Abstract
Shiga toxin-producing Escherichia coli (STEC) strains (n = 194) representing 43 serotypes and E. coli K-12 were examined for clustered regularly interspaced short palindromic repeat (CRISPR) arrays to study genetic relatedness among STEC serotypes. A subset of the strains (n = 81) was further analyzed for subtype I-E cas and virulence genes to determine a possible association of CRISPR elements with potential virulence. Four types of CRISPR arrays were identified. CRISPR1 and CRISPR2 were present in all strains tested; 1 strain also had both CRISPR3 and CRISPR4, whereas 193 strains displayed a short, combined array, CRISPR3-4. A total of 3,353 spacers were identified, representing 528 distinct spacers. The average length of a spacer was 32 bp. Approximately one-half of the spacers (54%) were unique and found mostly in strains of less common serotypes. Overall, CRISPR spacer contents correlated well with STEC serotypes, and identical arrays were shared between strains with the same H type (O26:H11, O103:H11, and O111:H11). There was no association identified between the presence of subtype I-E cas and virulence genes, but the total number of spacers had a negative correlation with potential pathogenicity (P < 0.05). Fewer spacers were found in strains that had a greater probability of causing outbreaks and disease than in those with lower virulence potential (P < 0.05). The relationship between the CRISPR-cas system and potential virulence needs to be determined on a broader scale, and the biological link will need to be established.
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) has been recognized as a human pathogen since the early 1980s, when two consecutive outbreaks of STEC serotype O157:H7 in contaminated beef patties sickened 47 people in the United States (1). To date, over 400 additional serotypes have been associated with bacterial gastroenteritis worldwide (2), and there are over 175,000 estimated cases of STEC infections each year in the United States alone (3). Depending on the ability to cause outbreaks and/or severe disease, Karmali et al. (4) classified STEC serotypes into seropathotypes (SPTs) A to E: SPT A causes outbreak and disease at high rates, and SPT E has not been linked to outbreaks or severe disease.
Clustered regularly interspaced short palindromic repeats (CRISPRs) were first discovered in E. coli in 1987 (5) and have now been found in ∼45% of bacteria and ∼90% of archaea (6–8). CRISPRs function as heritable and adaptive immune systems against mobile genetic elements (phages and plasmids, etc.) (9–11) and are made of three components: a leader sequence that carries a promoter for transcription, CRISPR-associated genes (cas) encoding proteins with multiple functions, and CRISPR arrays formed by repeats and spacers (12). While most repeats are typically indistinguishable in size and sequence within a defined locus, they are intercalated by nonrepeated short sequences called spacers, which are of a constant number of nucleotides and are unique and hypervariable within a locus (13). They may originate from mobile and invasive genetic elements incorporated into the array and subsequently could serve as the sequence-specific recognition portion of the immune system (14–16).
Four CRISPR loci have been described in E. coli (17–19). CRISPR1 and CRISPR2 have identical consensus repeats (20) and are located between iap and cysH and between ygcF and ygcE, respectively (17, 19). CRISPR1 cas genes form the I-E CRISPR subtype (18). CRISPR3 and CRISPR4 also share identical consensus repeats (20), and both loci are located between clpA and infA. CRISPR3 cas genes form the I-F CRISPR subtype (17, 19, 20). Array size and content vary among CRISPR types and strains. It is not common that the four loci are present in a single E. coli isolate, but CRISPR1 and CRISPR2 are most frequently found in E. coli (19, 21).
CRISPR arrays evolve by polarized acquisition of novel spacers and represent a chronological record of infectious assault on a bacterium from viral and other genetic elements. Distal spacers from the leader sequence are older and are shared among strains, while newer spacers are closer to the leader and more strain specific. Occasionally, sporadic deletions of internal spacers do occur (22). Differences in spacer content would indicate variations in the host environment and geographical locations and may be useful in evolutionary and epidemiological studies (12). This variability makes CRISPR arrays suitable genetic markers for bacterial subtyping.
A primary biological role of CRISPR-cas systems is to provide acquired immunity to protect bacterial cells against mobile genetic elements such as viruses and plasmids (10, 11). Conversely, evolution of pathogenic strains is attributed to the acquisition of elements such as transposons, phages, genomic islands, and plasmids through lateral gene transfer (23, 24). For example, genomic analysis of STEC strains of serogroups O26, O103, O111, and O157 revealed that they have much larger genomes than nonpathogenic E. coli, mainly due to a large content of prophages and other integrative elements (25). It is expected that strains containing functional CRISPR systems restrict the acquisition of mobile genetic elements and that strains with the most complex and active CRISPR systems have a lower susceptibility to invasion by mobile genetic elements (19). However, studies on the relationship of CRISPR systems and the acquisition of genetic mobile elements resulted in different findings. While an inverse relationship between the presence of cas and virulence factors in Enterococcus spp. was reported, no correlation was found between CRISPR and the presence of plasmids containing antimicrobial resistance genes in E. coli (26, 27). We hypothesize that CRISPR arrays are a suitable marker for STEC serotyping and that there could be a correlation between the presence of CRISPR elements and virulence determinants in STEC. To test this hypothesis, we described CRISPR arrays of 194 STEC strains of 43 serotypes, investigated array relationships among serotypes, and explored the potential relationship between CRISPR elements and virulence genes.
MATERIALS AND METHODS
STEC strains.
A set of 190 STEC strains from our collection were analyzed, including 30 O26, 30 O103, 41 O111, 6 O45, 4 O121, 6 O145, and 12 O157 strains and a variable number of strains of other serogroups (see Table S1 in the supplemental material). The strains were isolated from a variety of geographical locations and sources, including humans, cattle and beef products, sheep, goat, deer, okapi, and produce. Collection dates range from 1976 to 2010.
DNA isolation.
Genomic DNA was extracted from a pure culture after streaking onto LB agar and incubation at 35°C for 24 h, using Instagene matrix (Bio-Rad, Hercules, CA). Briefly, 1 to 2 colonies were suspended in 1 ml of ultrapure water and centrifuged. The supernatant was discarded, and 200 μl of Instagene matrix was added, followed by incubation at 56°C for 15 min and at 94°C for 8 min. After centrifugation, the supernatant containing DNA was stored at −20°C until use.
PCR and DNA sequencing.
CRISPR array sequences were obtained through PCR and Sanger sequencing using previously described primers (21). PCR mixtures consisted of 1 μl of bacterial DNA mixed with HotStarTaq Plus Master mix (12.5 μl) (Qiagen, Valencia, CA), 10 pM forward and reverse primers, and water to reach a final reaction mixture volume of 25 μl. PCR parameters included an initial denaturation step at 95°C for 5 min and 10 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 90 s for 10 cycles, followed by 25 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 90 s plus a 10-s cycle elongation for each successive cycle and a final extension step at 72°C for 10 min (21). PCR products were sequenced by MCLAB (South San Francisco, CA) from both ends using Applied Biosystems fluorescent dye terminator technology in an ABI 3730xl sequencer with the same PCR primers.
CRISPR array sequence analysis.
Sequences were assembled with Geneious software v. 6.0.5 (New Zealand). Arrays were extracted by using the “clean sequence tool” enclosed in a macro script/database provided by DuPont, as previously described (28). The tool detected repeats listed in a repeat database and automatically separated repeats and the intercalated short sequences—spacers—into different columns. Data were subsequently formatted to a graphic representation of each spacer and repeat based on their sequence (28). To corroborate array sequences, each sequence was tested by using the CRISPRfinder program (http://crispr.u-psud.fr/Server/) (29). In addition, CRISPR sequences of four major STEC serogroups (O26, O103, O111, and O157) and E. coli K-12 were obtained from the NCBI and included in the analysis (see Table S1 in the supplemental material).
To analyze arrays, strains were arranged based on the presence of common consecutive spacers from the distal end to the leader sequence. Strains with the longest series of spacers on their array were designated “anchors,” which were used as a guide for organizing strains into clusters.
Protospacer analysis.
Spacer identity was determined by using a stand-alone blast program (blast+ 2.2.27) against the NCBI nonredundant (nr) nucleotide collection. Protospacers were defined as homologous sequences with an E value of <1.10e−5 and <10% difference in sequence length (21). Self-matches to E. coli CRISPR locus sequences were omitted.
Subtype I-E cas screening.
A seropathotype (4)-balanced subset of 81 strains was selected based on our previous study (30) to screen for the presence of cas1 and cas2, which are markers of the I-E system (see Table S1 in the supplemental material). Primers cas1FW (5′-CGCCTGCATTATGCTCGAAC-3′), cas1REV (5′-CATTTTGCGCACCACCTTCA-3′), cas2FW (5′-ATGAGCATGGTCGTGGTTGT-3′), and cas2REV (5′-CCCATCCAAATCCACCGGAA-3′) were designed based on whole-genome sequencing (WGS) of 24 strains by using Geneious v. 6.0.5. In separate reactions for subtype I-E cas1 and cas2 genes, 12.5 μl of HotStarTaq Plus Master mix (Qiagen) was mixed with 10 pM forward and reverse primers (Invitrogen, Carlsbad, CA), 1 μl of bacterial DNA, and water for a final reaction mixture volume of 25 μl. PCR parameters were an initial denaturation step at 95°C for 5 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 90 s; and a final extension step at 72°C for 10 min.
Subtype I-E cas analysis.
A maximum likelihood phylogenetic tree was constructed based on the concatenated sequence of subtype I-E cas system genes (cas1, cas2, cas3′, cse1, cse2, cas6e, cas7, and cas5) (20) of 16 STEC strains sequenced previously (31) and 8 publically available E. coli sequences (GenBank) (see Table S1 in the supplemental material). The tree was constructed by using Mega 5.1 (32) with 1,000 bootstrap iterations, and E. coli K-12 was used as the outgroup. A pairwise distance matrix was calculated based on a total of 1,014 single-nucleotide polymorphisms (SNPs) to display the evolutionary divergence between different groups on the phylogenetic tree (Mega 5.1 with 1,000 bootstrap replications).
Virulence gene screening.
The presence of selected virulence genes, stx1, stx2, eae, hlyA, pagC, sen, nleB, efa-1, efa-2, terC, ureC, iha, aidA-I, nle2-3, nleG6-2, nleG5-2, irp2, and fyuA, was determined for a subset (see Table S1 in the supplemental material) of strains from our previous studies (30).
Statistical analysis.
Data were analyzed with SSPS v20. Analysis of variance (ANOVA) or a Kruskal-Wallis test was performed, when suitable. P values of <0.05 were considered statistically significant.
Nucleotide sequence accession numbers.
Sequences identified were submitted to GenBank with accession numbers KF522692 to KF523262.
RESULTS
In the current work, we screened and characterized CRISPR arrays of 194 STEC strains of 43 representative serotypes and also evaluated the potential association between CRISPRs and virulence genes.
CRISPR arrays.
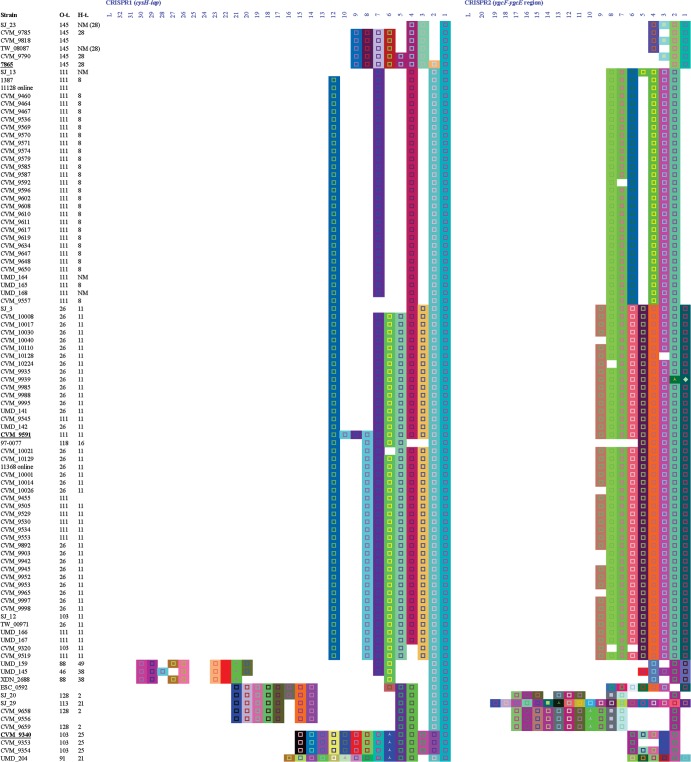
Four types of CRISPR arrays (18) were identified among the 194 STEC strains and E. coli K-12. CRISPR1 and CRISPR2 were present in all strains tested. One strain (95_3322) also had both CRISPR3 and CRISPR4, whereas 193 strains displayed a short, combined array, CRISPR3-4 (Table 1). The length of CRISPR1 and CRISPR2 arrays varied from 1 to 20 spacers, with most having 5 or 7 spacers. Strain 95_3322 CRISPR3 and CRISPR4 arrays were 11 and 6 spacers in length, respectively, whereas the combined array CRISPR3-4 typically had only 1 spacer (Table 1). Nearly 90% of STEC strains (173/195) carried an additional array in the I-E system located 0.5 kb from CRISPR2 (17–19). This array, CRISPR2b, had one spacer, and its sequence was conserved among strains (18) (see Table S3 in the supplemental material).
TABLE 1.
General characteristics of CRISPR arrays from E. coli (n = 195)
| Characteristic | Value for array |
||||||
|---|---|---|---|---|---|---|---|
| CRISPR1 | CRISPR2a | CRISPR2b | CRISPR3 | CRISPR4 | CRISPR3-4 | Total | |
| No. of isolates with array | 195 | 195 | 186 | 1 | 1 | 193 | 771 |
| No. of unique arrays | 78 | 79 | 6 | 1 | 1 | 6 | 171 |
| No. of spacers in array | |||||||
| Range | 1–20 | 1–20 | 0–1 | 11 | 6 | 1–13 | |
| Avg | 9 | 7 | 1 | 11 | 6 | 1 | |
| Mode | 5 | 7 | 1 | 11 | 6 | 1 | |
| Total no. of spacers | 1,612 | 1,349 | 157 | 11 | 6 | 218 | 3,353 |
| No. of different spacers | 258 | 230 | 1 | 11 | 6 | 22 | 528 |
| No. of unique spacers | 128 | 123 | 0 | 11 | 6 | 15 | 283 |
| Spacer length (bp) | |||||||
| Avg | 32 | 32 | 32 | 32 | 32 | 32 | |
| Min | 31 | 30 | 32 | 32 | 32 | 28 | |
| Max | 34 | 35 | 32 | 32 | 33 | 34 | |
| No. of protospacers detected | 4 | 4 | 0 | 1 | 1 | 0 | 10 |
CRISPR1 was less polymorphic than CRISPR2. Most CRISPR1 arrays (94%; 184/195) shared an ancestral (first) spacer, and 64% (125/195) also shared the second-oldest spacer (Fig. 1), indicating a common origin. However, CRISPR2 arrays did not share the first spacer, and many shared only the second spacer. Both loci showed numerous deletions of spacers, mostly of 2 or 3 spacers. Interestingly, despite the observation that the older spacer of CRISPR1 was shared by 184 strains, the first repeat was shared by only 151 strains (see Table S4 in the supplemental material). For most of the combined arrays, CRISPR3-4 had only one spacer (95%; 180/195), and this same spacer was present in 145 strains across different serotypes, reflecting a common origin.
FIG 1.
CRISPR1 and CRISPR2 arrays of STEC strains. The left block represents CRISPR1 and the right block represents CRISPR2 for the same strains in the same order. Only spacers are shown and are represented by colored squares. A same color/figure combination represents identical nucleotide sequence. Spacers on the right are older than those on the left. Column L indicates the leader sequence position. Strains underlined and in boldface type are anchor strains. Sequences were extracted by using a proprietary macro designed by DuPont (28). The same software was used for the representation of spacers and repeats. Except for E. coli K-12, all 194 strains were STEC (stx1, stx2, or stx1 and stx2 positive). t., type; NM, nonmotile; OR and ONT, O-antigen nontypeable; UN, nontypeable.
Spacer diversity.
A total of 3,353 spacers were identified, of which 528 were distinct. The average length of a spacer was 32 bp, ranging from 30 to 35 bp. Approximately one-half of the 528 spacers (54%) were unique (Table 1) and were found in strains of less common serotypes (Fig. 1). Many strains shared spacers in the same CRISPR loci, but no spacers were shared between CRISPR loci (Fig. 1).
Ten of the 528 spacers had identities with sequences from plasmids of Salmonella enterica or E. coli (i.e., protospacers). These spacers were observed in 13 strains. Additionally, one spacer showed identity to bacteriophage P7 and was present in 12 of the 13 strains (see Table S2 in the supplemental material). Most spacers (8/10) with known protospacers formed part of CRISPR1, and some strains (7/13) had more than one of these spacers in their array (Table 2). For example, strains XDN_4854 and XDN_5545 contained five and four of these spacers in CRISPR1, respectively. Strain 95_3322 carried one of the spacers in arrays CRISPR1, CRISPR3, and CRISPR4. The locations of these spacers in the array were random, from positions 1 to 19. Most strains harboring these spacers were of uncommon serotypes, and five of them were not even serologically typeable (Table 2). The sequence homology of spacers with phage and plasmid is consistent with the role that CRISPR plays in resisting mobile genetic elements, as previously described (9, 10).
TABLE 2.
Location of spacers with protospacers in STEC strain arrays
| Strain | Serotype | CRISPR | No. of spacers in the array | No. of spacers with protospacers | Position in the array (from leader sequence) in cluster: |
|||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |||||
| 90_0327 | O22:H8 | 1 | 11 | 2 | 2 | 7 | ||
| 95_3322 | O22:H5 | 1 | 9 | 1 | 2 | |||
| 3 | 11 | 1 | 1 | |||||
| 4 | 6 | 1 | 6 | |||||
| ESC_0589 | NTa | 1 | 7 | 2 | 2 | 3 | ||
| ESC_0608 | O73:H18 | 1 | 12 | 1 | 2 | |||
| ESC_0613 | O168:H8 | 1 | 7 | 1 | 2 | |||
| UMD_131 | OR:H9 | 2 | 12 | 3 | 10 | 11 | 12 | |
| XDN_2746 | O83:H8 | 1 | 3 | 1 | 2 | |||
| XDN_4854 | ONT:H10 | 1 | 19 | 5 | 2 | 8 | 17 | 19 |
| 2 | 8 | 1 | 8 | |||||
| XDN_5545 | ONT:H7 | 1 | 19 | 4 | 2 | 7 | 17 | 19 |
| XDN_5578 | ONT:H46 | 1 | 9 | 2 | 2 | 7 | ||
| XDN_11682 | O83:H8 | 1 | 3 | 1 | 2 | |||
| XDN_15432 | O83:H8 | 1 | 10 | 2 | 7 | 2 | ||
| XDN_23765 | ONT:H2 | 1 | 6 | 1 | 2 | |||
NT, nontypeable.
Array organization by serotype.
CRISPR arrays were organized based on the spacer content of anchor strains, which are defined as those strains containing all spacers for a group/cluster in the correct order representing ancestral strains. Although a universal anchor was not identified, four clusters were established, each one with one anchor (Fig. 1). The first cluster was formed by O145 strains and anchored by a human isolate, 07865 (O145:H28). The second group was anchored by CVM 9591 (O111:H11), isolated from a cow in 1995. The cluster included two subgroups: O111:H8 and O111:NM in a block and O26:H11, O103:H11, and O111:H11, among others, in a second group. The third cluster was more diverse, formed by several serotypes, including O45:H2, O103:H2, O103:H25, O91:H21, and O91:H14. This group was anchored by CVM 9340 (O103:H25), isolated from humans. The last group was also very diverse, anchored by 08023 (O121:H19). Strains of less common serotypes did not form clusters. Since CRISPR1 and CRISPR2 coclustered, the same arrangement was achieved by using either one as a guide (Fig. 1). This was consistent with a parallel evolution of the two CRISPR loci over time.
Strain clustering based on CRISPR spacer content correlated well with STEC serotype. For instance, serotype O111:H8 formed a large cluster of 29 strains that had almost identical spacer contents with only a few minor deletions of 1 or 2 spacers in CRISPR1 and CRISPR2. Similar findings were observed among serotypes O26:H11, O103:H2, and O157:H7. Unique, long CRISPR arrays were present in less common STEC serotypes (Fig. 1).
It was notable that spacer content seemed to correlate well with strains retaining the same H-antigen type but not necessarily with strains having the same O group. For example, O103:H2 did not share any spacer in CRISPR2 with O103:H11, although they did have common ancestral spacers in CRISPR1 (3/12 strains). However, O103:H2 clustered together with O45:H2 and contained identical spaces in CRISPR1 up to the fourth spacer, where O103:H2 had an additional eight spacers. Similarly, O45:H2 and O103:H2 differed only by one spacer deletion in CRISPR2 (Fig. 1). On the other hand, only 9 of 17 spacers were shared between O111:H8 and O111:H11 strains, whereas strains of O26:H11 and O103:H11 had practically identical arrays, forming a subcluster based on antigen H11 (Fig. 1). Taken together, these data may point to H-antigen loci as being more phylogenetically stable, while O-antigen alleles appear to be shuffled in the evolution of some STEC clades (31).
Correlation between CRISPR content and occurrence of virulence genes.
Previous reports indicated an inverse correlation between the presence of virulence genes and the distribution of cas genes in Enterococcus faecalis (27). Therefore, we analyzed a subset of strains (n = 81) of different STEC seropathotypes (see Table S1 in the supplemental material) for virulence genes (30) and the presence of subtype I-E cas genes. While most strains (91%) had cas1, all STEC strains carried cas2. Because of such high positive rates, there was no significant difference in the presence of subtype I-E cas among different seropathotypes. Similarly, no association between the presence of subtype I-E cas and virulence genes was observed.
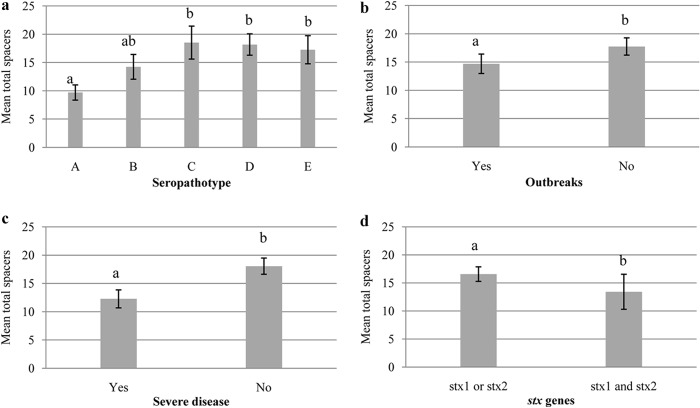
A significant difference in the total content of spacers between strains of different seropathotypes was observed (P < 0.05) (Fig. 2a). Fewer spacers were found in strains that had greater potential of causing outbreaks (SPTs A and B) than in those with lower virulence potential (SPTs C, D, and E) (P < 0.05) (Fig. 2b). Similarly, strains with a higher potential for causing severe disease (SPTs A, B, and C) had fewer spacers than did those with lower potential (SPTs D and E) (P < 0.05) (Fig. 2c). An association between the number of spacers and the presence of certain virulence genes was also observed. For example, eae-positive strains had significantly fewer spacers than eae-negative strains (P < 0.05). Other virulence genes, including pagC, sen, terC, ureC, nleB, nle2-3, nleG6-2, and nleG5-2, also showed the same significant relationship with the number of spacers. However, the opposite relationship was seen with the fyuA and irp2 genes, and no association was detected between the number of spacers and the presence of hlyA, aidA-I, iha, efa-1, efa-2, stx1, and stx2 (data not shown). Interestingly, strains containing both stx genes showed significantly fewer spacers than did those with only one of them (P < 0.05) (Fig. 2d).
FIG 2.
Association of total spacer content with seropathotype, the ability to cause outbreaks and severe disease, and stx genes. Bars represent total spacer counts (CRISPR1, CRISPR2a, CRISPR2b, CRISPR3-4, CRISPR3, and CRISPR4). (a) Seropathotypes A to E; (b) potential ability to cause an outbreak; (c) potential ability to cause severe disease based on the classification of Karmali et al. (4); (d) stx genes. Vertical lines represent ±2 standard errors. Statistical tests revealed significant differences (P < 0.05) between seropathotypes and between the ability to cause outbreaks and severe disease and stx gene content (P < 0.05). Different letters above the bars indicate significant differences.
Subtype I-E cas phylogeny.
To investigate the relationship between CRISPR and the evolutionary history of strains, we reconstructed a maximum likelihood phylogenetic tree based on the concatenated sequence of the subtype I-E cas system genes extracted from 24 E. coli strains (Fig. 3). The strains were grouped into four major clades, except for E. coli K-12, which was used as the outgroup. All O157:H7 strains formed a single clade, whereas O103:H2 strains belonged to another cluster. However, an O103:H25 strain (CVM 9340) appeared in a separated clade. Interestingly, the remaining strains of serotypes O111:H11, O111:H8, and O26:H11 were clustered together, indicating a closer phylogenetic relationship and more conserved subtype I-E cas alleles among them.
FIG 3.
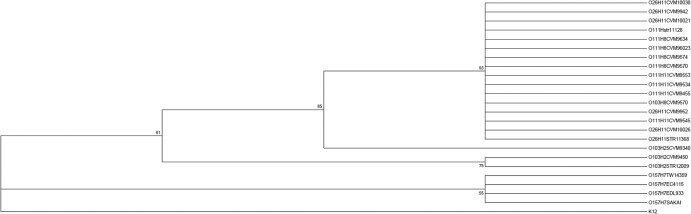
Maximum likelihood phylogenetic tree based on concatenated sequences of type I-E cas genes in 24 STEC strains. Concatenated sequences of type I-E cas genes (cas1, cas2, cas3′, cse1, cse2, cas6e, cas7, and cas5) were obtained from our previous study on 16 STEC strains (31) and publically available sequences of 8 E. coli strains. The maximum likelihood phylogenetic tree was generated by using Mega 5.1 (32), with 1,000 bootstrap replications. E. coli K-12 was used as the control outgroup strain.
Additionally, a pairwise distance matrix of SNP differences (see Table S5 in the supplemental material) supported phylogeny results of the maximum likelihood analysis. For example, the number of SNP differences between the group formed by H8 and H11 and groups O103:H25, O103:H2, and O157:H7 were 14, 74, and 100 SNPs, respectively (Fig. 3).
DISCUSSION
In this study, we determined the occurrence and content of CRISPR loci in STEC strains and observed conservation among strains of the same serotype (O- and H-antigen type combination) but not between serogroups (i.e., only O-antigen type). However, in some cases, strains of different O groups but with the same H type shared identical CRISPR sequences, suggesting that such serotypes might have common ancestors (Fig. 1). This may provide a genetic basis for the specific detection and tracking of particular E. coli strains in the environment or in the food supply. In addition, a significant negative association was observed between the number of spacers (an indicator of CRISPR system activity) and the pathogenic potential of STEC strains, as indicated by their seropathotype (4), a finding heretofore undescribed among STEC strains.
Other studies also demonstrated the relationship between CRISPR array content and serotypes. Delannoy et al. (33, 34) reported the presence of specific CRISPR polymorphisms related to O:H serotypes of STEC O26:H11, O45:H2, O103:H2, O104:H4, O111:H8, O145:H28, and O157:H7, which were useful to differentiate these serotypes. However, they reported numerous cross-reactions: primers for O145:H28 reacted with O28:H28 strains, and primers detecting O103:H2 and O45:H2 altogether also cross-reacted with O128:H2 and O145:H2 strains, among others (33). Similarly, Yin et al. (18) confirmed a relationship between CRISPR polymorphisms and serotypes and also described a strong conservation of CRISPR arrays within isolates of the same H type, including H7, H2, and H11. Our data showed similar findings: strains of different O types shared identical arrays with strains of the same H antigen (O26:H11, O103:H11, and O111:H11 as well as O45:H2 and O103:H2), but arrays of strains of the same O group with different H types seemed unrelated (O103:H2 and O103:H11), further underscoring the linkage between CRISPR arrays and H-antigen alleles (Fig. 1). A previous study on the evolutionary history of non-O157 STEC by WGS showed that O26:H11 and O111:H11 grouped together, also suggesting that strains with the same H antigens may have common ancestors (31). Furthermore, we could not distinguish between strains of serotypes O26:H11, O111:H8, and O111:H11 based on the concatenated sequences of their subtype I-E cas genes, reflecting a close relatedness of these serotypes. Ju et al. (31) also demonstrated that strains with H8/H11 antigens formed a major clade on a genome-wide phylogenetic tree but displayed closer relatedness with O103:H2 strains. In contrast, group H8/H11was closer to O103:H25 strains than to O103:H2 strains based on subtype I-E cas sequences. Thus, concatenated subtype I-E cas genes could not be used to determine the same phylogenies as those found in genomic comparisons among serotypes (35).
Four CRISPR arrays have been identified in E. coli but are rarely found in a single isolate (17, 19). Similarly to what was reported by Yin et al. (18), our data showed that the type I-E CRISPR-cas system (CRISPR1 and CRISPR2) was most widely distributed in STEC strains. One strain (95_3322), however, carried the four arrays, and the remaining strains carried a shorter, combined CRISPR3-4 array, as previously described (19), which is associated with the fusion of the remaining sections of loci 3 and 4 when subtype I-F genes, originally located between the two loci, are deleted (19). To confirm the absence of subtype I-F cas genes, we sequenced the region between primers C3Fw (clpA target) and C4 Rev (infA target). In most cases (179/190), the fragment produced was ∼800 bp instead of the expected ∼3,000 bp when subtype I-F cas genes were present (19). The absence of cas genes and repeats among these motifs suggests a relatively minor role for CRISPR system I-F in STEC “immune” function.
We identified 10 protospacers among STEC spacers. Most protospacers (9/10) were located in plasmids from Salmonella and E. coli, including multiple protospacers from the same plasmid (see Table S2 in the supplemental material), for both CRISPR1 and CRISPR2. Additionally, a spacer that had sequence identity with bacteriophage P7 was found in 12 of the 13 strains where matching protospacers were identified (see Table S2 in the supplemental material). Yin et al. (18) also observed that multiple spacer sequences originated from the same origin, and Datsenko et al. (36) demonstrated that a mutated motif stimulated the acquisition of more spacers from the same target to strengthen immunity against the element.
Longer CRISPR arrays reflect larger numbers of immunization events and can be evidence of more active CRISPR systems. In this work, we postulated that these events may have contributed to preventing the uptake and acquisition of virulence genes. The role of CRISPR as an immune system against mobile genetic elements was previously reported (9, 37). Since many virulence determinants are acquired through mobile genetic elements (25), it is expected that strains with more active CRISPR systems carry fewer virulence genes and other mobile genetic elements, but studies on the role of CRISPR systems in acquisition of virulence determinants showed contradictory results. One study found that CRISPR-cas systems were inversely correlated with the presence of acquired antibiotic resistance in Enterococcus faecalis (38). Similarly, an inverse correlation between the presence of two virulence genes and the distribution of cas genes in Enterococcus spp. was reported, and fewer virulence genes were detected when cas genes were present (27). In contrast, the acquisition of plasmids carrying antimicrobial genes was not related to the presence of the CRISPR system in E. coli (26). A recent study showed that uropathogenic E. coli strains seemed less likely to have CRISPR loci than nonuropathogenic E. coli strains from the same patient, suggesting that CRISPR may have a role in the acquisition of phage and plasmids and serves as an adaptive advantage for the group (39). In the present study, we found that subtype I-E cas genes were not related to the presence of virulence markers in STEC (30); however, statistical differences indicated that seropathotypes historically associated with outbreaks and severe disease had fewer spacers than others (Fig. 2), suggesting a negative correlation between CRISPR array length, an indicator of CRISPR-cas system activity, and the propensity for pathogenic trait acquisition. Moreover, while the presence of some virulence genes (9/18) was inversely related to a lower spacer content, strains with both stx genes had significantly fewer spacers than did those having only one (P < 0.05). These findings were consistent with the documented role of CRISPR-cas immune systems in limiting the uptake of genetic material derived from mobile and invasive elements such as phages and plasmids, yet experiments have failed to prove that wild-type E. coli CRISPR systems actively function as immune systems (21, 37, 40). Recent evidence indicates that CRISPR systems are involved in bacterial virulence, but this role may not be directly related to an immune system function. For example, cas9 from Francisella novicida indirectly regulated genes to prevent host recognition (41), and Legionella pneumophila cas2 was required for intracellular infection of amoebae, an amplification step in their life cycle (42). Our data did not demonstrate that the CRISPR system acted as an immune system in the STEC strains. However, the inverse relationship between CRISPR array length and virulence genes may be associated with other attributes. For instance, Louwen et al. suggested an association between more pathogenic Campylobacter jejuni and the presence of nonfunctional CRISPR, which was likely due to an indirect relationship between the production of gangliosides (linked to Guillain-Barré syndrome) and a higher level of resistance to phage, resulting in a lower evolutionary pressure on the CRISPR system selecting against them (43). Similarly, Bikard et al. (44) showed that Streptococcus pneumoniae selected against CRISPR arrays. Therefore, some relationships (direct and/or indirect) exist between CRISPR systems and virulence, and further studies on a broad range of bacteria are needed to assess such relationships.
The current study provides insights into the occurrence and role of CRISPR-cas systems in STEC serogroups (O26, O103, and O111) as well as several additional uncommon serotypes. CRISPR array sequence analysis suggests that H antigen might have been acquired more ancestrally than O antigen since arrays are shared by strains with the same H antigen but not by strains with the same O antigen. Alternatively, stability among H antigens in STEC strains may also point to a more vertical inheritance pattern and less promiscuity than O-antigen evolution, known to be dappled by numerous horizontal gene transfer events throughout its radiation in E. coli (45). Also, the relationship between CRISPR elements and pathogenicity traits in STEC needs to be studied to determine whether they have a causal relationship or whether a formal balancing selection drives the acquisition of the two. Further studies using additional and genetically diverse strains would provide a better understanding of the CRISPR-cas system in STEC and E. coli as a whole. CRISPR arrays and other genetic markers could be used to differentiate high-risk STEC from low-risk strains, thereby providing useful tools for the control of STEC infections and insights into their genetic content and phenotypic traits.
Supplementary Material
ACKNOWLEDGMENTS
We thank Philippe Horvath at DuPont for providing the CRISPR macro and Edward Dudley at Pennsylvania State University for sharing his new proposed nomenclature of the CRISPR2 array. We are also thankful to Ruth Timme, James Pettengill, and Peter Evans at CFSAN for their comments and insights.
Footnotes
Published ahead of print 13 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03018-13.
REFERENCES
- 1.Wells JG, Davis BR, Wachsmuth IK, Riley LW, Remis RS, Sokolow R, Morris GK. 1983. Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J. Clin. Microbiol. 18:512–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanco JE, Blanco M, Alonso MP, Mora A, Dahbi G, Coira MA, Blanco J. 2004. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from human patients: prevalence in Lugo, Spain, from 1992 through 1999. J. Clin. Microbiol. 42:311–319. 10.1128/JCM.42.1.311-319.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15. 10.3201/eid1701.09-1101p1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karmali MA, Mascarenhas M, Shen S, Ziebell K, Johnson S, Reid-Smith R, Isaac-Renton J, Clark C, Rahn K, Kaper JB. 2003. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41:4930–4940. 10.1128/JCM.41.11.4930-4940.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. 1987. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169:5429–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrangou R. 2013. CRISPR-Cas systems and RNA-guided interference. Wiley Interdiscip. Rev. RNA 4:267–278. 10.1002/wrna.1159 [DOI] [PubMed] [Google Scholar]
- 7.Barrangou R, Horvath P. 2012. CRISPR: new horizons in phage resistance and strain identification. Annu. Rev. Food Sci. Technol. 3:143–162. 10.1146/annurev-food-022811-101134 [DOI] [PubMed] [Google Scholar]
- 8.Bhaya D, Davison M, Barrangou R. 2011. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45:273–297. 10.1146/annurev-genet-110410-132430 [DOI] [PubMed] [Google Scholar]
- 9.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- 10.Garneau JE, Dupuis M, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71. 10.1038/nature09523 [DOI] [PubMed] [Google Scholar]
- 11.Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845. 10.1126/science.1165771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170. 10.1126/science.1179555 [DOI] [PubMed] [Google Scholar]
- 13.Al-Attar S, Westra ER, van der Oost J, Brouns SJJ. 2011. Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol. Chem. 392:277–289. 10.1515/BC.2011.042 [DOI] [PubMed] [Google Scholar]
- 14.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151(Part 8):2551–2561. 10.1099/mic.0.28048-0 [DOI] [PubMed] [Google Scholar]
- 15.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60:174–182. 10.1007/s00239-004-0046-3 [DOI] [PubMed] [Google Scholar]
- 16.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964. 10.1126/science.1159689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Touchon M, Rocha EP. 2010. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One 5:e11126. 10.1371/journal.pone.0011126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin S, Jensen MA, Bai J, Debroy C, Barrangou R, Dudley EG. 2013. The evolutionary divergence of Shiga toxin-producing Escherichia coli is reflected in clustered regularly interspaced short palindromic repeat (CRISPR) spacer composition. Appl. Environ. Microbiol. 79:5710–5720. 10.1128/AEM.00950-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Díez-Villaseñor C, Almendros C, García-Martínez J, Mojica FJ. 2010. Diversity of CRISPR loci in Escherichia coli. Microbiology 156(Part 5):1351–1361. 10.1099/mic.0.036046-0 [DOI] [PubMed] [Google Scholar]
- 20.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9:467–477. 10.1038/nrmicro2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Touchon M, Charpentier S, Clermont O, Rocha EP, Denamur E, Branger C. 2011. CRISPR distribution within the Escherichia coli species is not suggestive of immunity-associated diversifying selection. J. Bacteriol. 193:2460–2467. 10.1128/JB.01307-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190:1390–1400. 10.1128/JB.01412-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence JG. 2005. Common themes in the genome strategies of pathogens. Curr. Opin. Genet. Dev. 15:584–588. 10.1016/j.gde.2005.09.007 [DOI] [PubMed] [Google Scholar]
- 24.Coombes BK, Gilmour MW, Goodman CD. 2011. The evolution of virulence in non-O157 Shiga toxin-producing Escherichia coli. Front. Microbiol. 2:90. 10.3389/fmicb.2011.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, Oshima K, Kodama T, Abe H, Nakayama K, Kurokawa K, Tobe T, Hattori M, Hayashi T. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:17939–17944. 10.1073/pnas.0903585106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Touchon M, Charpentier S, Pognard D, Picard B, Arlet G, Rocha EP, Denamur E, Branger C. 2012. Antibiotic resistance plasmids spread among natural isolates of Escherichia coli in spite of CRISPR elements. Microbiology 158(Part 12):2997–3004. 10.1099/mic.0.060814-0 [DOI] [PubMed] [Google Scholar]
- 27.Lindenstrauß AG, Pavlovic M, Bringmann A, Behr J, Ehrmann MA, Vogel RF. 2011. Comparison of genotypic and phenotypic cluster analyses of virulence determinants and possible role of CRISPR elements towards their incidence in Enterococcus faecalis and Enterococcus faecium. Syst. Appl. Microbiol. 34:553–560. 10.1016/j.syapm.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 28.Horvath P, Romero DA, Coûté-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190:1401–1412. 10.1128/JB.01415-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a Web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35:W52–W57. 10.1093/nar/gkm360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ju W, Shen J, Toro M, Zhao S, Meng J. 2013. Distribution of pathogenicity islands OI-122, OI-43/48, and OI-57 and a high-pathogenicity island in Shiga toxin-producing Escherichia coli. Appl. Environ. Microbiol. 79:3406–3412. 10.1128/AEM.03661-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ju W, Cao G, Rump L, Strain E, Luo Y, Timme R, Allard M, Zhao S, Brown E, Meng J. 2012. Phylogenetic analysis of non-O157 Shiga toxin-producing Escherichia coli strains by whole-genome sequencing. J. Clin. Microbiol. 50:4123–4127. 10.1128/JCM.02262-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delannoy S, Beutin L, Fach P. 2012. Use of clustered regularly interspaced short palindromic repeat sequence polymorphisms for specific detection of enterohemorrhagic Escherichia coli strains of serotypes O26:H11, O45:H2, O103:H2, O111:H8, O121:H19, O145:H28, and O157:H7 by real-time PCR. J. Clin. Microbiol. 50:4035–4040. 10.1128/JCM.02097-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delannoy S, Beutin L, Burgos Y, Fach P. 2012. Specific detection of enteroaggregative hemorrhagic Escherichia coli O104:H4 strains by use of the CRISPR locus as a target for a diagnostic real-time PCR. J. Clin. Microbiol. 50:3485–3492. 10.1128/JCM.01656-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao G, Meng J, Strain E, Stones R, Pettengill J, Zhao S, McDermott P, Brown E, Allard M. 2013. Phylogenetics and differentiation of Salmonella Newport lineages by whole genome sequencing. PLoS One 8:e55687. 10.1371/journal.pone.0055687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. 2012. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 3:945. 10.1038/ncomms1937 [DOI] [PubMed] [Google Scholar]
- 37.Edgar R, Qimron U. 2010. The Escherichia coli CRISPR system protects from λ lysogenization, lysogens, and prophage induction. J. Bacteriol. 192:6291–6294. 10.1128/JB.00644-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmer KL, Gilmore MS. 2010. Multidrug-resistant enterococci lack CRISPR-cas. mBio 1(4):e00227-10. 10.1128/mBio.00227-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dang TN, Zhang L, Zöllner S, Srinivasan U, Abbas K, Marrs CF, Foxman B. 2013. Uropathogenic Escherichia coli are less likely than paired fecal E. coli to have CRISPR loci. Infect. Genet. Evol. 19:212–218. 10.1016/j.meegid.2013.07.017 [DOI] [PubMed] [Google Scholar]
- 40.Swarts DC, Mosterd C, van Passel MW, Brouns SJ. 2012. CRISPR interference directs strand specific spacer acquisition. PLoS One 7:e35888. 10.1371/journal.pone.0035888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. 2013. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 497:254–257. 10.1038/nature12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gunderson FF, Cianciotto NP. 2013. The CRISPR-associated gene cas2 of Legionella pneumophila is required for intracellular infection of amoebae. mBio 4(2):e00074-13. 10.1128/mBio.00074-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Louwen R, Horst-Kreft D, de Boer AG, van der Graaf L, de Knegt G, Hamersma M, Heikema AP, Timms AR, Jacobs BC, Wagenaar JA, Endtz HP, van der Oost J, Wells JM, Nieuwenhuis EE, van Vliet AH, Willemsen PT, van Baarlen P, van Belkum A. 2013. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barré syndrome. Eur. J. Clin. Microbiol. Infect. Dis. 32:207–226. 10.1007/s10096-012-1733-4 [DOI] [PubMed] [Google Scholar]
- 44.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. 2012. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe 12:177–186. 10.1016/j.chom.2012.06.003 [DOI] [PubMed] [Google Scholar]
- 45.Tarr PI, Schoening LM, Yea YL, Ward TR, Jelacic S, Whittam TS. 2000. Acquisition of the rfb-gnd cluster in evolution of Escherichia coli O55 and O157. J. Bacteriol. 182:6183–6191. 10.1128/JB.182.21.6183-6191.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.