP in cloud and precipitation processes. These included whether they are the sole source of natural atmospheric ice nucleators warmer than about −15°C, whether they are sufficiently abundant to trigger precipitation directly and/or via secondary ice multiplication, which operates principally in the range of −3 to −8°C (24, 25, 26), whether there are large seasonal and regional variations in their emissions, and whether their numbers can be defined and their sources identified. This work reports progress toward answering the first and last priorities through the development and application of a quantitative PCR (qPCR) test for the ice nucleation-active (INA) bacteria, the abundant and highly active source of biological INP (12).
To date, around a dozen species of INA bacteria have been identified, isolated mainly from plant surfaces. They are spread among three orders of the Gammaproteobacteria and include Pseudomonas syringae, Ps. fluorescens, Pantoea agglomerans, and Xanthomonas campestris (27). All express isoforms of the same Ina protein which, when bound together in the outer membrane, can trigger nucleation at up to −1.5°C (28, 29). More typically, the upper limit is −2 to −4°C (15). They are particularly abundant on crops (7) but are also plentiful on many nonagricultural plants and in habitats such as freshwater and associated biofilms (7, 15, 30). Possession of ice nucleation activity may confer several advantages (11, 30, 31), including the ice nucleation of cloud droplets as an active deposition and dissemination mechanism (8, 15, 30, 32).
INA bacteria have been detected in air above crops under dry conditions and were enhanced ∼30-fold during rainfall (33, 34), and they were relatively abundant in air downwind of harvesting (35). They have also been isolated from cloud water (18, 36, 37), from ice and rain at up to 2,500 m above a wheat field (8), and from ∼50% of rain and snow samples (15). A typically large reduction in INP active at >−12°C after heat treatment or cell wall digestion suggests their ubiquity in precipitation (38, 39); rainfall itself stimulates the release of biological INP from vegetation and the soil surface (33, 34, 40, 41).
Direct, sensitive detection of INA bacteria in atmospheric samples is required, because environmental populations are a mixture of culturable, viable but nonculturable (VBNC), moribund, and dead cells. All members of this continuum may possess ice nucleation activity. Detection of VBNC cells may be particularly important, because many environmental factors reduce culturability (42, 43). For example, aerosolization of Gram-negative bacteria originally isolated from air raised their VBNC-to-viable ratios >250-fold in only 20 min (44), and Attard et al. (37) observed a significant loss of culturability, but not ice nucleation activity, after exposure of INA bacteria to UV-A for ≥20 h. Furthermore, not all INA bacteria can be readily culturable on commonly used agars (45).
Amplification of the ina gene in INA bacteria will detect culturable, VBNC, and even a subset of dead cells. However, detection of the gene does not equate to these cells also being efficient INP, since aggregates of the protein required for high-temperature activity are naturally infrequent (46, 47) and are “controlled by complex and interacting factors such as genotype, environment, and host plant species” (46). At colder temperatures, clusters of two to three proteins will trigger freezing (48), so that by −10 to −12°C the nucleation frequency may be 1 in every 10 cells or higher (28, 31). Conversely, dead INA bacteria that have digested their own genome but which retain the Ina protein will go undetected; some INA Pseudomonas strains possess active, thermostable DNAses (T. C. J. Hill, unpublished data).
Successful qPCR of the ina gene relies primarily upon the design of the primers. They must amplify as many ina gene alleles as possible with both high specificity and sensitivity. However, due to the diversity of the gene (49), ina primers have, to date, been designed to amplify only single alleles or clades (45). This study describes the design of the first broad-spectrum ina gene primers and their use in qPCR to quantify INA bacteria on plants and in snow and hail.
MATERIALS AND METHODS
Vegetation sampling.
INA bacterial populations on plants were measured using qPCR of the ina gene in DNA extracted from leaf washings. Crops and a border grass were sampled in July 2010 at an irrigated field watered with a linear-move sprinkler at the University of Wyoming's Sustainable Agriculture Research and Extension Center (SAREC), near Lingle, WY. Grasses, shrubs, and trees were sampled the following month from roadside pasture and among native vegetation in the Laramie Mountains. Details of sites, sampling dates, and samples are given in Table S1 in the supplemental material.
Using nitrile gloves, a bulked sample was obtained from each vegetation type, each comprising leaves and seed heads (cereals and grasses) from many plants. Samples were stored in new resealable zippered storage bags at 4°C for 1 to 3 days before processing. Each sample was placed in a 1-liter conical flask that had been decontaminated by rinsing for 1 h with 5% H2O2, followed by two rinses with deionized water and autoclaving. To this, 250 ml of 0.45-μm-diameter-pore filtered 0.01 M sodium phosphate buffer (pH 7) with 0.1% Difco proteose peptone no. 3 (BD, Franklin Lakes NJ) was added (7), and the flask was shaken at 200 rpm for 1 h. Washings were poured through a sterile 100-μm-mesh cell strainer (BD Falcon, Franklin Lakes, NJ) into sterile, polypropylene 50-ml Falcon centrifuge tubes (Corning, Tewksbury, MA). One was frozen at −20°C for INP determinations, and the remainder, containing 200 ml, was centrifuged at 3,000 × g for 30 min. From these, pellets were combined, resuspended in a total volume of 1.25 ml, and stored at −70°C for DNA extraction. A blank extraction was also performed.
Snow and hail sampling.
Snow was sampled in two general locations, within Laramie and ∼15 km to the southeast in the Laramie Mountains. It was collected either during snowfalls or, when snowfalls continued into the night, the following morning. Details of sites, snowfall, sampling dates, and samples are given in Table S1 in the supplemental material. Nitrile gloves were worn and, unless the snow was still falling, surface snow was brushed away to expose a fresh surface for sampling. Using a zippered storage bag from a freshly opened pack, the top ∼1 cm was then shaved from the surface until 1 to 2 kg was collected. The sample was double bagged and stored at −20°C until being processed.
A single hail sample was also analyzed. In Laramie, at 9:35 p.m. on 10 June 2010, hail up to 1.5 cm in diameter fell from a thunderstorm that suddenly activated while overhead. This was collected over the few minutes of the hailswath into a new autoclave bag held open using nitrile gloves. The bag was immediately placed in a freezer and the mostly frozen hail stored at −20°C.
Snow and hail samples were melted at room temperature in a class II biosafety laminar flow cabinet (Labconco, Kansas City, MO), which was reserved for extractions from aerosols and precipitation samples and located in a separate and otherwise unused laboratory from that used for PCR. The cabinet's UV lamp was turned on for several hours before it was used to inactivate any potentially contaminating DNA. Melted samples were filtered through 0.2-μm-diameter-pore Nuclepore track-etched polycarbonate membranes (Whatman, GE Healthcare Life Sciences, Piscataway, NJ) that had been decontaminated by soaking in 15% H2O2 for 10 min, followed by two rinses in deionized water (18 MΩ, autoclaved at 135°C for 30 min to fragment DNA and stored in dedicated Pyrex bottles) and drying on foil. Membranes were mounted into 47-mm-wide, 0.45-μm-diameter-pore Nalgene sterile filter units (Thermo Scientific, Rochester, NY). After filtering, the filters were removed with flamed forceps and stored in sterile 50-ml Falcon tubes at −20°C until being processed.
To resuspend the filtered material, 6 or 7 ml of the deionized water, detailed above, was added to each tube, and tubes were shaken at 400 rpm for 20 min. Half of each sample was removed for immediate DNA extraction, while the remainder was frozen for measurement of INP. The DNA subsample was centrifuged at 22,000 × g for 5 min at 18°C in a 2-ml sterile screw-cap microcentrifuge tube by removing the supernatant and refilling the tube in several steps until ∼100 μl remained.
DNA extraction.
For DNA extraction from leaf washings, 50 or 100 μl of the 1.25 ml of leaf wash concentrate was processed using the standard protocol of the PowerLyzer UltraClean microbial DNA isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA). Homogenization was performed using a FastPrep bead beater (BIO 101 Inc., Carlsbad, CA) at setting 4 for 5 min. The same method was used to obtain DNA from pure cultures of INA bacteria.
For the snow samples, we modified the extraction protocol to increase recoveries (in consultation with MO BIO). We omitted the step to precipitate non-DNA organic and inorganic material (solution MD2), added a second ethanol wash (solution MD4), and added a second DNA elution (solution MD5). This increased DNA recoveries from ∼25% with the standard method to ∼90%, tested using 120 and 1,200 Ps. syringae Cit7 cells added directly to extraction tubes. Correction for the losses from each method in the qPCR is described below.
Various precautions were taken to prevent contamination of snow and hail samples. Extractions were performed in the biosafety laminar flow cabinet described above. Its UV lamp was turned on for several hours before use, and equipment within it was rotated to irradiate all surfaces. The microcentrifuge head was soaked in 5% H2O2, rinsed in deionized water, and dried overnight in the cabinet, and the inside surfaces of the microcentrifuge and bead beater were sponged liberally with 5% H2O2 and also dried overnight. The MO BIO kit used was reserved for snow and hail samples. For PCR, tubes containing aliquots of the qPCR master mix were brought into this separate laboratory for dispensing of the DNA. Finally, several blank extractions were performed, including of filters before and after peroxide treatment, and of a full extraction blank for which autoclaved deionized water was used in place of snow melt.
Isolation, INP production, and identity of INA isolates.
A diverse collection of INA bacterial isolates, several obtained during this study, were used to design and validate the primers. Details of their origin, activity, and identity are given in Table 1. Dilution series of the leaf washings were plated onto low-INP agar made using 0.45-μm-diameter-pore-filtered nutrient broth, 2.5% glycerol, and 15 g liter−1 agarose (certified molecular biology agarose; Bio-Rad, Hercules, CA). Dilution plates were incubated at 18°C for 3 to 5 days, after which a small quantity from 32 or 64 randomly chosen colonies was transferred and suspended in 50 μl of 0.45-μm-pore-diameter filtered PO4/peptone buffer. Suspensions that froze at warmer temperatures than the blanks (a pipette tip touched to an area of uncolonized agar and then inserted into buffer) were streak plated onto fresh agar and retested before being selected. Initially, wooden toothpicks were used for picking colonies, but these were found to release INP active at ∼−13°C. In later tests, using sterile plastic pipette tips, blanks started nucleating at −15 to −16°C.
TABLE 1.
Origin, activity, identity, and closest ina allele affiliation of isolates
| Isolate | Source | Location | Ice nucleation activitya (°C) | 16S rRNA |
ina |
||
|---|---|---|---|---|---|---|---|
| Closest isolate(s) and GenBank accession no. | Similarity (%) | Closest isolate, ina gene allele/gene name, and GenBank accession no. | Similarity (%) | ||||
| Cit7b | Navel orange leaf | Near Exeter, CAb | −3.5 (−2.5) | Ps. syringae (AY574914) | 100 | Ps. syringae inaZ (X03035) | 99.2 |
| BXIN4 | Bean pod or leaf lesion | Lingle, WY; 42.1315 N, 104.392 W | −3.0 (−2.5) | Ps. syringae (AB680547) | 100 | Ps. syringae inaQ (EU360731) | 99.5 |
| Ps. congelans (NR_028985) | 100 | ||||||
| PCa2a | Cabernet sauvignon grapevine cane lesion | Coonawarra, South Australia; 37.361 S, 140.838 E | −3.0 | Ps. syringae (AB680547), Ps. congelans (NR_028985) | 100 100 | Ps. syringae inaQ (EU360731) | 97.4 |
| HCh1a | Chardonnay grapevine shoot, healthy | Hallston, Victoria, Australia; 38.3465 S, 146.028 E | −3.0 | Ps. syringae (AJ889841) | 100 | Ps. syringae inaQ (EU360731) | 97.2 |
| BXIN3 | Bean pod or leaf lesion | Lingle, WY; 42.1315 N, 104.392 W | −3.5 (−2.5) | Ps. syringae (CP000075), Ps. congelans (NR_028985) | 100 100 | Ps. syringae Psyr1608 (CP000075) | 99.2 |
| GCh5Fc | Chardonnay grapevine shoot killed by frost | Glenlofty, Victoria, Australia; 37.120 S, 143.217 E | −4.0 (−3.5) | Ps. syringae (AB680547), Ps. congelans (NR_028985) | 100 100 | Ps. syringae inaV (AJ001086) | 100 |
| Sco1009b | Corn (maize) leaves, senescent | Lingle, WY; 42.1315 N, 104.3965 W | −2.5 (−2.0) | Ps. syringae pv. atropurpurea (AB001440) | 100 | Ps. syringae inaV (AJ001086) | 83 |
| PCa2bi | Cabernet sauvignon grapevine cane lesion | Coonawarra, South Australia; 37.361 S, 140.838 E | −3.0 | Ps. viridiflava (AY574912) | 100 | Ps. syringae inaV (AJ001086) | 85 |
| GrFc | Grape | CAc | −4.0 (−3.0) | Pa. ananatis (CP001875) | 99.6 | Pa. ananatis iceA (AF387802) | 99 |
| SBPci | Bean pod lesion | Lingle, WY; 42.1315 N, 104.392 W | −3.5 (−3.0) | Pa. agglomerans (FJ756354) | 99.9 | Pa. ananatis inaA (CP001875) | 76 |
| Sba1007a | Barley green leaves and heads | Lingle, WY; 42.1315 N, 104.395 W | −3.0 | X. campestris pv. campestris (CP000050) | 100 | X. campestris pv. raphani XCR 4000 (CP002789) | 92 |
| Sba1007bi | Barley green leaves and heads | Lingle, WY; 42.1315 N, 104.395 W | −3.5 | X. campestris pv. campestris (CP000050) | 100 | X. campestris pv. raphani XCR 4000 (CP002789) | 92 |
| Sbr1009a | Smooth brome (B. inermis) green leaves | Lingle, WY; 42.1320 N, 104.395 W | −2.0 | X. translucens (NR_036968) | 99.9 | X. campestris pv. translucens inaX (X52970) | 97 |
| MU26d | Wood frog (Rana sylvatica) gut | Adams County, OHd | −3.0 | Ps. fluorescens (JF327445) | 99.9 | Ps. fluorescens inaW (X04501) | 89 |
| BF81Fbe | Air during rain in ponderosa pine forest | Manitou Forest, CO; 39.1028 N, 105.104 W | −4.0 (−2.5) | Ps. koreensis (NR_025228) Ps. fluorescens (JN679853), Ps. putida (HM217118) | 99.9 99.9 99.9 | Ps. fluorescens inaW (X04501) | 86 |
| LSb | Peat moss (S. capillifolium) | Bog, Lewis, Outer Hebrides, United Kingdom; 58.1 N, 6.7 W | −3.0 | Pseudomonas sp. strain LD002 (HQ713573), Pseudomonas sp. strain NZ099 (AF388207)f | 100 99.9 | Ps. fluorescens inaW (X04501) | 87 |
| MM3b | Dwarf birch (B. nana) and bog rosemary (A. polifolia) | Martimoaapa mire, Finland; 65.81 N, 25.22 E | −3.0 | Pseudomonas sp. strain NZ099 (AF388207)f | 100 | Ps. fluorescens inaW (X04501) | 87 |
| χ16 | Snow | Mt. Parnassos ski center, Greece; 38.5393 N, 22.60645 E | −3.5 | Ps. auricularis (AB681727) | 99.9 | Ps. fluorescens inaW (X04501) | 87 |
| χ17 | Snow | Mt. Parnassos ski center, Greece; 38.5393 N, 22.60645 E | −3.0 | Ps. poae (GU188955) | 99.9 | Ps. fluorescens inaW (X04501) | 86 |
| GraPa8 | Grass | Mt. Parnitha, Greece; 38.172444 N, 23.728622 E | −3.5 | Pseudomonas sp. strain JCM5484 (AB685689), Ps. auricularis (AB681727) | 99.9 99.8 | Ps. fluorescens inaW (X04501) | 86 |
The warmest temperature of activity from ∼107 cells (tested in 0.5°C increments). Numbers in parentheses are the warmest temperatures of activity, if higher, in other tests using LNP medium but where numbers of bacteria were not determined.
Data are according to Lindow (71). Provided courtesy of S. E. Lindow (University of California, Berkeley). The genome has been sequenced (72).
From Phelps et al. (73). Provided courtesy of C. E. Morris (INRA, Montfavet, France).
From Lee et al. (74). Provided courtesy of M. R. Lee (Miami University, Oxford, OH).
Provided courtesy of J. Fröhlich-Nowoisky (Max Planck Institute for Chemistry, Mainz, Germany).
Classified as part of the Ps. putida lineage.
To fully assess INP production, selected isolates were grown in the low nitrogen and phosphorus liquid medium (LNP) recommended by Nemecek-Marshall et al. (50). LNP contained 50 mM morpholinepropanesulfonic acid (MOPS) at pH 7.2, 25 mM KCl, 2 mM NH4Cl, 10 mM Na2SO4, 10 mM NaCl, 1 mM MgCl2, 0.1 mM KH2PO4, 0.1 mM CaCl2, 0.01 mM FeCl3, and galactose as a carbon source at 0.4% (wt/vol). Galactose was chosen because it produced both a high frequency of type 1 nuclei (i.e., active warmer than −5°C) and, overall, the most INP out of 10 carbon sources compared (50). LNP was made with 0.2-μm-pore-size-filter-sterilized deionized water (blanks nucleated at <−15°C). Isolates were grown at 32°C for 3 days and then incubated at 16°C for 1 day to allow full development of high-temperature INP (51). Testing for INA was performed using up to 32 aliquots of 30 to 80 μl of the culture dispensed into sterile polypropylene PCR trays as described below. Cell numbers were determined using dilution series on nutrient agar amended with 2.5% glycerol, and the number of wells containing a total of ∼107 cells was back calculated.
Isolates were identified from their nearly full-length 16S rRNA genes sequences. Each PCR mixture contained 1 U of GoTaq hot start polymerase (Promega, Madison, WI) in 1× Colorless GoTaq Flexi buffer (no Mg in buffer), 0.5 μM primer 27f (5′ AGAGTTTGATCMTGGCTCAG 3′), 0.5 μM primer 1492r (5′ GGTTACCTTGTTACGACTT 3′), 0.2 mM each deoxynucleoside triphosphate (dNTP), 1.5 mM MgCl2, 10 ng genomic DNA, and deionized water to a total volume of 25 μl. Cycling conditions were an initial denaturation at 95°C for 2 min, followed by 35 cycles of 94°C for 15 s, 48°C for 15 s, and 72°C for 45 s. Products were sequenced in both directions at the University of Wyoming's Nucleic Acid Facility and identified using a National Center for Biotechnology Information BLAST search.
Measurement of ice-nucleating particles.
Concentrations of INP in leaf washings and filter-concentrated melt samples were derived using the drop freezing method. Samples were rapidly thawed and vortexed, and then 24 or 32 aliquots of 50 μl were dispensed into sterile 96-well polypropylene PCR trays (μCycler; Life Science Products Inc., Frederick, CO). The tray was covered with a silicone sealing mat for PCR and placed in a thermal cycler (PTC-200; MJ Research) with the block held at 4°C. The cycler was programmed to descend in 1°C increments from −2°C to its limit at −9°C. After 5 min at each temperature, the number of frozen wells was visually counted and the temperature lowered to the next increment. Temperature variation across the block was ±0.2°C, measured using a thermistor verification probe (VPT-0300; Bio-Rad, Hercules, CA). The tray was then transferred to an aluminum incubation block for PCR plates (VWR, Radnor, PA) within a foam box, cooled in a freezer to ∼−12°C (∼−15°C for testing of isolates). The thermistor was inserted into a side well, and after 10 min the block temperature and number of frozen wells were recorded. Serial dilutions in deionized water (INP free until <−15°C) were used to measure INP concentrations across the temperature range. To test the sensitivity of the INP in leaf washings to moderate heat, trays were thawed and then heated in the thermal cycler to 60°C for 20 min and then retested. To test the sensitivity of the INP to a denaturing heat, 1.8 ml of the washings or melt samples was aliquoted into a 2-ml screw-cap microcentrifuge tube and incubated at 105°C for 10 min (after allowing time for equilibration). The temperatures of 60 and 105°C were chosen after tests of the heat sensitivity of a range of local isolates (Cit7, BXIN3, BXIN4, Sco1009b, SBPci, and Sbr1009a). Isolates were grown in LNP medium, and then aliquots were heated and freeze tested. Non-heat-treated aliquots froze at ≥−3°C. Heating within the range of 55 to 80°C lowered the onset of nucleation to a new plateau of −7 to −9°C (−5.5°C for Sbr1009a), while heating to 90 to 95°C eliminated ice nucleation activity (tested to −18°C) in all isolates except Sbr1009a, for which heating to 105°C lowered its onset to −10°C. Thus, in all isolates, 60°C degraded the INP from class A aggregates to the less efficient class B structures (52), while 105°C denatured the protein in all isolates except Sbr1009a (a xanthomonad containing the inaX allele), which was reduced to possessing class C structures (i.e., individual proteins).
Cumulative numbers of INP per ml test solution were estimated using the formula −ln(f)/V, where f is the proportion of droplets not frozen and V is the volume of each aliquot (53), and then converted to INP per gram of fresh weight of leaf or per gram of snow or hail.
Choice of region for primer design within the ina gene.
Amino acids in the core region of the ina gene are arranged into repeating sequences with nested periodicities of 8, 16, and 48 (49), with the 16-mers being the most faithfully repeated (54). Conserved sequences such as these should normally aid the design of broad-spectrum primers, but in this case their repetition promotes primer binding at multiple sites, leading to multiple PCR products (e.g., see references 55 and 56). If multiple products are limited to two or three, they can still be used for qPCR so long as the reaction is efficient and repeatable (they can even be useful for identification of the allele in the postreaction analyses). However, their occurrence tends to reflect imperfect primer binding and, hence, inefficient amplification and consequent underestimation of gene copy number.
The core of the ina gene can be subdivided into four blocks, with the fourth having the least-exact pattern of repetition (57). Warren and Corotto (57) noted, however, that this greater deviation from perfect periodicity in block 4 was also conserved, indicating that it plays a functional role. Block 4's lower level of internal homology, combined with the conservation of its irregularities, make it a preferred region for priming. Also, of all the blocks it varies least in its length (49), potentially enabling all PCR products to be of equal length, which is useful for confirmation of PCR product identity when gelled. Ahern (58) tested numerous primer combinations within blocks 2, 3, and 4 and the C terminus and found that those targeting block 4 were indeed the most successful.
Sequencing of block 4 of the ina gene in isolates.
DNA sequences of block 4 of the ina gene were obtained from the 20 isolates. Initially, a selection of forward and reverse primers was manually designed and tested based on an alignment of all published ina genes. One pair, 3076f (5′ AGYTCGCTGATTGCGGGNC 3′) and 3463r (5′ STGTAVCKTTTNCCGTCCCA 3′), consistently amplified a product (425 bp) in all isolates. This spanned the latter half of block 4 and nine bases of the C terminus. Frequent mispriming on the ina gene prevented it from being used for qPCR. Each PCR mixture contained 1 U of GoTaq hot start polymerase (Promega, Madison, WI) in 1× colorless GoTaq Flexi buffer (no Mg in buffer), 0.7 μM 3076f, 0.9 μM 3463r, 0.2 mM each dNTP, 1.25 mM MgCl2, 10 ng genomic DNA, and deionized water to a total volume of 20 μl. Cycling conditions were an initial denaturation at 95°C for 2 min, followed by 35 cycles of 94°C for 15 s and 53°C for 30 s (combined primer annealing and extension). Products were sequenced in both directions. The new sequences were aligned with those already available, and the expanded set was used for primer design.
Phylogenetic analyses.
Phylogenetic analyses were conducted using MEGA version 5 (59). Alignment of positions 3078 to 3462 (base numbering according to that of inaZ) was performed manually, since all were of equal length. To illustrate the sequence differences between alleles, a phylogenetic tree was initially constructed using proportional distances between nucleotide sequences. To more faithfully represent evolutionary distance (although based on only a limited portion of the gene), a second phylogenetic tree was derived from the amino acid sequences using the Jones-Taylor-Thornton model (60), applying a gamma-shaped parameter of 0.49 to adjust for nonuniformity of evolutionary rates among sites and assuming no positions were invariable. This model was chosen using MEGA's model selection facility (employing the maximum likelihood method and optimizing a neighbor-joining tree) as that which best described the substitution pattern. Neighbor joining was used to construct both the DNA- and amino acid-derived trees, with confidence levels of branch placement in the latter phylogram obtained using the interior branch test on 1,000 bootstrap replications.
Primer design within block 4.
Primers for qPCR were designed manually. Initially, the alignment was examined for conserved sequences of amino acids. Those sequences composed of amino acids possessing, overall, the fewest alternative codons were chosen, since this would minimize degeneracy in the primers. Since most of these sequences were partially or wholly repeated along the block, only those with a unique DNA sequence at the 3′ end of the primer were selected in order to minimize the likelihood of them producing multiple products. Several primers at each of several positions were designed, further minimizing their degeneracy where possible by exploiting G:T bonds and neutral G:A mismatches. Combinations of primer pairs were then tested on isolates. The two best-performing pairs were 3308f (5′ GGCGATMGVAGCAAACTSAC 3′) with 3463r (5′ STGTAVCKTTTNCCGTCCCA 3′) and 3341fb (5′ AHTGTRYBYTSATGGCBGGVGA 3′) with 3462r1 (5′ TGTAVCKTTTSCCGTCCCAG 3′). Product sizes were 194 and 162 bp, respectively.
Quantitative PCRs.
Real-time qPCRs were optimized. For primer 3308f, used with 3463r and 3462r1, each reaction mixture contained 1 U of GoTaq hot start polymerase (Promega, Madison, WI) in 1× colorless GoTaq Flexi buffer (no Mg in buffer), 0.9 μM each primer, 0.2 mM each dNTP, 1.25 mM MgCl2, 4% dimethylsulfoxide (DMSO), 0.5× EvaGreen (Biotium Inc., Hayward, CA), 1 to 8 μl DNA extract, and deionized water to a total volume of 25 μl. Cycling conditions were an initial denaturation at 95°C for 2 min, followed by 43 cycles of 94°C for 15 s, 51 to 54°C for 25 s (combined primer annealing and extension, with a higher temperature to reduce mispriming if required), and then a hold at 79°C for 3 s before signal acquisition at 515 to 530 nm (as for Sybr green 1). After amplification, a melt of the PCR products was performed. Ps. syringae Cit7's amplicon melted at 83.5°C, while those of the other species and strains had melting temperature (Tm) values of 81.5 to 87°C.
For primers 3341fb and 3462r1, each reaction mixture contained 1 U GoTaq hot start polymerase, 1× Flexi buffer (no Mg), 1.75 μM 3341fb, 1.3 μM 3462r1, 0.2 mM each dNTP, 1.5 mM MgCl2, 5% DMSO, 0.5× EvaGreen, 1 to 8 μl DNA extract, and deionized water to a total volume of 25 μl. Cycling conditions were an initial denaturation at 95°C for 2 min, followed by 43 cycles of 94°C for 15 s, annealing and extension at 53 to 55°C for 30 s, and then a hold at 80°C for 3 s before signal acquisition. Annealing and extension at 55°C gave equal coverage and produced fewer nonspecific products in environmental samples but delayed signal appearance by 1 to 2 cycles. Pseudomonas syringae Cit7's amplicon melted at 82°C, while the Tm of others ranged from 82 to 86.5°C.
Amplification was performed on a Bio-Rad DNAEngine fitted with a Chromo4 real-time PCR detector (Hercules, CA). Two to four replicate reactions were used. Products were confirmed by electrophoresis in 1.5% MetaPhor agarose gels (Cambrex, Rockland, MN) in 1× sodium borate buffer (20× stock contained 47 g liter−1 boric acid and 8 g liter−1 NaOH) (to give a pH of 8.2) at 200 V for up to 35 min, using ethidium bromide for visualization. A 50-bp ladder (G4521; Promega, Madison, WI) was used for sizing. Some products were sequenced to identify the principal ina allele in the amplicon.
Conveniently, there is a direct correlation between the number of ina genes in a sample and the number of INA bacterial cells; in the two Ps. syringae, four Pa. ananatis, and four X. campestris pathovars so far fully sequenced, there is one ina gene copy per genome. For qPCR standards, we used DNA extracted from Ps. syringae Cit7 cells. To automatically adjust for the different losses associated with each extraction method, we produced two standard series of DNA extracted from known numbers of cells (enumerated by dilution plating). Aliquots of 102 to 107 cells were added to MicroBead tubes and processed using either the standard or the high-efficiency methods. The former was used for qPCR of leaf washings and the latter for qPCR of snow samples.
Testing for inhibition was necessary with precipitation samples due to the omission of a purification step during their extraction and the use of up to 12 μl of each sample's DNA extract per reaction (in 50-μl PCRs). Inhibition was assessed by spiking replicate qPCRs with 5 to 10 copies of the ina gene. When inhibition was apparent in the spikes, smaller amounts of DNA were tested, in combination with spiking, to find the noninhibitory dose. One sample, a 5-cm snowfall over Laramie on 3 February 2012 that was part of a large system that tracked from the southwestern United States, contained high levels of inhibitory humics (after filtering, fine organic particles turned the filter chocolate brown). Thus, quantification of the ina gene was achieved using a presence/absence system with reactions initiated with only 0.2 μl DNA extract. Thirteen PCRs were run and examined for the presence of a product. Some products were clearly positive, while some produced faint amplicons of the correct size. These indistinct reactions were used to initiate a seminested PCR, replacing primer 3308f with 3341fb, and then gelled to confirm gene presence/absence. Two to eight negative controls were included in each PCR. Seven of the 13 PCRs were positive (four were sequenced), and all were assumed to have contained a single ina gene copy, except from one that sequencing revealed was a mix of two alleles. These data were then used to estimate ina gene copy numbers in this sample.
Nucleotide sequence accession numbers.
Sequences have been deposited in GenBank under accession numbers KC311253 to KC311272 for 16S rRNA sequences and KC311273 to KC311292 for ina genes; sequences for the same isolate differ by 20 digits (e.g., for Cit7 they are KC311253 and KC311273, respectively).
RESULTS
Phylogeny.
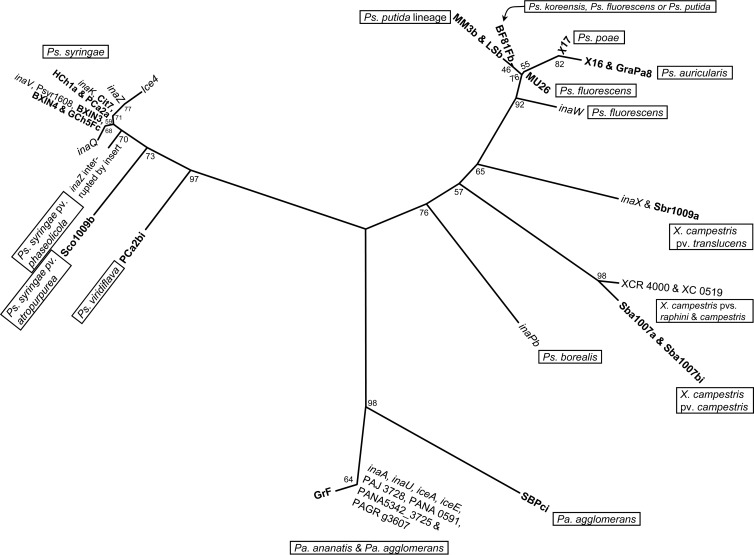
Partial sequencing of the ina gene (385 bp of block 4 of the core region) revealed 18 new alleles among the 20 isolates sequenced (Fig. 1 and Table 1). Alleles ranged from minor variations on established branches to new and deeply branching clades. To illustrate the former, all of the new alleles clustering around the inaZ-inaK-inaV-inaQ node of Ps. syringae were silent mutations, translating into amino acid sequences identical to either inaV or inaK (Fig. 1 and 2). In contrast, the allele in isolate Sco1009b, identified as Ps. syringae pv. atropurpurea from its 16S rRNA sequence, possessed only 82 to 84% similarity to all other Ps. syringae alleles (amino acid residue similarity was ∼94%). Interestingly, an INA strain of Ps. syringae pv. atropurpurea has recently been isolated from cloud water at the puy de Dôme in the Massif Central region of France (36, 37).
FIG 1.
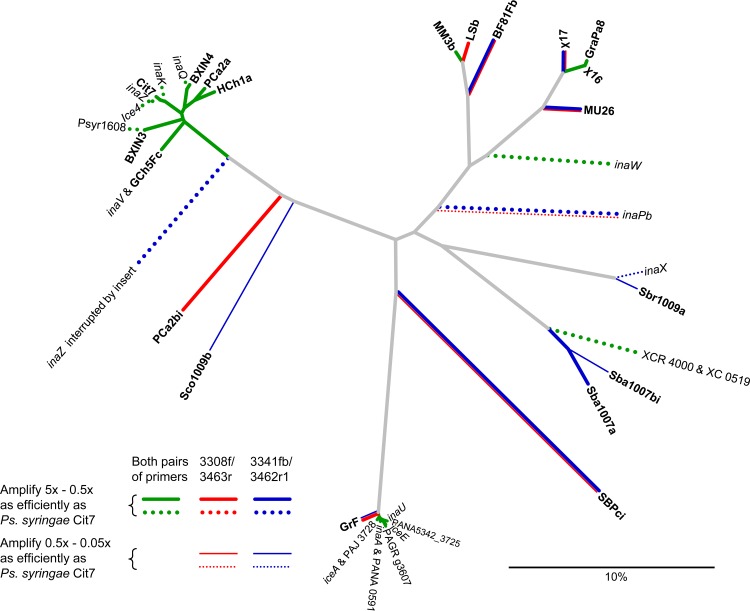
Unrooted phylogram illustrating sequence differences between ina alleles based on 385 nucleotides of the core of the gene. Isolates from this study are given in boldface. The tree was generated using neighbor-joining based on proportional distance (the scale bar shows percent dissimilarity). Also shown are the demonstrated (solid lines) and predicted (dashed lines) strengths of amplification using the two primer pairs.
FIG 2.
Phylogenetic relatedness among alleles using distances derived from amino acid sequences and the Jones-Taylor-Thornton model (60). Because only 128 amino acid residues of the gene were used, this phylogram provides only an approximate representation of evolutionary distances. Confidence levels are expressed as percent support for each branch placement and were derived using the interior branch test. GenBank accession numbers for reference sequences are given in the supplemental material.
Other distinct alleles included those in PCa2bi, identified as Ps. viridiflava from its 16S rRNA; SBPci, a Pa. agglomerans strain; Sba1007a and Sba1007bi, two strains of X. campestris pv. campestris; and the clades containing MM3b, LSb, BF81Fb, χ16, χ17, GraPa8, and MU26, a mix of pseudomonads from nonagricultural sources which included Ps. putida (Fig. 1 and 2). Pseudomonas viridiflava and Ps. putida have long been known to contain INA strains (61, 62), but their ina genes have not been sequenced; Castrillo et al. (63) amplified a nearly full-length product in Ps. putida using primers for inaZ but did not sequence it. While the Greek isolates χ16, χ17, and GraPa8 were provisionally identified as Ps. fluorescens, from their possession of cytochrome c oxidase and a lack of induction of a hypersensitive response in tobacco (D. G. Georgakopoulos, unpublished data), both their 16S rRNA and ina gene sequences suggest they belong to other pseudomonad species, species not previously known to be INA.
The most notable changes in topology, when comparing trees constructed using DNA to amino acid residues, are a much closer association of isolate Sco1009b with other Ps. syringae alleles and the tighter grouping of nonagricultural pseudomonads and inaW.
Primer testing on isolates.
Each of the two primer pairs amplified most alleles of the ina gene (Fig. 3; also see Fig. S1 and Table S2 in the supplemental material). Taken together, they should amplify all known alleles, including those in untested isolates based on the DNA sequences at their priming sites.
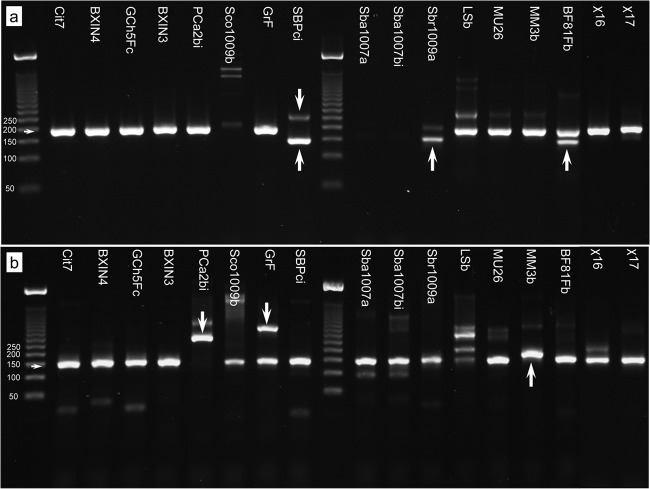
FIG 3.
Comparison of qPCR performance for primer pairs 3308f/3463r (a) and 3341fb/3462r1 (b). Horizontal arrows indicate the sizes of intended products, upward-pointing arrows show unintended but workable misprimes off the ina gene, and downward-pointing arrows show confounding misprimes off other genes.
Primer pair 3308f/3463r amplified most ina genes strongly and with good specificity (Fig. 3a; also see Fig. S1a in the supplemental material). The pair successfully amplified all alleles from Ps. syringae, Ps. viridiflava, and all pseudomonads from nonagricultural sources, although the latter with generally lower efficiency. For example, isolate χ17 amplified cleanly but after a significant delay, possibly caused by the obstruction of primer 3308f by a secondary fold incorporating the latter half of the priming site (modeled using mfold; The RNA Institute, College of Arts and Sciences, University of Albany [http://mfold.rna.albany.edu/?q=mfold] [64]).
Mispriming occurred with some isolates. It particularly delayed the amplification of the Pa. agglomerans isolate SPBci and the X. translucens isolate Sbr1009a (Fig. 3a; also see Fig. S1a in the supplemental material), causing 20- to 200-fold underestimations of their populations compared to Ps. syringae Cit7 (see Table S2). Amplification of Sbr1009a was improved by replacing 3463r with 3462r1, since it binds more strongly at the 3′ end. Mispriming in SBPci, Sbr1009a, and BF81FB occurred as a result of primer 3308f also binding at base 3356 (base numbering of inaZ), the result of both a mismatch at its intended target and atypical complementarity with two bases at the incorrect site.
There were also failures. The primers did not amplify the ina gene in the Ps. syringae pv. atropurpurea isolate Sco1009b or the new INA strains of X. campestris (isolates Sba1007a and Sba1007bi). Various modifications to 3308f to correct for the mismatches did not improve this, suggesting the problem lay with the reverse primer. However, no mismatches of consequence occur with either primer in the sequenced strains of X. campestris pv. raphini and X. campestris pv. campestris (GenBank accession numbers AE008922, AM920689, CP000050, and CP002789), suggesting this primer pair successfully amplifies some INA xanthomonad strains.
Primer pair 3341fb/3462r1 was often complementary to 3308f/3463r. That is, it was generally more successful with alleles problematic for 3308f/3463r but was also more prone to mispriming (Fig. 3b; also see Fig. S1b and Table S2 in the supplemental material). For example, it failed with Ps. viridiflava due to a mismatch with 3462r1 at its 3′ end but it amplified Ps. syringae pv. atropurpurea (isolate Sco1009b). Unlike the other primer pair, it also cleanly amplified the Pa. agglomerans isolate SPBci and also strongly amplified most alleles in the nonagricultural pseudomonads. Finally, it also amplified the new INA strains of X. campestris, although with some delay. Mispriming occurred with the Ps. putida isolate MM3b (Fig. 3b) but without ill effect, and it was caused by 3341fb binding at base 3317, the result of its sequence partly repeating at that position combined with stronger binding with two degenerate bases in the primer.
More serious were misprimes off genes other than the ina gene in several isolates. These could cause overestimation or false positives if not confirmed by gel electrophoresis. Two products, a 292-bp product in PCa2bi and a 373-bp product in GrF (Fig. 3b, arrows), were sequenced. Analysis of their sequences (see Table S2 in the supplemental material) indicated that mispriming resulted from regions of amino acid sequence similarity to both primers. In both cases, almost complete complementarity occurred along the first 9 to 11 bases back from their 3′ ends. To attempt to prevent this, the combined annealing/extension temperature was raised or its time reduced to starve these longer products of time for formation, but both modifications also caused reduced amplification efficiency of other alleles. Isolate LSb also suffered from mispriming, but the reason for it was not apparent.
With both primer pairs the response to ina gene copy number was linear over six orders of magnitude (up to 106 added). The efficiency of amplification was 84 to 86% for both. Sensitivity was affected by the choice of reverse primer and the propensity of primer 3341fb to misprime. In tests using Ps. syringae Cit7 standards, both 3308f/3462r1 and 3341fb/3462r1 detected 1 to 3 gene copies, with the uncertainty being due to binomial sampling variation. 3308f/3463r sensitivity was often limited to 5 to 10 gene copies (see Fig. S2 in the supplemental material) due to the production of primer-dimers.
qPCR of INA bacteria in environmental samples.
All blank extractions and PCR negative-control reactions were negative. Table 2 summarizes results of the qPCR of ina genes in leaf washings and precipitation using the two primer pairs. For crops, INA bacterial populations on dicots ranged from ∼105 to ∼107 g−1 fresh weight of vegetation, while on monocots the range was roughly an order of magnitude higher, reaching ∼108 g−1 fresh weight on wheat and barley. Sequencing of the qPCR amplicons from wheat and barley showed that Ps. syringae isolate BXIN4 predominated on both. This allele is a variant of Ps. syringae inaQ and was the most frequently occurring in the region. It was also found in isolates recovered from bean lesions and leaf washings of sugar beet and barley at the irrigated field, as well as dominating the qPCR products of leaf washings of wheat, corn, and ley field vegetation from an organic farm in Colorado (unpublished data from the same sites used by Garcia et al. [35]). Much smaller INA bacterial populations were found on naturally occuring grasses than on irrigated grasses, and none at all were detected on Quaking Aspen (Populus tremuloides) or Lodgepole Pine (Pinus contorta). An unexpectedly high population occurred on mountain mahogany (Cercocarpus montana), a shrub that possesses small, thorn-protected, verdant leaves. While there was generally good correspondence between ina gene counts obtained with both 3308f and 3341fb primers on the irrigated crops, in naturally occurring vegetation, primer 3341fb was more prone to mispriming.
TABLE 2.
Quantification of ina genes in leaf washings and precipitation using two primer pairsb
| Plant or date of precipitation (mo/day/yr) |
ina gene copies g−1 (fresh wt) vegetation or precipitation |
Closest isolate or allele of any sequenced qPCR amplicons (% similarity) |
||
|---|---|---|---|---|
| 3308f/3462r1 or 3463r | 3341fb/3462r1 | 3308f/3462r1 or 3463r | 3341fb/3462r1 | |
| Irrigated crops | ||||
| Alfalfa | 1.1 × 106 | 2.5 × 106 | ||
| Corn | 1.0 × 105 | 5.9 × 105 | ||
| Beans | 3.3 × 105 | 7.6 × 105 | ||
| Potato | 9.6 × 105 | MP | ||
| Sugar beet | 7.2 × 106 | 4.7 × 106 | ||
| Barley | 7.6 × 107 | 1.4 × 108 | Ps. syringae BXIN4 (100) | Ps. syringae BXIN4 (100) and X. campestris inaX (95) |
| Smooth brome grass | 2.1 × 106 | 2.9 × 106 | ||
| Wheat | 1.0 × 108 | 1.8 × 108 | Ps. syringae BXIN4 (100) | Ps. syringae BXIN4 (100) |
| Naturally occurring vegetation | ||||
| Crested wheat grass | 4.0 × 103 | 2.1 × 104 | ||
| Smooth brome grass | 2.7 × 104 | MP | ||
| Western wheatgrass | 3.8 × 104 | 1.4 × 105 | ||
| Common timothy | 7.6 × 102 | MP | ||
| Redtop bentgrass | 3.1 × 103 | ND | ||
| Aspen | ND | ND | ||
| Mountain mahogany | 9.2 × 103a | 6.4 × 105 | ||
| Lodgepole pine | ND | ND | ||
| Sagebrush | 8.4 × 102a | MP | ||
| Hail | ||||
| 6/10/2010 C | 1.2 × 100 | Ps. syringae BXIN4 (100) | ||
| Snow | ||||
| 1/25/11 M | 8 × 10−2 | Ps. syringae BXIN4 (100) | ||
| 1/25/11 C | 1 × 10−1 | Pa. agglomerans SBPci (96) | ||
| 1/31/11 M | ND | |||
| 1/31/11 C | 2 × 10−2 | Not sequenced | ||
| 2/5/11 and 2/6/11 C | 2 × 10−2 | Ps. syringae BXIN4 (99.5) | ||
| 2/24/11 and 2/25/11 C | ND | |||
| 3/8/11 M | ND | |||
| 3/12/11 and 3/13/11 C | ND | |||
| 3/28/11 and 3/29/11 C | 1 × 10−2 | Ps. syringae BXIN4 (100) | ||
| 1/22/12 C | ND | |||
| 2/2/12 and 2/3/12 M | 3 × 10−1 | Ps. poae χ17 (100) | ||
| 2/2/12 and 2/3/12 C | 4.5 × 10−1 | Ps. syringae BXIN4 (100), Ps. syringae Cit7 (99), mix of Ps. syringae BXIN4 and Cit7 | Ps. syringae BXIN4 (99) | |
| 2/12/12 and 12/13/12 M | ND | |||
| 2/28/12 C site 1 | 9 × 10−3 | Ps. syringae inaV (98) | ||
| 2/28/12 C site 2 | ND | |||
| 4/15/12 and 4/16/12 M | 4 × 10−3 | X. campestris XCR 4000 (98.5) | ||
Underestimation due to mispriming.
C and M indicate samples collected in Laramie city and in the Laramie Mountains, respectively. ND, no ina gene product; MP, misprime preventing quantification.
Only primer pairs 3308f/3462r1 and 3308f/3463r were used with precipitation (Table 2), since initial tests showed they had higher sensitivity and rarely misprimed with these samples. The highest concentration of ina genes, 1.2 g−1 melt, occurred in hail that fell at the start of an evening thunderstorm. Sequencing of the qPCR product showed it was principally the common BXIN4 allele. ina genes were detected in 9/16 snow samples at concentrations ranging from <0.01 to 0.45 ina genes g−1 snowmelt. Sequencing of these qPCR products revealed a wide range of INA bacteria: Ps. syringae allele BXIN4 predominated in about half, while other principal sequences comprised two other alleles from the Ps. syringae clade, one Pa. agglomerans allele, one X. campestris pv. campestris allele, and one from the Ps. fluorescens/putida group identical to an isolate obtained from snow in Greece. Other partially readable sequences in the snow samples included alleles related to Cit7, inaV, SBPci, inaA-inaE-inaU, and inaX.
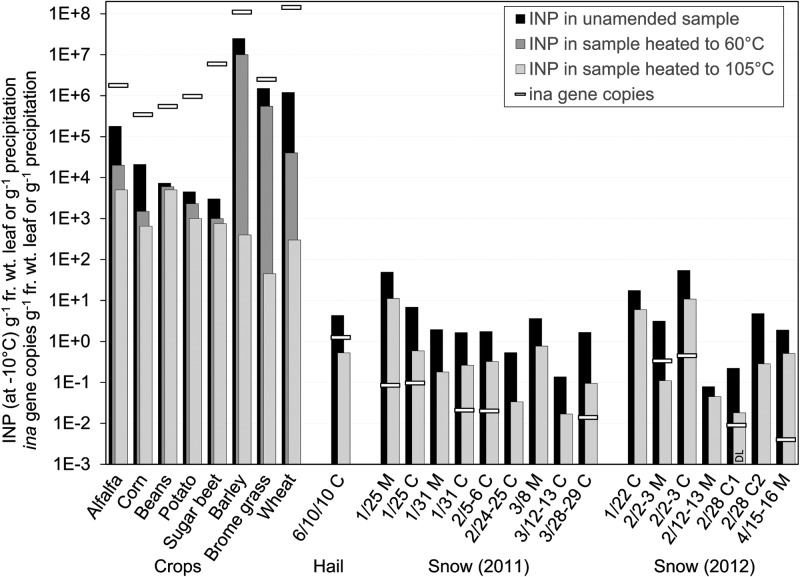
Comparisons between ina gene copy numbers and numbers of INP in the same samples are summarized in Fig. 4. In leaf washings, the ratio of ina gene copies to INP active at −10°C ranged from 2 to 2,000 (median, ∼50). These values are comparable, considering the lower test temperature, to ratios of 15 to 20,000 (median, 500) found for INA Ps. syringae to INP active at −5°C on plants grown in 40% relative humidity (RH) (46) and to ratios of 100 to 12,500 (median, ∼1,000) for INA bacteria to INP active at −3°C to −4°C on field-grown oat leaves (47). Heat treatment to denature the biological fraction in leaf washings reduced the INP active at −10°C by >75% in all plant species other than sugar beet. In the three monocots, heating to 105°C essentially eliminated all ice nucleation activity; even at −18°C the INP number was reduced by >95% compared to that of untreated samples (see Fig. S3 in the supplemental material). This suggests that the prodigious INA bacterial populations in the three monocots were the principal sources of these INP. The resistance of 30 to 40% of the INP to deactivation by 60°C heat treatment in barley and smooth brome grass may reflect the abundance of INA xanthomonads in these populations; as noted above, an INA xanthomonad isolated from smooth brome was unusually resistant to heat inactivation.
FIG 4.
Comparison of numbers of ice-nucleating particles (measured at −10°C) and ina gene copies in leaf washings and, when detected, in precipitation samples (full spectra are given in Fig. S3 and S4 in the supplemental material). C and M indicate samples collected in Laramie city and in the Laramie Mountains, respectively. DL indicates the lower detection limit in a sample in which no INP were detected after heat treatment. fr. wt., fresh weight.
In contrast, the ratio of ina gene copies to INP active at −10°C in precipitation samples was <1 in all samples. The summer hail sample possessed the highest ratio of ∼0.3. For snow, they ranged from below the detection limit to 0.1 ina genes per INP active at −10°C, with a mean ratio of 0.025 in samples in which the gene was detected. It is probable that some of these are underestimates given the inability of primer pair 3308f/3462r1 or 3308f/3463r to detect some alleles and their lower efficiency with others. However, the consistency of the low levels suggests the INA bacteria were generally infrequent. Even so, biological INP were still the predominant source of INP in 15/16 snow samples (to the limit of measures at −12°C; see Fig. S4 in the supplemental material). On average, ∼85% of INP active at −10°C were destroyed by heating. The one snow sample that was atypical (see Fig. S4, 2/12&13/12 M) stained the filter a light tan color, had the lowest INP concentration overall, and possessed a log-linear relationship between INP and temperature indicative of minerals (19).
DISCUSSION
Taken together, the two primer pairs developed in this work amplified all tested alleles of the ina gene, the gene that codes for the ice-nucleating protein in INA bacteria. They should also amplify all published alleles for which we lacked representative isolates based on the DNA sequences at their priming sites. Previously, due to the high diversity of the ina gene, primers have been designed to amplify only single alleles or clades, such as inaZ (56, 63, 65), inaA (56, 65), inaW (55, 56, 58, 63), or inaX (56). Primers targeting the core and able to amplify multiple alleles have been developed (C. Guilbaud and C. Morris, personal communication) but have not to our knowledge been published.
Both primer pairs have potentially high sensitivity. Primer combination 3308f/3462r1 was able to detect spikes with a mean of 4 ina gene copies added to atmospheric aerosol samples (35), and both 3308f/3462r1 and 3308f/3463r were able to detect spikes of 6 ina gene copies in snow samples (the 95% confidence ranges for these spikes were 0 to 8 and 1 to 10 copies, respectively). The presence/absence method used to estimate ina gene number in the humic snow sample of 2 to 3 February 2012, in which each PCR amplified the DNA extracted from the equivalent of only ∼5 g snow, suggests that single gene copies can be amplified.
However, neither of the primer pairs was able to amplify all alleles individually, and they needed to be used in combination to obtain complete coverage. Figure 1 summarizes their strengths and weakness and illustrates this complementarity. Apart from the mismatches to primers, the impedance of primer annealing by DNA secondary structure also seems to have contributed to inefficient amplification of some isolates (e.g., Sbr1009a, LSb, and χ17). Secondary structure is promoted throughout the core of the ina gene, because the first two amino acids of each octapeptide are typically alanine and glycine, with codons GCN and GGN, respectively, creating G/C-rich sequences every 24 bases that often form strongly bound stems (58). DMSO was a necessary addition to the PCR mixes (66), especially for efficient amplification of inaX-related alleles using 3341fb and of inaW-related alleles using 3308f.
Since each primer set amplifies a subset of the total diversity of ina alleles, each will underestimate the total population of INA bacteria. This will be significant if alleles that either primer pair fails to amplify comprise a significant proportion of the population. Although they possess complementarity, the overlap in their coverage is too great to allow summing of both results to obtain a total. Therefore, the more accurate estimate will be provided by the primer set that, in the absence of any mispriming, gives the higher value. This was typically 3341fb/3462r1, by a factor of ∼2 to 5, due to its broader coverage (Table 2), as was demonstrated by sequencing qPCR amplicons from barley. When using 3341fb/3462r1, the qPCR estimate for barley was 1.4 × 108 ina gene copies g−1 fresh weight. Sequencing of the qPCR product revealed two approximately codominant alleles, one identical to isolate BXIN4 and the other related to inaX in X. campestris pv. translucens. In contrast, when primer pair 3308f/3463r was used, the estimated ina gene copy number was halved and the sequence of the qPCR amplicon was dominated solely by the BXIN4 sequence. Although providing wider coverage, primer 3341fb was also more prone to mispriming at levels high enough to nullify the test (Table 2 and Fig. 3b). In contrast, 3308f often produced more conservative estimates but had better specificity (35).
Both sets amplified as-yet undescribed ina gene alleles (Table 2). Sequencing of qPCR products from leaf washings of other sampled crops (35 and Hill, unpublished) detected numerous ina alleles from the Pantoea that were not found in isolates screened from the same leaf washings, suggesting these strains were not readily cultured. A higher failure rate would also be expected for new ina alleles. Thus, it is promising that all ina genes in the diverse collection of INA bacteria assembled during this study could be amplified by one or both primer pairs.
For the cereals, the qPCR estimates corresponded closely to populations of viable INA Ps. syringae found on barley in the neighboring state, Montana (67). On these fully grown plants, peak populations of INA Ps. syringae across all varieties were similarly high, at 0.4 × 108 to 1 × 108 CFU per gram fresh weight of leaf (converted from their values of 0.8 × 107 to 4 × 107 CFU per leaf by using 0.2 g per leaf and adjusting for relative abundance of INA Ps. syringae in the total population). Unexpectedly, viable counts of INA bacteria in our barley leaf washings equated to only 1.2 × 106 g−1 leaf, only a small percentage of the number found using qPCR, suggesting lower viability on our late-season crop. Smaller populations of INA bacteria were found on cereals by Lindow et al. (7), whose estimates ranged from none detected to <103 CFU g−1 fresh weight, while Lindemann et al. (33) found modest levels of ∼105 INA bacteria g−1 on wheat in mid-June. These smaller populations may have been due simply to the samples being taken earlier in the growing season (67). Among noncereal crops, the qPCR values found in this study, and in Garcia et al. (35), were consistently 10 to 1,000 times higher than values found using culturing (7, 33). However, it is possible that, once again, seasonal and local environmental influences accounted for many of the differences, in addition to likely differences in viability and high spatial variability in populations. Hirano et al. (68) found INA bacterial populations ranged from 4 × 103 to 2 × 106 on individually tested corn leaves. Lack of detection of ina genes on Quaking Aspen (Populus tremuloides) and Lodgepole Pine (Pinus contorta) matches the findings of Lindow et al. (7), who recorded ∼103 INA bacteria g−1 fresh weight of leaf on Ps. tremuloides and none detected on any of three pine species tested.
We detected ina genes in 9/16 snow samples, quite similar to the proportion of 3/9 fresh snow samples from which Ps. syringae was recovered using culturing (15). In contrast, while we detected only <0.01 to 0.45 ina genes g−1 snowmelt, Morris et al. (15) recovered 0.15 to 130 Ps. syringae CFU g−1 (all strains tested from fresh snow were INA). Although the snow samples contained diverse ina genes, most were from the Ps. syringae clade, presumably a reflection of their prevalence on the region's agricultural crops.
INA bacteria, or at least INA bacteria with residual DNA, contributed a minor proportion of INP active at −10°C in precipitation, even though most INP, from their susceptibility to heat, had a biological source. This relative scarcity of the INA bacteria among a predominantly biological source of warm-temperature INP mirrors the characteristics of total aerosols sampled above crops in September and October in Colorado (35). An exception was the June thunderstorm hail, which contained ∼1.2 ina genes g−1 hail and ∼0.5 ina genes per hail stone, indicating a substantial potential contribution to the warmest INP. On a per-weight basis, this was 10 times the abundance of INA Ps. syringae found in ice and rain collected 180 to 2,500 m above a wheat field in Montana (8). As in Sands et al. (8), Ps. syringae was also the predominant allele in the hail. Vali (69) concluded that storm INP were of local surface origin, and significant epiphytic populations of INA bacteria would have been present on the green rangeland vegetation that surrounded Laramie at the time.
The numbers of INP active at −10°C in snow were up to eight times higher than those found in other studies in adjoining states (17, 39). This is likely due to the larger number of samples surveyed here, as well as the relatively low measurable cap on values in the method used by Christner et al. (39). Reductions in INP observed after heating broadly match those of Christner et al. (39), although they were not as pronounced; they observed complete loss of ice nucleation activity, to −9°C, in all 10 snow samples from Montana. The concentration of INP active at −10°C in the hail sample was within the range of ∼3 to 100 g−1 found in northeast Colorado by Vali (69).
Development of universal ina gene primers for qPCR of the INA bacteria requires substantially more ina genes to be sequenced to reveal its true phylogenetic diversity. In this study, we found 18 new ina alleles, several of which are highly divergent from known clades. Direct sequencing of qPCR products also indicated significantly more diversity than is currently represented by isolates, particularly among the Pantoea. The sensitivity and coverage of both qPCR tests could also be significantly improved, and biases caused by differences in allele amplification efficiencies avoided, if the primers are combined with a probe and used in droplet digital PCR (70).
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by NSF grants 0841542 (T.H. and G.F.) and 0841602 (P.D. and D.G.).
We especially thank Nathan Scarlett (Rathbone Wine Group) as well as Sonja Needs and Nicky Cooley (University of Melbourne) for selection of diseased grapevine canes and sampling in Australia and valuable advice and assistance with isolations and screening. We are grateful to Jamie Freeman (University of East London) for collection of vegetation from peat bogs in Scotland and Finland. We also acknowledge Helen Ahern's approach and persistence in designing and testing earlier core region ina gene primers. Revisions suggested by two anonymous reviewers greatly improved the manuscript.
Footnotes
Published ahead of print 6 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02967-13.
REFERENCES
- 1.Vali G. 1968. Ice nucleation relevant to hail formation. Stormy Weather Group, Scientific Report MW-58. McGill University, Montreal, Canada [Google Scholar]
- 2.Schnell RC, Vali G. 1972. Atmospheric ice nuclei from decomposing vegetation. Nature 236:163–165 [Google Scholar]
- 3.Maki LR, Galyan EL, Chang-Chien M-M, Caldwell DR. 1974. Ice nucleation induced by Pseudomonas syringae. Appl. Microbiol. 28:456–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnell RC, Vali G. 1976. Biogenic ice nuclei: Part I. Terrestrial and marine sources. J. Atmos. Sci. 33:1554–1564 [Google Scholar]
- 5.Vali G, Christensen M, Fresh RW, Galyan EL, Maki LR, Schnell RC. 1976. Biogenic ice nuclei. II. Bacterial sources. J. Atmos. Sci. 33:1565–1570 [Google Scholar]
- 6.Maki LR, Willoughby KJ. 1978. Bacteria as biogenic sources of freezing nuclei. J. Appl. Meteorol. 17:1049–1053 [Google Scholar]
- 7.Lindow SE, Arny C, Upper CD. 1978. Distribution of ice nucleation-active bacteria on plants in nature. Appl. Environ. Microbiol. 36:831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sands DC, Langhans VE, Scharen AL, de Smet G. 1982. The association between bacteria and rain and possible resultant meteorological implications. J. Hungarian Meteorol. Serv. 86:148–152 [Google Scholar]
- 9.Hoose C, Kristjánsson JE, Burrows SM. 2010. How important is biological ice nucleation in clouds on a global scale? Environ. Res. Lett. 5:024009. 10.1088/1748-9326/5/2/024009 [DOI] [Google Scholar]
- 10.Upper CD, Vali G. 1995. The discovery of bacterial ice nucleation and its role in the injury of plants by frost, p 29–39 In Lee RE, Warren GJ, Gusta LV. (ed), Biological ice nucleation and its applications. APS Press, St. Paul, MN [Google Scholar]
- 11.Szyrmer W, Zawadzki I. 1997. Biogenic and anthropogenic sources of ice-forming nuclei: a review. Bull. Am. Meteorol. Soc. 78:209–228 [Google Scholar]
- 12.Morris CE, Georgakopoulos DG, Sands DC. 2004. Ice nucleation active bacteria and their potential role in precipitation. J. Phys. IV France 121:87–103. 10.1051/jp4:2004121004 [DOI] [Google Scholar]
- 13.Möhler O, DeMott PJ, Vali G, Levin Z. 2007. Microbiology and atmospheric processes: the role of biological particles in cloud physics. Biogeosciences 4:1059–1071. 10.5194/bg-4-1059-2007 [DOI] [Google Scholar]
- 14.Möhler OM, Georgakopoulos DG, Morris CE, Benz S, Ebert V, Hunsmann S, Saathoff H, Schnaiter M, Wagner R. 2008. Heterogeneous ice nucleation activity of bacteria: new laboratory experiments at simulated cloud conditions. Biogeosciences 5:1425–1435. 10.5194/bg-5-1425-2008 [DOI] [Google Scholar]
- 15.Morris CE, Sands DC, Vinatzer BA, Glaux C, Guilbaud C, Buffière A, Yan S, Dominguez H, Thompson BM. 2008. The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J. 2:321–334. 10.1038/ismej.2007.113 [DOI] [PubMed] [Google Scholar]
- 16.Phillips VTJ, DeMott PJ, Andronache C. 2008. An empirical parameterization of heterogeneous ice nucleation for multiple chemical species of aerosol. J. Atmos. Sci. 65:2757–2783. 10.1175/2007JAS2546.1 [DOI] [Google Scholar]
- 17.Bowers RM, McLetchie S, Knight R, Fierer N. 2011. Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J. 5:601–612. 10.1038/ismej.2010.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delort A-M, Vaïtilingom M, Amato P, Sancelme M, Parazols M, Mailhot G, Laj P, Deguillaume L. 2010. A short overview of the microbial population in clouds: potential roles in atmospheric chemistry and nucleation processes. Atmos. Res. 98:249–260. 10.1016/j.atmosres.2010.07.004 [DOI] [Google Scholar]
- 19.Conen F, Morris CE, Leifeld J, Yakutin MV, Alewell C. 2011. Biological residues define the ice nucleation properties of soil dust. Atmos. Chem. Phys. 11:9643–9648. 10.5194/acp-11-9643-2011 [DOI] [Google Scholar]
- 20.Després VR, Huffman JA, Burrows SM, Hoose C, Safatov AS, Buryak G, Fröhlich-Nowoisky J, Elbert W, Andreae MO, Pöschl U, Jaenicke R. 2012. Primary biological aerosol particles in the atmosphere: a review. Tellus B 64:015598. 10.3402/tellusb.v64i0.15598 [DOI] [Google Scholar]
- 21.Christner BC. 2012. Cloudy with a chance of microbes. Microbe 7:69–75 [Google Scholar]
- 22.Fröhlich-Nowoisky J, Burrows SM, Xie Z, Engling G, Solomon PA, Fraser MP, Mayol-Bracero OL, Artaxo P, Begerow D, Conrad R, Andreae MO, Després VR, Pöschl U. 2012. Biogeography in the air: fungal diversity over land and oceans. Biogeosciences 9:1125–1136. 10.5194/bg-9-1125-2012 [DOI] [Google Scholar]
- 23.DeMott PJ, Prenni AJ. 2010. New directions: need for defining the numbers and sources of biological aerosols acting as ice nuclei. Atmos. Environ. 44:1944–1945. 10.1016/j.atmosenv.2010.02.032 [DOI] [Google Scholar]
- 24.Hallett J, Mossop SC. 1974. Production of secondary ice particles during the riming process. Nature 249:26–28. 10.1038/249026a0 [DOI] [Google Scholar]
- 25.Mossop SC, Hallett J. 1974. Ice crystal concentration in cumulus clouds: influence of the drop spectrum. Science 186:632–634. 10.1126/science.186.4164.632 [DOI] [PubMed] [Google Scholar]
- 26.Mossop SC. 1976. Production of secondary ice particles during the growth of graupel by riming. Quart. J. Roy. Meteorol. Soc. 102:45–47. 10.1002/qj.49710243104 [DOI] [Google Scholar]
- 27.Hirano SS, Upper CD. 1995. Ecology of ice nucleation-active bacteria, p 41–61 In Lee RE, Warren GJ, Gusta LV. (ed), Biological ice nucleation and its applications. APS Press, St. Paul, MN [Google Scholar]
- 28.Kim HK, Orser C, Lindow SE, Sands DC. 1987. Xanthomonas campestris pv. translucens strains active in ice nucleation. Plant Dis. 71:994–997. 10.1094/PD-71-0994 [DOI] [Google Scholar]
- 29.Lindow SE, Lahue E, Govindarajan AG, Panopoulos NJ, Gies D. 1989. Localization of ice nucleation activity and the iceC gene product in Pseudomonas syringae and Escherichia coli. Mol. Plant Microbe Interact. 2:262–272. 10.1094/MPMI-2-262 [DOI] [PubMed] [Google Scholar]
- 30.Morris CE, Sands DC, Vanneste JL, Montarry J, Oakley B, Guilbaud C, Glaux C. 2010. Inferring the evolutionary history of the plant pathogen Pseudomonas syringae from its biogeography in headwaters of rivers in North America, Europe, and New Zealand. mBio 1:e00107-10. 10.1128/mBio.00107-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirano SS, Upper CD. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae–a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624–653. 10.1128/MMBR.64.3.624-653.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamilton WD, Lenton TM. 1998. Spora and Gaia: how microbes fly with their clouds. Ethol. Ecol. Evol. 10:1–16. 10.1080/08927014.1998.9522867 [DOI] [Google Scholar]
- 33.Lindemann J, Constantinidou HA, Barchet WR, Upper CD. 1982. Plants as sources of airborne bacteria, including ice nucleation-active bacteria. Appl. Environ. Microbiol. 44:1059–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Constantinidou HA, Hirano SS, Baker LS, Upper CD. 1990. Atmospheric dispersal of ice nucleation-active bacteria: the role of rain. Phytopathology 80:934–937. 10.1094/Phyto-80-934 [DOI] [Google Scholar]
- 35.Garcia E, Hill TCJ, Prenni AJ, DeMott PJ, Franc GD, Kreidenweis SM. 2012. Biogenic ice nuclei in boundary layer air over two U.S. High Plains agricultural regions. J. Geophys. Res. 117:D18209. 10.1029/2012JD018343 [DOI] [Google Scholar]
- 36.Amato P, Parazols M, Sancelme M, Laj P, Mailhot G, Delort A-M 2007. Microorganisms isolated from the water phase of tropospheric clouds at the Puy de Dôme: major groups and growth abilities at low temperatures. FEMS Microbiol. Ecol. 59:242–254. 10.1111/j.1574-6941.2006.00199.x [DOI] [PubMed] [Google Scholar]
- 37.Attard E, Yang H, Delort A-M, Amato P, Pöschl U, Glaux C, Koop T, Morris CE. 2012. Effects of atmospheric conditions on ice nucleation activity of Pseudomonas. Atmos. Chem. Phys. 12:10667–10677. 10.5194/acp-12-10667-2012 [DOI] [Google Scholar]
- 38.Christner BC, Morris CE, Foreman CM, Cai R, Sands DC. 2008. Ubiquity of biological ice nucleators in snowfall. Science 319:1214. 10.1126/science.1149757 [DOI] [PubMed] [Google Scholar]
- 39.Christner BC, Cai R, Morris CE, McCarter KS, Foreman CM, Skidmore ML, Montross SN, Sands DC. 2008. Geographic, seasonal, and precipitation chemistry influence on the abundance and activity of biological ice nucleators in rain and snow. Proc. Natl. Acad. Sci. USA 105:18854–18859. 10.1073/pnas.0809816105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prenni AJ, Tobo Y, Garcia E, DeMott PJ, McCluskey C, Kreidenweis SM, Prenni J, Huffman A, Pöschl U, Pöhlker C. 2013. The impact of rain on ice nuclei populations. Geophys. Res. Lett. 40 10.1029/2012GL053953 [DOI] [Google Scholar]
- 41.Huffman JA, Pöhlker C, Mason R, Robinson NH, Prenni A, Gochis D, Day D, DeMott P, Després V, Fröhlich J, Gallagher M, Garcia E, Harris E, Müller I, Schmer B, Sinha B, Sun A, Tobo Y, Andreae MO, Kreidenweis S, Smith J, Guenther A, Jimenez J, Bertram A, Pöschl U. 2013. High concentrations of biological aerosol particles and ice nuclei during and after rain. Atmos. Chem. Phys. Discuss. 13:1767–1793. 10.5194/acpd-13-1767-2013 [DOI] [Google Scholar]
- 42.Burrows SM, Elbert W, Lawrence MG, Pöschl U. 2009. Bacteria in the global atmosphere. Part 1. Review and synthesis of literature data for different ecosystems. Atmos. Chem. Phys. 9:9263–9280. 10.5194/acp-9-9263-2009 [DOI] [Google Scholar]
- 43.Wilson M, Lindow SE. 1992. Relationship of total viable and culturable cells in epiphytic populations of Pseudomonas syringae. Appl. Environ. Microbiol. 58:3908–3913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heidelberg J, Shahamat M, Levin M, Rahman I, Stelma G, Grim C, Colwell R. 1997. Effect of aerosolization on culturability and viability of gram-negative bacteria. Appl. Environ. Microbiol. 63:3585–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Georgakopoulos DG, Després V, Fröhlich-Nowoisky J, Psenner R, Ariya PA, Pósfai M, Ahern HE, Moffett BF, Hill TCJ. 2009. Biological, physical and chemical characterization of aerosol particles. Biogeosciences 6:721–737. 10.5194/bg-6-721-2009 [DOI] [Google Scholar]
- 46.O'Brien RD, Lindow SE. 1988. Effect of plant species and environmental conditions on ice nucleation activity of Pseudomonas syringae on leaves. Appl. Environ. Microbiol. 54:2281–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirano SS, Baker LS, Upper CD. 1985. Ice nucleation temperature of individual leaves in relation to population sizes of ice nucleation active bacteria and frost injury. Plant Physiol. 77:259–265. 10.1104/pp.77.2.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Govindarajan AG, Lindow SE. 1988. Size of bacterial ice-nucleation sites measured in situ by radiation inactivation analysis. Proc. Natl. Acad. Sci. USA 85:1334–1338. 10.1073/pnas.85.5.1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warren GJ. 1995. Identification and analysis of ina genes and proteins, p 85–99 In Lee RE, Warren GJ, Gusta LV. (ed), Biological ice nucleation and its applications. APS Press, St. Paul, MN [Google Scholar]
- 50.Nemecek-Marshall M, Laduca R, Fall R. 1993. High level expression of ice nuclei in a Pseudomonas syringae strain is induced by nutrient limitation and low temperature. J. Bacteriol. 175:4062–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruggles JA, Nemecek-Marshall M, Fall R. 1993. Kinetics of appearance and disappearance of classes of bacterial ice nuclei support an aggregation model for ice nucleus assembly. J. Bacteriol. 175:7216–7221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turner MA, Arellano F, Kozloff LM. 1990. Three separate classes of bacterial ice nucleation structures. J. Bacteriol. 172:2521–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vali G. 1971. Quantitative evaluation of experimental results on the heterogeneous freezing nucleation of supercooled liquids. J. Atmos. Sci. 28:402–409 [Google Scholar]
- 54.Warren G, Corotto L, Wolber P. 1986. Conserved repeats in diverged ice nucleation structural genes from two species of Pseudomonas. Nucleic Acids Res. 14:8047–8060. 10.1093/nar/14.20.8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahern HE, Walsh KA, Hill TCJ, Moffett BF. 2007. Fluorescent pseudomonads isolated from Hebridean cloud and rain water produce biosurfactants but do not cause ice nucleation. Biogeosciences 4:115–124. 10.5194/bg-4-115-2007 [DOI] [Google Scholar]
- 56.Nejad P, Ramstedt M, Granhall U, Roos S, McIvor I. 2006. Biochemical characterization and identification of ice-nucleation-active (INA) willow pathogens by means of BIOLOG MicroPlate, INA gene primers and PCR-based 16S rRNA-gene analyses. J. Plant Dis. Protect. 113:97–106 [Google Scholar]
- 57.Warren G, Corotto L. 1989. The consensus sequence of ice nucleation proteins from Erwinia herbicola, Pseudomonas fluorescens and Pseudomonas syringae. Gene 85:239–242. 10.1016/0378-1119(89)90488-5 [DOI] [PubMed] [Google Scholar]
- 58.Ahern HE. 2007. Ph.D. thesis. Cloud borne bacteria: community composition and potential impact on atmospheric nucleation. University of East London, London, United Kingdom [Google Scholar]
- 59.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comp. Appl. Biosci. 8:275–282 [DOI] [PubMed] [Google Scholar]
- 61.Paulin J-P, Luisetti J. 1978. Ice nucleation activity among phytopathogenic bacteria, p 725–731 In Proceedings of the 4th International Conference on Plant Pathological Bacteria. Station de Pathologie Végétale et Phytobactériologie, Institute National Recherche Agronomique, Beaucouzé, France [Google Scholar]
- 62.Lee MR, Lee RE, Strong-Gunderson JM. 1991. Isolation of ice nucleation active bacteria from a freeze-tolerant frog: identification of Pseudomonas putida strains active in ice nucleation. In 5th International Conference on Biological Ice Nucleation. University of Wisconsin, Madison, WI [Google Scholar]
- 63.Castrillo LA, Lee RE, Jr, Lee MR, Rutherford ST. 2000. Identification of ice-nucleating active Pseudomonas fluorescens strains for biological control of overwintering Colorado potato beetles (Coleoptera: Chrysomelidae). J. Econ. Entomol. 93:226–233. 10.1603/0022-0493-93.2.226 [DOI] [PubMed] [Google Scholar]
- 64.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dimou D, Kallimanis A, Perisynakis A, Chatziloukas E, Drainas C. 2006. A contribution to the analysis of the genetic diversity of bacterial ice nucleation genes in Hellenic niches. HSBMB Newsl. 53:66–67 [Google Scholar]
- 66.Frackman S, Kobs G, Simpson D, Storts D. 1998. Betaine and DMSO: enhancing agents for PCR. Promega notes 65:27. Promega, Madison, WI: http://www.promega.com/resources/articles/pubhub/promega-notes-1998/betaine-and-dmso-enhancing-agents-for-pcr/ [Google Scholar]
- 67.Georgakopoulos DG, Sands DC. 1992. Epiphytic populations of Pseudomonas syringae on barley. Can. J. Microbiol. 38:111–114. 10.1139/m92-018 [DOI] [PubMed] [Google Scholar]
- 68.Hirano SS, Nordheim EV, Arny DC, Upper CD. 1982. Lognormal distribution of epiphytic bacterial populations on leaf surfaces. Appl. Environ. Microbiol. 44:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vali G. 1978. Freezing nucleus content of hail and rain in NE Colorado. Am. Meteorol. Soc. Monogr. 38:93–105 [Google Scholar]
- 70.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang S, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW. 2011. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83:8604–8610. 10.1021/ac202028g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lindow SE. 1985. Ecology of Pseudomonas syringae relevant to the field use of Ice− deletion mutants constructed in vitro for plant frost control, p 23–35 In Halvorson HO, Pramer D, Rogul M. (ed), Engineered organisms in the environment: scientific issues. American Society for Microbiology, Washington, DC [Google Scholar]
- 72.Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, Cherkis K, Roach J, Grant SR, Jones CD, Dangl JL. 2011. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathol. 7:e1002132. 10.1371/journal.ppat.1002132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Phelps P, Giddings TH, Prochoda M, Fall R. 1986. Release of cell-free ice nuclei by Erwinia herbicola. J. Bacteriol. 167:496–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee MR, Lee RE, Strong-Gunderson JM, Minges SR. 1995. Isolation of ice-nucleating active bacteria from the freeze-tolerant frog Rana sylvatica. Cryobiology 32:358–365. 10.1006/cryo.1995.1036 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.