Abstract
Conjugation activity of plasmid pLS20 from Bacillus subtilis subsp. natto is induced when cells are diluted into fresh medium and diminishes as cells enter into stationary-phase growth. Transcriptional profiling shows that during mid-exponential growth, more than 5% of the host genes are affected in the presence of the plasmid, in contrast to the minor changes seen in freshly diluted and stationary-phase cells. Changes occurred in many metabolic pathways, although pLS20 does not confer any detectable burden on its host cell, as well as in membrane and cell wall-associated processes, in the large motility operon, and in several other cellular processes. In agreement with these changes, we found considerable alterations in motility and enzyme activity and increased resistance against several different forms of stress in cells containing the plasmid, revealing that the presence of pLS20 has a broad impact on the physiology of its host cell and increases its stress resistance in multiple aspects. Additionally, we found that the lack of chromosomal gene yueB, known to encode a phage receptor protein, which is upregulated in cells containing pLS20, strongly reduced conjugation efficiency, revealing that pLS20 not only increases fitness of its host but also employs host proteins for efficient transfer into a new cell.
INTRODUCTION
Conjugation is a major mechanism for the transmission of genetic material between bacterial cells (1, 2). Conjugative elements comprise autonomously replicating plasmids as well as conjugative transposons. Both types of elements carry genes encoding a type IV secretion system (T4SS) mediating the direct transfer of substrate DNA into neighboring cells (3–5).
Plasmid pLS20 was originally isolated from B. subtilis subsp. natto IF0335 and is a self-replicative plasmid vertically maintained by its endogenously encoded segregation system (Alp7AR) (6). pLS20 is self-transmissible and able to transfer to Bacillus species such as B. anthracis, B. cereus, B. licheniformis, B. megaterium, B. pumilus, and B. thuringiensis (7). The plasmid encodes 84 identifiable open reading frames (8). Transfer activity increases after inoculation into rich medium and rapidly decreases while cells move toward stationary-phase growth. We have shown that the transfer-proficient state is accompanied by the assembly of conserved conjugation proteins at the cell pole or at the lateral cell membrane, as exemplified by the conserved T4SS ATPase VirB4, which is absent during periods of growth in which transfer does not occur (8).
The transcriptional analysis of host and plasmid gene expression in different Escherichia coli or Pseudomonas putida strains containing large conjugative plasmids revealed host genotype-specific differences in gene expression on both the plasmid and the chromosome (9, 10). These studies did not address whether the conjugative plasmid decreases fitness of host cells. It is known that conjugative plasmids carrying drug resistance genes can create fitness costs to their host, but costs can decrease over many generations and at the same time hosts can gain fitness, which continues even in strains cured of the plasmid (11). The underlying mechanism of this phenomenon is not yet clear.
Recently, it has been shown that the nonconjugative plasmid pBS32 found in the undomesticated (i.e., nonlaboratory adapted) B. subtilis strain NCIB3610 interferes with competence development via ComI and with sporulation and biofilm formation via RapP (12). pLS20 also inhibits competence by a plasmid-encoded repressor of ComK (RokLS20) (13), the transcriptional activator of competence genes. Taken together, these findings reveal several effects of conjugative plasmids with their host cell under nutrient- or growth-limiting conditions.
We wondered if the presence of the large conjugative plasmid pLS20 would alter the physiological state of its host in rich medium, and if cells gain additional benefits from carrying a conjugative plasmid under these conditions. Indeed, we found that pLS20 transiently alters the transcriptional activity of more than 5% of the genes from the B. subtilis genome, accompanied by considerable changes in the physiology of the cell, proving that plasmid effects on host cells are not restricted to stationary growth in a bacterium.
MATERIALS AND METHODS
Bacterial strains and plasmids, strain construction, and media.
Bacterial strains, plasmids, and primers used to perform this study are listed in Table 1 and in Table S2 in the supplemental material. The sequence of pLS20 can be found under accession number NC_015148.1 (for an annotation, see Fig. 4A and B). The Escherichia coli strain DH5α was used for molecular cloning and propagation of plasmids. All Bacillus strains (Table 1) were freshly streaked on LB agar plates the day before inoculation into liquid medium and grown overnight either at 30°C or 37°C. During overnight incubation and on plates, the selective pressure was always kept with appropriate antibiotics at the following final concentrations: chloramphenicol (5 μg/ml), spectinomycin (100 μg/ml), tetracycline (20 μg/ml), kanamycin (10 μg/ml), lincomycin (25 μg/ml), and erythromycin (1 μg/ml). No selective pressure was applied after inoculation of cells into fresh medium to perform the individual assays, which was also the case when cells were harvested for RNA extraction.
TABLE 1.
Strains used in this study
| Strain | Descriptiona | Source or reference |
|---|---|---|
| PY79 | Prototrophic laboratory strain | R. Losick, Harvard University, USA |
| NCIB 3610 | Prototrophic isolate, reference sequence NC_000964.3 (Bacillus subtilis strain 168) | Bacillus Genetic Stock Center, Ohio State University, USA |
| BEST40401 | RM125 (hsdR hsdM leu arg) pLS20cat | 14 |
| SB505 | 3610 tasA::spc | 15 |
| DS1796 | PY79 motAB::tet | D. Kearns, Indiana University, USA |
| GP397 | 3610 yueB::tet | J. E. Gonzalez-Pastor laboratory |
| GP399 | 3610 yukBA::tet | J. E. Gonzalez-Pastor laboratory |
| DS313 | PY79 sigD::tet | 16 |
| AKR04 | PY79 pLS20neo | 8 |
| TCR06 | PY79 thrC::mls | 8 |
| TCR07 | 3610 pLS20cat | This study |
| TCR08 | 3610 tasA::spc pLS20cat | This study |
| TCR09 | PY79 motAB::tet pLS20neo | This study |
| TCR10 | 3610 yueB::tet pLS20cat | This study |
| TCR11 | 3610 yukBA::tet pLS20cat | This study |
| TCR12 | PY79 sigD::tet pLS20neo | This study |
| TCR13 | PY79 pLS20neo amyE::Pxyl-yueB-yfp | This study |
| TCR15 | PY79 pLS20neo yukBA::tet amyE::Pxyl-yueB-yfp | This study |
| LH1 | PY79 thrC::Physpank::cds7 pLS20neo | This study |
| LH2 | PY79 thrC::Physpank::cds8 pLS20neo | This study |
| WG 8 | PY79 thrC::Physpank::rapLS20 | This study |
cat, chloramphenicol; spc, spectinomycin; tet, tetracycline; neo, neomycin-kanamycin; mls, lincomycin-erythromycin.
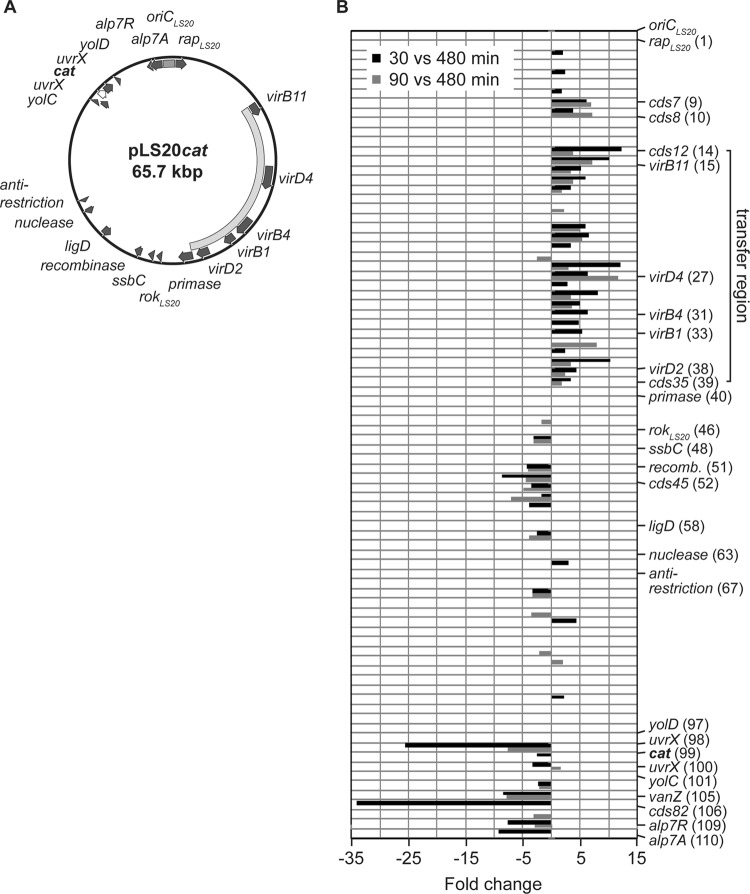
FIG 4.
Transcriptome analysis of the conjugative plasmid pLS20. (A) Circular map of plasmid pLS20cat. Genes showing significant homologies to genes with known or identified functions are illustrated as arrows in dark gray. The transfer region responsible for the conjugative ability of pLS20 is indicated by a curved bar in light gray and the origin of replication of the plasmid (oriCLS20) by a gray bar. The cat gene marked in boldface and shown as a white arrow with a black outline was inserted via the unique SalI restriction site on pLS20, interrupting the uvrC gene (14). (B) Transcriptional profile of genes carried by pLS20 correlated with their linear location on the plasmid for two different time points during exponential growth. Displayed are the expression ratios as fold changes (P < 0.3) of RNA derived from early exponential growth (30 versus 480 min; black bars) and of RNA derived from mid-exponential growth (90 versus 480 min; gray bars) compared to RNA derived from stationary phase. The putative transfer region is indicated by a solid line and comprises the conserved core components virB1, virB4, virB11, virD2, and virD4 found on pLS20. Additionally, genes already indicated in panel A and genes which are significantly altered in their expression and described in the text were also included in the figure. Numbers in parentheses correspond to the locus tags of the NCBI reference sequence NC_015148.1.
To establish wild strain 3610 containing pLS20cat (TCR07), equal amounts of NCIB 3610 and BEST40401 were mixed together as described before (14). To select for transconjugants and to counterselect against the auxotroph genetic background of BEST40401, the mixture was spread on MSgg minimal medium (15) solidified with 1.5% Bacto agar and supplemented with chloramphenicol. To generate strains TCR08, TCR10, and TCR11 containing plasmid pLS20cat, mating of TCR07 with SB505, GP397, or GP399 was performed. To generate strains TCR9 and TCR12 containing plasmid pLS20neo, the donor strain AKR04 was mixed with DS1796 and DS313. Mixtures of donor and recipient cells were plated on LB agar plates with appropriate antibiotics selecting for the resulting strains.
To generate the xylose-inducible amyE::Pxyl-yueB-yfp construct, yueB was amplified by PCR using primer pair 1/2 and chromosomal DNA of PY79 as the template, digested with ApaI and XhoI, and cloned into correspondingly digested pSG1193, creating plasmid pTCR06.
For the generation of isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible constructs, genes encoding cds7, cds8, and rapLS20 were PCR amplified using primer pairs 3/4, 5/6, and 7/8 and pLS20neo as the template. The fragments were digested with NheI and SphI and ligated into NheI- and SphI-digested pDP150 (16), creating plasmids pLH1, pLH2, and pWG8 (thrC::Physpank::rapLS20).
Transformation of PY79 or PY79 containing pLS20neo (AKR04) with plasmids pTCR06, pLH1, pLH2, and pWG8 resulted in strains LH1, LH2, and WG8. To create strain TCR15, TCR13 was transformed with chromosomal DNA of GP399.
Conjugation assays.
To test mutant strains for their conjugation efficiency, mating experiments were performed as described before (14). Only donor cells or recipients cells were directly compared, which derived either from the wild strain NCIB3610 or from the domesticated laboratory strain PY79. Two-sided independent-sample t tests for every comparison were performed using GraphPad Prism version 4.02 for Windows.
Swimming motility assay.
Cells were grown for 90 min at 37°C and 200 rpm in LB medium, harvested, and resuspended in phosphate-buffered saline (PBS) buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4) to the same optical density at 600 nm (OD600). Cells were centrally spotted onto freshly prepared soft agar plates (10 g/liter tryptone, 5 g/liter NaCl) containing 0.3% Bacto agar and were analyzed for colony spreading after 5 h of growth at 37°C. To compare the pLS20-containing strain to the pLS20-free strain, a two-sided dependent sample t test was performed using GraphPad Prism version 4.02 for Windows.
Viability assays.
Cultures were grown for 90 min at 37°C and 200 rpm in LB medium and exposed to 5% (wt/vol) potassium chloride, 10% (vol/vol) ethanol, acid stress at pH 4.5 or alkaline stress at pH 9, and cold shock at 15°C or heat shock at 54°C, which is the maximum temperature at which B. subtilis can survive. Growth was monitored by measuring the OD600, and appropriate dilutions were plated before and 1 h, 2 h, and 3 h after stress induction on LB agar plates to determine cell viability. Cell numbers before stress induction served as the reference for the estimation of cell viability after exposure to different stress conditions. To analyze the survival of pLS20-containing cells over time compared to pLS20-free cells, a 2-way analysis of variance (ANOVA) with a Bonferroni posttest for pairwise comparisons at each time point was performed using GraphPad Prism version 4.02 for Windows.
Sporulation assay.
To determine the sporulation efficiency of plasmid-free cells, plasmid-containing as well as plasmid-free cells overexpressing RapLS20 in independent cultures were grown in Difco sporulation medium (DSM). To induce expression of RapLS20, 1 mM IPTG was added to the cultures. After growth for 24 h at 37°C and 200 rpm, the cells were plated in appropriate dilutions on LB agar plates before and after incubation at 80°C for 20 min to count viable cells and cells deriving from spores. To compare the sporulation efficiency of the different strains, a two-sided independent sample t test was performed using GraphPad Prism version 4.02 for Windows.
Lactate dehydrogenase (LDH) and malate dehydrogenase assay.
Cells were grown for 90 min in LB medium at 37°C, washed 3 times with sodium phosphate buffer (50 mM, pH 6.75), resuspended in lysis buffer (50 mM sodium phosphate buffer, 1 mM dithiothreitol [DTT], 0.5 mg/ml lysozyme, 1× Complete protease inhibitor cocktail [Roche]; pH 6.75), and kept on ice for 30 min. Cells were lysed by sonication, centrifuged at 5,000 rpm at 4°C for 5 min, and then centrifuged at 13,000 rpm at 4°C for 30 min. The total protein concentration was determined spectrophotometrically by using Bradford reagent.
Lactate dehydrogenase activity was assayed by incubation of cleared lysates with a reaction mixture containing 100 mM morpholinepropanesulfonic acid (MOPS), 5 mM MgCl2, 5 mM DTT, and 0.5 mM NADH. Malic enzyme activity was determined by incubation of cleared lysates with a reaction mixture containing 50 mM Tris-HCl (pH 7.5) and 2 mM NAD+. Addition of 5 mM pyruvate or 50 mM malate started the reaction, and absorbance at 365 nm was monitored to assess NADH oxidation or NAD+ reduction. To compare enzyme activities of the different strains, a two-sided independent sample t test was performed using GraphPad Prism version 4.02 for Windows.
Chromosomal transcriptome analysis.
Plasmid pLS20-containing cells and control cells were harvested from three independently growing cultures after 30, 90, and 480 min of growth (t30, t90, and t480, respectively) in LB medium at 37°C. Total RNA was extracted by using the hot acid-phenol isolation procedure as detailed before (17). The isolated RNA samples were assayed for integrity and purity by agarose gel electrophoresis, and the concentration was measured via absorbance at 260 nm. Reverse transcription of RNA, cDNA labeling, and hybridization of the probes were performed as described before (17). From each time point at least three replicates of every comparison were hybridized to a B. subtilis microarray (18, 19) and included in the analysis. Slides were scanned on a GMS 418 apparatus (Genetic Microsystems, Inc.). Separate images were acquired for Cy3 and Cy5 and then processed and analyzed with GenePix Pro 6.0 software (Axon Instruments, Inc.).
Data analysis.
After background subtraction, signal intensities for each replica were normalized and statistically analyzed using the Lowess intensity-dependent normalization method (20) included in the Almazen System software (Alma Bioinformatics S.L.). P values were calculated with the Student t test algorithm based on the differences between log2 ratio values for each biological replicate. Genes were considered differentially expressed when they fulfilled the following filter parameters: expression ratio, ≥1.8; P ≤ 0.2.
Plasmid transcriptome analysis.
The plasmid microarray contains the 84 open reading frames of pLS20 and, as controls, 6 randomly designed oligomers as well as 7 oligomers representing Escherichia coli genes. Oligonucleotides with a length of 65 bp were designed and purchased from Sigma-Aldrich Chemie GmbH and attached in duplicates to Ultra-GAPS-coated glass slides (Corning Life Sciences). RNAs deriving from pLS20-containing cells were used to compare the time points after 30 min and 90 min to the time point after 480 min. Each hybridization was performed in biological triplicates and scanning of slides as well as image and data analysis were performed as described for the chromosomal transcriptome analysis.
Quantitative RT-PCR.
Two independent cultures of pLS20-carrying and pLS20-free cells were harvested after 90 min of growth in LB medium at 37°C, and pellets were stored at −80°C. Total RNA was isolated using a PureLink RNA Minikit (Ambion), and isolated RNAs were subjected to DNase I treatment using a Turbo DNA-free kit (Ambion). Reverse transcription was performed with a ProtoScript II first-strand synthesis kit, random hexamers, and 500 ng of DNase I-treated RNA. For quantitative reverse transcription-PCR (RT-PCR), cDNAs were diluted 1/10 in nuclease-free water and added to SsoAdvanced SYBR green supermix (Bio-Rad) containing 300 nM gene-specific primers (see Table S2 in the supplemental material). Quantitative RT-PCR was performed on a CFX96 touch real-time PCR detection system (Bio-Rad) according to the recommended protocol. CFX Manager Software (Bio-Rad) was used to analyze the data, and the mean relative expression ratio of the genes was calculated by the comparative threshold cycle (CT) method (17). The Relative Expression Software Tool V2.0.13 was used to determine the statistical significance. Quantitative RT-PCRs were performed in duplicate for each biological replicate.
RESULTS AND DISCUSSION
Plasmid pLS20 substantially changes gene transcription of the host genome during mid-exponential growth.
We have noticed that cells carrying pLS20 show differences in their handling in the laboratory, e.g., pLS20 cells from the mid-exponential phase form much tighter pellets during centrifugation than pLS20-free cells of the same strain background (data not shown). We wondered if the presence of the plasmid and/or the accompanied conjugation activity of the plasmid during exponential growth alters the physiology of cells by changing the transcription of genes on the host chromosome. Therefore, we performed transcriptional profiling using whole genome arrays of B. subtilis. RNA was isolated from cells carrying pLS20cat (here termed pLS20 for simplicity), a version with a cat resistance cassette integrated into a UV resistance gene for the selection of transfer and, as reference, from cells lacking the plasmid that have grown under identical conditions. To exclude effects of metabolic burden or indirect consequences of differing growth rates on the transcription profile, we investigated strain NCIB3610 as the host for pLS20. Whereas cells of the laboratory strain PY79 containing pLS20 grew mildly but noticeably slower than pLS20-free cells, the wild-type isolate NCIB3610 did not show any difference in growth rate or in attainment of final optical density in rich or in minimal medium (Fig. S1A in the supplemental material and data not shown), revealing that 3610 is a suitable host to study growth rate-independent effects due to the presence of pLS20. Of note, NCIB naturally contains another plasmid, pBS32 (GenBank accession number NC_02809.1). We chose three time points for our analysis, 30 min after addition of fresh medium to stationary-phase cells, where conjugation activity commences, after 90 min, where conjugation activity is maximal (OD of 0.4) (Fig. S1B in the supplemental material), and 8 h postinduction, when cells entered stationary-phase growth. DNA arrays were run in parallel and were repeated 3 times (three independent experiments using three cultures per strain); only genes that were consistently more than 1.8-fold up- or downregulated in each experiment were judged as truly altered in expression due to the presence of pLS20.
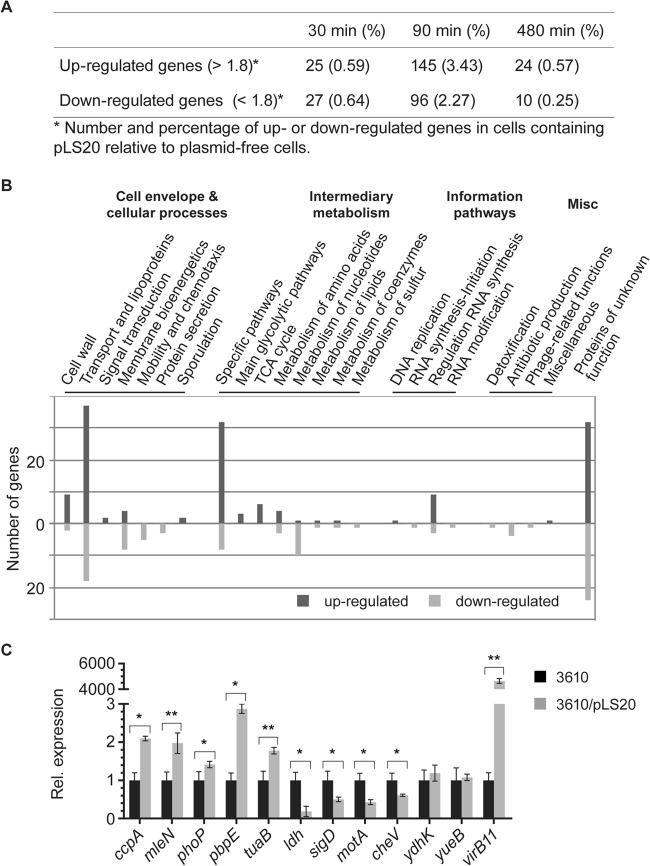
We found a total of 145 genes that were consistently induced at t90 and 96 genes whose synthesis was repressed at this time point. At t30, only 52 genes in total showed differential expression, and at t480, 34 genes showed significant changes in expression (Fig. 1A; also see Table S1 in the supplemental material). Thus, during mid-exponential growth and maximal conjugational activity of pLS20, almost 5% of the genes on the host genome show differential expression, whereas only minor changes take place at the other time points under nutrient-rich conditions. There was very little overlap of induced or repressed genes between 30 and 90 or 90 and 480 min (see Table S1 in the supplemental material). Only 7 genes were induced at t30 and also at t90, showing a sharp incline of the induction/repression of the 241 genes at t90. For a majority of the affected genes, induction rates were altered around 2-fold (see Table S1 in the supplemental material for P values), but some genes (e.g., ldh, lctP, and yvfV) showed more than 7-fold differences in their expression (see Table S1 in the supplemental material).
FIG 1.
Transcriptional profiling of cells containing the conjugative plasmid pLS20. (A) Table showing overall changes in transcription levels between cells carrying or lacking pLS20. (B) Summary of transcriptional changes due to the presence of pLS20 in Bacillus subtilis strain 3610 during mid-exponential growth. Chromosomal genes affected in plasmid-carrying cells were grouped and listed according to functional categories of the SubtiList database (http://genolist.pasteur.fr/SubtiList/). The presence of pLS20 mainly affects the expression of genes that encode proteins belonging to the cell envelope and cellular processes, such as transporters, lipoproteins, and cell wall and membrane bioenergetics, as well as to the intermediary metabolism. Misc, miscellaneous. (C) Quantitative RT-PCR analysis of selected genes affected by the presence of the conjugative plasmid pLS20 during mid-exponential growth. Shown is the relative (Rel.) expression of genes from pLS20-carrying cells compared to that of pLS20-free cells. The expression level of the indicated genes was internally normalized to the transcript levels of sigA and rpsJ. RNA was isolated from two independently growing cultures of each strain, and quantitative RT-PCRs were performed in duplicate for each culture (n = 4). Errors bars represent standard deviations from the means, and statistical significance was calculated using the Relative Expression Software Tool V2.0.13 (*, P < 0.05; **, P < 0.01).
To verify the changes observed in the microarrays during mid-exponential growth, we performed quantitative RT-PCR on 12 representative genes. For 10 genes, we found similar tendencies and ranges of induction or repression, respectively (Fig. 1C), while for 2 genes, we found no significant change in transcription. Thus, the differences in transcription levels observed in the arrays largely represent actual changes in RNA levels.
A majority of changes affect central metabolic pathways and the cell envelope.
A large fraction of the genes identified with altered transcriptional patterns falls into the group of proteins with functions in the cell envelope, such as various transporters and lipoproteins, enzymes involved in cell wall synthesis, or proteins involved in membrane bioenergetics (Fig. 1B; also see Table S1 in the supplemental material). A marked increase in the transcription of genes responsible for the synthesis of teichuronic acids may reflect the need for an altered cell wall structure, which also occurs under circumstances of phosphate-limiting conditions. When Bacillus cells are starved for phosphate, the phosphate-rich teichoic acids are replaced by phosphate-free teichuronic acids (21). Additionally, the plasmid-containing cells showed an upregulation of 10 other genes that respond to phosphate starvation and that are under the control of PhoP (see Table S1 in the supplemental material). Therefore, we examined the growth of cells that were starved for phosphate but could not observe any difference in growth rate between the cells lacking or containing pLS20 (data not shown).
A second large group of genes is involved in metabolism, mainly in the utilization of alternative carbon sources, but also in carbon core metabolism and in metabolism of nucleotides or amino acids (Fig. 1B; also see Table S1 in the supplemental material). Remarkably, there is a correlation between the upregulated ABC- or phosphotransferase system (PTS)-dependent transporters and the enzymes that further degrade the transported carbohydrates into utilizable monosaccharides. LB medium provides only a small amount of carbohydrates such that cells like Escherichia coli mainly gain energy through the utilization of amino acids (19). It appears that pLS20-containing cells change their metabolism by increasing the expression of at least 12 alternative carbohydrate transporters and the corresponding enzymes to make these sugars accessible to the pentose-phosphate pathway and further the tricarboxylic acid (TCA) cycle. Additionally, plasmid-containing cells seem to use overflow carbohydrates such as lactate or acetoin to either antiport against malate or to redirect acetoin to the TCA cycle via the synthesis of acetyl-coenzyme A (CoA).
A third large group of differently expressed genes are those acting in signal transduction and regulatory pathways (Fig. 1B), most notably several transcriptional regulators (see Table S1 in the supplemental material). Associated with this group are flotillin T and its associated NfeD2 protein (22), which are involved in signaling via the organization of proteins, including a histidine kinase, within defined places in the membrane (23), emphasizing the importance of membrane-associated changes due to pLS20. Among the upregulated proteins that are transcriptional regulators are DnaA and CcpA (see Table S1 in the supplemental material). DnaA, the replication initiator protein, also acts as a transcription factor and directly regulates the expression of at least 20 operons and, as a result, more than 50 genes (24). Presumably, DnaA acts as an initiator of pLS20 replication as well, since pLS20 does not possess an endogenous replication protein, as is the case for RepN from plasmid pLS32, and shows perfect consensus DnaA-binding sequences in its oriCLS20 region (25, 26). Catabolite control protein A, CcpA, is involved in the regulation of carbohydrate metabolism and mediates catabolite repression but also catabolite activation (27). Indeed, CcpA targets nearly 30% of the genes which are upregulated after 90 min of growth but only 6% of the genes that are downregulated. However, 51 genes of the 58 upregulated genes under the control of CcpA are also controlled by additional factors, e.g., AbrB (regulating 19 out of the 51 genes) or CodY, such that a clear determination of the connection between increased CcpA levels and changes in transcription of genes identified in this analysis is not straightforward. Moreover, up- and downregulated genes identified include the transcription factor PhoP (see Table S1), which may also have an effect on several changes that were observed. Interestingly, the expression of TasA was increased. TasA is an amyloid-like protein that supposedly builds up filaments connecting B. subtilis cells in biofilms (15). We tested if tasA mutant cells are less efficient in conjugational activity but did not find a difference from nonmutant cells (Fig. 2).
FIG 2.
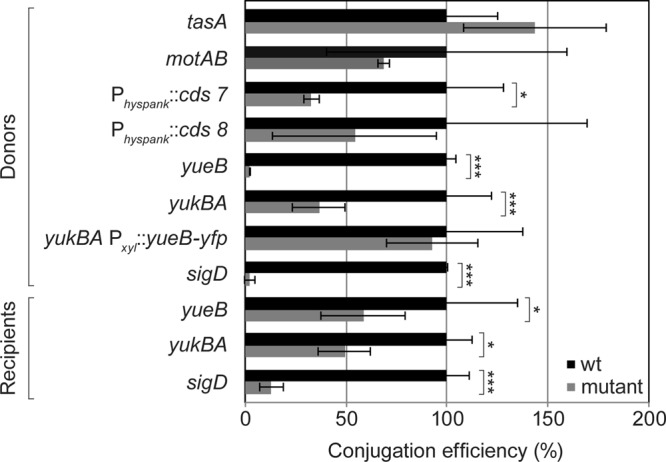
Relative conjugational transfer efficiencies of plasmid pLS20. The efficiencies were assessed in donor mutant strains and recipient mutant strains (gray) and compared to those of wild-type donors and wild-type recipients (black) from the same genetic background. To test for donor efficiency, the recipient strain was always kept the same and the genetic background of the donor strain varied. To test the ability to receive the plasmid from the wild-type strain, the donor strain was kept the same and the genetic background of the recipients varied. Each experiment was performed at least in triplicate, and the conjugation efficiency was calculated as the number of transconjugants (CFU) per number of donors (CFU). For comparing relative levels, the efficiency of wild-type donor or wild-type recipient cells were set to 100%. Error bars represent standard deviations. Statistical significances were computed using two-sided independent sample t test. P < 0.05 (*) and P < 0.001 (***) compared to the pLS20-free strain.
Among downregulated genes are those involved in motility, whose transcription is controlled by the alternative transcription factor sigma D (sigD, cheV, mcpA, mcpB, motA, motB, lytA, and others) and of genes involved in pyrimidine biosynthesis (Fig. 1B; also see Table S1 in the supplemental material). Interestingly, sigma D mutant cells showed a 50-fold reduction in conjugation efficiency as donors but also as recipients (Fig. 2). Because the loss of activity is also seen in recipient cells, it is probably an indirect effect, most likely through the structure of the cell wall, because in sigD mutant cells, major cell wall-modifying enzymes are not expressed (16). However, changes in sigD-regulated genes were between 2- and 4-fold (see Table S1 in the supplemental material), which are unlikely to account for the strong effect of the sigD deletion on conjugation. Thus, conjugational activity of pLS20 is accompanied by considerable changes in gene expression that will affect envelope structure and metabolism, as well as a variety of other physiological parameters of the host cell.
Altered transcription patterns are reflected in changes in host physiology, most markedly through increased stress resistance.
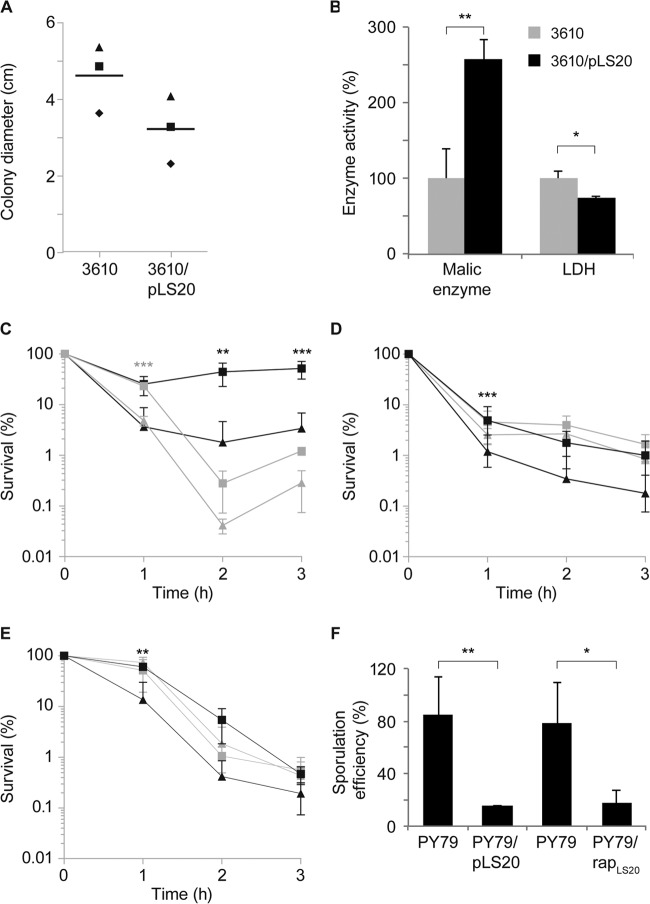
Based on the data from the transcriptional profiling of pLS20-containing cells, we wondered if changes in gene expression lead to noticeable changes in cellular physiology. We tested the phenotypes associated with some of the genes showing altered transcription during mid-exponential growth. As motility genes were downregulated, we performed motility assays on soft agar plates. Cells carrying pLS20 were moderately less motile than pLS20-free cells (Fig. 3A). A reduction in motility may increase the contact time between host and recipient cells, increasing transfer efficiency. To test this, we assayed motAB mutants, which are completely immobile (28), for conjugation activity but did not observe a decrease in conjugation efficiency (Fig. 2). However, an increased contact time in motAB mutant cells may be counteracted by their inability to move at all. To test this point, we investigated if different shaking conditions affect conjugational activity of pLS20. The highest transfer efficiency was seen in nonshaking and mildly shaking cultures, while an increased rate of shaking of the culture strongly reduced conjugation rates (see Fig. S2 in the supplemental material). Thus, the decrease in motility appears to be an effect generated through mating pair formation by the pLS20-encoded T4SS. Likewise, it has been reported that a derepressed derivative of the F-like plasmid R1, R1drd19, which conjugates constitutively, strongly reduces motility in E. coli K-12 due to increased cell aggregation (29).
FIG 3.
Conjugative plasmid pLS20 changes the cellular physiology of its host. (A) The conjugative plasmid pLS20 slows down the swimming motility behavior of its host. Cells were grown to the mid-exponential growth phase and adjusted to the same OD, and the same volumes of cultures were centrally spotted on soft agar plates containing 0.3% (wt/vol) agar and incubated at 37°C for 5 h. The diameter of colony spreading was determined from at least 10 plates for each experiment, and the means of each experiment are plotted in corresponding symbols for each strain. A horizontal line shows the means of the three experiments. The P value was calculated by using a two-sided dependent sample t test. P = 0.0045 compared to the pLS20-free strain. (B) Malic enzyme and lactate dehydrogenase (LDH) activity of plasmid-free strains and strains containing the conjugative plasmid pLS20. Cells were grown to the mid-exponential-growth phase and harvested, and the lysate was tested for the specific activity of malic enzymes or LDH by following the absorption of NADH at 365 nm. Error bars denote standard deviations. P values were calculated by using two-sided independent sample t tests. P < 0.05 (*) and P < 0.01 (**) compared to the pLS20-free strain. (C to E) Survival of cells containing pLS20 (squares) or lacking pLS20 (triangles) under different stress conditions. Cells were grown to the mid-exponential growth phase and were treated with 5% (wt/vol) sodium chloride (black) or 10% (vol/vol) ethanol (gray) (C), exposed to acid stress at pH 4.5 (black) or alkaline stress at pH 9 (gray) (D), and exposed to cold shock at 15°C (black) or heat shock at 54°C (gray) (E). Survival was assayed by plating appropriate dilutions after 1 h, 2 h, and 3 h. Each time point is the average from three independent experiments, and error bars denote standard deviations. P values were calculated by using two-way ANOVA. Significant differences in survival of the pLS20-containing strain compared to the pLS20-free strain were found under conditions of salt, ethanol, acid, and cold stress (P < 0.005). Bonferroni's posttest was used for pairwise comparisons. P < 0.01 (**) and P < 0.001 (***) compared to the plasmid-free strain. (F) The conjugative plasmid pLS20 inhibits sporulation of its host. Cells were grown in Difco sporulation medium (DSM) for 24 h at 37°C and plated in triplicate. PY79/rapLS20 denotes induction of RapLS20 expression from an ectopic site on the chromosome in cells lacking pLS20. Data were derived from three independent experiments, and error bars denote standard deviations. P values were calculated by using two-sided independent sample t tests. P < 0.05 (*) and P < 0.01 (**) compared to strain PY79.
The expression of lactate dehydrogenase (ldh), converting pyruvate to l-lactate, was reduced during mid-exponential growth in plasmid-carrying cells. We tested cells 90 min after addition of fresh medium for LDH activity, which was markedly lower in cells carrying pLS20 than in plasmid-free cells (Fig. 3B). We also found a more than 2-fold increase in the activity of malic enzyme converting malate to pyruvate in cells carrying pLS20, which we tested because of the observed upregulation of malate transporters mleN and maeN and upregulation of the malic enzyme mleA in the strain carrying pLS20 (see Table S1 in the supplemental material). mleN and mleA are thought to be cotranscribed and were shown to exhibit higher transcriptional expression in rich medium than in malate or glucose minimal medium during exponential growth (30). mleN encodes a malate/lactate antiporter synchronously coupling Na+ efflux with H+ import (31). Taking these findings together, pLS20 seems to drive host metabolism toward enhanced utilization of malate, which is the preferred carbon source of B. subtilis in addition to glucose (32), while pLS20 appears to decrease the consumption of lactate in favor of antiport against malate. Given that malate is rapidly cometabolized with glucose (32) and is highly abundant in the soil due to its secretion by plants, it seems a rational design to maintain growth rates similar to those of the isogenic plasmid-free strains by the utilization of malate. Thus, altered transcription levels in pLS20-carrying cells can be seen to affect enzyme activities and to redirect metabolism of its host.
The most drastic change in cellular physiology was found in relation to stress resistance. Cells carrying pLS20 were slightly but measurably more resistant to the addition of 10% (vol/vol) ethanol (Fig. 3C) and, quite strikingly, were more resistant to salt stress than cells lacking pLS20. The addition of 5% (wt/vol) NaCl to exponentially growing cells led to pronounced cell death in cells devoid of pLS20, while in the presence of the plasmid, cells recovered 1 h after the addition of high salt and started to grow after 2 h (Fig. 3C). Increased stress resistance of plasmid-carrying cells was also seen for acid shock (Fig. 3D), while alkaline stress was not significantly altered (Fig. 3D). Additionally, pLS20 conferred cold stress resistance to its host (Fig. 3E) but had no significant effect on heat shock treatment (Fig. 3E). Thus, the activity of pLS20 increases fitness of cells in relation to several different stress conditions but with a certain specificity, for example, cold shock represses the induction of heat shock genes and vice versa (33), and different protection mechanisms are needed for acid or alkaline stress. To identify relevant stress-related genes, we analyzed the changes in transcription on the host genome. Transcription of ydhK, ybyB, and pbpE, which belong to the sigB and sigW regulons, were increased during mid-exponential growth. However, sigB mutant cells show only a minor growth defect upon survival after salt stress (34), so the reason for salt resistance and other stress resistances in pLS20-containing cells is likely multifactorial. These experiments show that pLS20 increases stress fitness of its host and leads to changes in enzyme activity and in motility. Thus, several changes observed at the transcriptional level translate into changes in the physiology of the cells.
pLS20 contains an orf encoding a protein similar to Rap proteins (named RapLS20), which downregulate sporulation via dephosphorylation of a response regulator within the phosphorelay (35). To test if the presence of pLS20 has an effect on sporulation efficiency, we tested plasmid-containing and pLS20-free cells for the ability to generate heat-resistant spores. Interestingly, pLS20-containing cells showed a more than 4-fold reduction in sporulation efficiency (Fig. 3F), showing that like a plasmid in another nondomesticated B. subtilis strain (36), pLS20 reduces sporulation capacity of host cells. To test if RapLS20 confers this effect or affects conjugation efficiency, we expressed the protein from an ectopic site on the B. subtilis chromosome in cells lacking pLS20. Induction of RapLS20 reduced sporulation efficiency to a degree very similar to that of the presence of pLS20 (Fig. 3F), indicating that RapLS20 is responsible for the effect. Conversely, overexpression of RapLS20 did not affect the efficiency of plasmid transfer (data not shown), suggesting that the protein does not play a role in the regulation of conjugation activity of pLS20. We speculate that pLS20 prefers to be present in cells that do not respond to environmental stress via sporulation to ensure the most rapid spreading while conferring some general stress resistance to the cells in order to better survive adverse environmental conditions.
Transcriptional analysis of plasmid-borne genes during exponential growth.
To investigate if the conjugation operon or any of the other genes present on the 65-kbp large pLS20 are subject to transcriptional control under the conditions tested above, we printed an array representing 84 open reading frames of pLS20. Total RNA was extracted at the three time points as described above from cells carrying pLS20. The abundance of RNA derived from early- and mid-exponentially growing cells (t30 and t90, respectively) was compared to that of RNA derived from stationary-phase cells (t480) in order to obtain relevant information of qualitative and quantitative changes in expression levels between these growth stages.
Figure 4 shows the transcriptional changes of the genes corresponding to their linear location on the plasmid for the two comparisons made (t30 versus t480 and t90 versus t480). Interestingly, the plasmid can be divided into two halves according to the transcriptional changes of the genes. The first half of the plasmid (with respect to oriCLS20) mainly represents genes that are upregulated during early- and mid-exponential growth and that can mostly be assigned to the transfer region of pLS20. The operon comprising the identified and putative conjugation proteins cds12 to cds35, including virB1, virB4, virB11, virD4, and virD2, was already induced 30 min after inoculation and was still induced 60 min later (t90) but to an overall lower extent, implying that it is transiently induced during early and mid-exponential growth. Thus, induction of the conjugation genes parallels the conjugation activity of pLS20, showing that the conjugation-proficient state is likely regulated at the transcriptional level.
Two genes (cds7 and cds8) lying directly upstream of the conjugation region coclustered with the operon (data not shown) and were included for further analysis. Homology searches revealed that cds7 bears a conserved H-E-X-X-H motif found in metalloproteases and has similarity to transcriptional regulators, while cds8 showed no homology to known genes. Nevertheless, we tested both for their role as potential transcriptional regulators and introduced additional copies into the chromosome. Overexpression of cds7 resulted in a mild reduction of conjugation efficiency compared to wild-type cells, whereas no effect could be seen for overexpression of cds8 (Fig. 2). Thus, CDS7 is unlikely to be a direct regulator of the conjugation operon of pLS20.
In contrast, in the second half of pLS20, several genes (e.g., the genes encoding the actin-like Alp7AR plasmid segregation system) were downregulated during early growth. In addition, the most negatively affected genes were cds54 (−25.6), coding for a UV damage repair protein, cds81 (−8.5), containing a conserved VanZ superfamily domain, cds82 (−34.1) and cds45 (−4.2), which do not share sequence homology to genes with known function, and cds44 (−8.5), a putative recombinase.
In conclusion, transcriptome analysis of pLS20 showed that changes on the host chromosome are accompanied by changes in transcription on pLS20 (mostly in the transfer region of the plasmid) but did not reveal a potential candidate explaining the changes seen on the host chromosome.
Conjugation efficiency is reduced in the absence of YueB in host cells.
Among the chromosomally induced genes was yueB, reported to be the receptor of phage SPP1 (37). Transcription of yueB was increased already after 30 min of inoculation, and the other genes of the operon, yukABCDE (gene order yukE-D-C-B-A-yueB), were also upregulated after 90 min (Table 1; also see Table S1 in the supplemental material). Being a receptor for a phage is not the primary function of yueB, and although YukBA displays similarity to SpoIIIE/FtsK-like DNA translocases and YukE to ESAT-6-like WXG100 proteins involved in virulence in Mycobacteria (38), no cellular function has yet been assigned to any of these genes. We found that conjugation efficiency was highly compromised in a yueB mutant strain (Fig. 2). Likewise, conjugation activity was strongly reduced in the absence of YukBA (formerly thought to be two genes), which is encoded upstream of yueB. Based on its similarity to DNA translocases SpoIIIE and SftA (39), YukBA could be used to export proteins or import DNA. To test for a polar effect of the yukBA deletion on yueB (which is at the end of the operon), we provided a yueB copy in trans on the chromosome. During ectopic expression of YueB, the yukBA deletion strain no longer showed a reduction in conjugation efficiency (Fig. 2), showing that YukBA does not play a role in conjugation, while YueB provides an important activity for this process. Conversely, the absence of YueB in recipient cells had a minor effect on conjugation efficiency (Fig. 2), while sigD mutant cells were very poor as donors as well as recipients cells for pLS20 (Fig. 2). These data suggest that YueB is involved in DNA transfer during conjugation and that pLS20 employs this host protein for efficient DNA transfer.
Conclusions.
We characterize an autonomously replicating and self-transferring plasmid which considerably affects global transcription of the host genome and its physiology without being a metabolic burden. It has been known before that conjugative plasmids can alter distinct aspects of their host cells, e.g., increase the efficiency of biofilm formation (40), sporulation, or competence (13, 36). However, conjugative plasmid pLS20 can induce considerable genome-wide changes in a Gram-positive host, even under nutrient-rich growth conditions. About 5% of the host genes are differentially transcribed during the conjugation-proficient growth phase, which in turn leads to marked changes in several cellular aspects. Major changes were seen in the expression of a variety of metabolic enzymes, of membrane proteins, as well as of proteins involved in cell wall synthesis. Most of the changes thus affected cellular metabolism and the protein composition of the cell membrane and wall. The large number of changes may reflect a reorganization of the cell envelope facilitating transfer of pLS20 into a recipient cell. Because this study is, to our knowledge, the first description of a global transcriptional response of a Gram-positive host to a conjugative plasmid, one cannot tell if this type of change is a general scheme for conjugative elements. On the phenotypic level, the presence of pLS20 led to reduced motility of cells, which we show is favorable for mating pair formation during conjugation, to changes in enzyme activities, and, most notably, to increased resistance to several different stress conditions. pLS20-containing cells were more resistant to salt, ethanol, cold, and acid stress than plasmid-free cells. Other stress conditions, like heat shock or alkaline stress, were not strongly affected, showing some degree of specificity of the generated stress resistance. Taken together, these factors increase host fitness and put B. subtilis into a somewhat different physiological state. Thus, conjugative plasmids provide interesting features for biotechnological applications, not only as mobile DNA transfer systems but also as factors increasing the fitness of cells during growth, at least in the case of pLS20.
Conjugation activity of pLS20 is paralleled by the induction of a large putative conjugation operon, which contains all conserved transfer proteins in addition to many genes of unknown function. Thus, as is the case for many conjugative plasmids, DNA transfer is regulated at the transcriptional level for pLS20. We do not yet know if the changes seen on the host chromosome are a direct consequence of the induction of the conjugation operon and if there is a master regulator governing the induction on pLS20. However, it remains an intriguing observation that the changes observed on the chromosome occur in the same time window as the maximal induction of the conjugation operon. Therefore, our work reveals that instead of being a metabolic burden to cells that gives some genetic advantage to host cells (e.g., antibiotic resistance), plasmids can broadly alter the properties and physiology of their host cell and be largely beneficial for the cells while at the same time inducing changes in the host that may be beneficial for their transfer. It will be interesting to investigate how the connection between transcriptional changes on the plasmid and on the host chromosome operates at a molecular level.
Supplementary Material
ACKNOWLEDGMENTS
We thank Carolina González de Figueras (CSIC-INTA) for crucial technical assistance, Ivan Berg for help with enzyme assays, and Mario Mergelsberg for help with the quantitative RT-PCR experiment (University of Freiburg). We thank Daniel B. Kearns (Indiana University) for providing us with a motAB::tet strain and Dieter Hauschke (University of Freiburg) for help with statistical analysis.
This work was supported by the German Research Foundation (DFG), by the Excellence Initiative of the German Research Foundation (GSC-4, Spemann Graduate School), by the Centre for Synthetic Microbiology, SYNMIRKO, in Marburg, Germany, and by Spanish Ministry of Science and Innovation grants CGL2012-39627-C03-02 and CONSOLIDER Ingenio 2010, CE-CSD2007-00005.
Footnotes
Published ahead of print 13 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03154-13.
REFERENCES
- 1.Cascales E, Christie PJ. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137–149. 10.1038/nrmicro753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Llosa M, Gomis-Ruth FX, Coll M, de la Cruz F. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1–8. 10.1046/j.1365-2958.2002.03014.x [DOI] [PubMed] [Google Scholar]
- 3.Fronzes R, Christie PJ, Waksman G. 2009. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 7:703–714. 10.1038/nrmicro2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grohmann E, Muth G, Espinosa M. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277–301. 10.1128/MMBR.67.2.277-301.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73:775–808. 10.1128/MMBR.00023-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derman AI, Becker EC, Truong BD, Fujioka A, Tucey TM, Erb ML, Patterson PC, Pogliano J. 2009. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol. Microbiol. 73:534–552. 10.1111/j.1365-2958.2009.06771.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koehler TM, Thorne CB. 1987. Bacillus subtilis (natto) plasmid pLS20 mediates interspecies plasmid transfer. J. Bacteriol. 169:5271–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer T, Rosch T, Itaya M, Graumann PL. 2011. Localization pattern of conjugation machinery in a Gram-positive bacterium. J. Bacteriol. 193:6244–6256. 10.1128/JB.00175-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyakoshi M, Shintani M, Terabayashi T, Kai S, Yamane H, Nojiri H. 2007. Transcriptome analysis of Pseudomonas putida KT2440 harboring the completely sequenced IncP-7 plasmid pCAR1. J. Bacteriol. 189:6849–6860. 10.1128/JB.00684-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyakoshi M, Nishida H, Shintani M, Yamane H, Nojiri H. 2009. High-resolution mapping of plasmid transcriptomes in different host bacteria. BMC Genomics 10:12. 10.1186/1471-2164-10-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dionisio F, Conceicao IC, Marques AC, Fernandes L, Gordo I. 2005. The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol. Lett. 1:250–252. 10.1098/rsbl.2004.0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konkol MA, Blair KM, Kearns DB. 2013. Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. J. Bacteriol. 195:4085–4093. 10.1128/JB.00696-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh PK, Ramachandran G, Duran-Alcalde L, Alonso C, Wu LJ, Meijer WJ. 2012. Inhibition of Bacillus subtilis natural competence by a native, conjugative plasmid-encoded comK repressor protein. Environ. Microbiol. 14:2812–2825. 10.1111/j.1462-2920.2012.02819.x [DOI] [PubMed] [Google Scholar]
- 14.Itaya M, Sakaya N, Matsunaga S, Fujita K, Kaneko S. 2006. Conjugational transfer kinetics of pLS20 between Bacillus subtilis in liquid medium. Biosci. Biotechnol. Biochem. 70:740–742. 10.1271/bbb.70.740 [DOI] [PubMed] [Google Scholar]
- 15.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 59:1229–1238. 10.1111/j.1365-2958.2005.05020.x [DOI] [PubMed] [Google Scholar]
- 16.Kearns DB, Losick R. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 19:3083–3094. 10.1101/gad.1373905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lioy VS, Machon C, Tabone M, Gonzalez-Pastor JE, Daugelavicius R, Ayora S, Alonso JC. 2012. The zeta toxin induces a set of protective responses and dormancy. PLoS One 7:e30282. 10.1371/journal.pone.0030282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zafra O, Lamprecht-Grandio M, de Figueras CG, Gonzalez-Pastor JE. 2012. Extracellular DNA release by undomesticated Bacillus subtilis is regulated by early competence. PLoS One 7:e48716. 10.1371/journal.pone.0048716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. 10.1093/nar/30.4.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eymann C, Mach H, Harwood CR, Hecker M. 1996. Phosphate-starvation-inducible proteins in Bacillus subtilis: a two-dimensional gel electrophoresis study. Microbiology 142:3163–3170. 10.1099/13500872-142-11-3163 [DOI] [PubMed] [Google Scholar]
- 22.Dempwolff F, Moller HM, Graumann PL. 2012. Synthetic motility and cell shape defects associated with deletions of flotillin/reggie paralogs in Bacillus subtilis and interplay of these proteins with NfeD proteins. J. Bacteriol. 194:4652–4661. 10.1128/JB.00910-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez D, Kolter R. 2010. Functional microdomains in bacterial membranes. Genes Dev. 24:1893–1902. 10.1101/gad.1945010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goranov AI, Katz L, Breier AM, Burge CB, Grossman AD. 2005. A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc. Natl. Acad. Sci. U. S. A. 102:12932–12937. 10.1073/pnas.0506174102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka T, Ishida H, Maehara T. 2005. Characterization of the replication region of plasmid pLS32 from the natto strain of Bacillus subtilis. J. Bacteriol. 187:4315–4326. 10.1128/JB.187.13.4315-4326.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meijer WJ, de Boer AJ, van Tongeren S, Venema G, Bron S. 1995. Characterization of the replication region of the Bacillus subtilis plasmid pLS20: a novel type of replicon. Nucleic Acids Res. 23:3214–3223. 10.1093/nar/23.16.3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonenshein AL. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5:917–927. 10.1038/nrmicro1772 [DOI] [PubMed] [Google Scholar]
- 28.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320:1636–1638. 10.1126/science.1157877 [DOI] [PubMed] [Google Scholar]
- 29.Barrios AF, Zuo R, Ren D, Wood TK. 2006. Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol. Bioeng. 93:188–200. 10.1002/bit.20681 [DOI] [PubMed] [Google Scholar]
- 30.Lerondel G, Doan T, Zamboni N, Sauer U, Aymerich S. 2006. YtsJ has the major physiological role of the four paralogous malic enzyme isoforms in Bacillus subtilis. J. Bacteriol. 188:4727–4736. 10.1128/JB.00167-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei Y, Guffanti AA, Ito M, Krulwich TA. 2000. Bacillus subtilis YqkI is a novel malic/Na+-lactate antiporter that enhances growth on malate at low protonmotive force. J. Biol. Chem. 275:30287–30292. 10.1074/jbc.M001112200 [DOI] [PubMed] [Google Scholar]
- 32.Kleijn RJ, Buescher JM, Le Chat L, Jules M, Aymerich S, Sauer U. 2010. Metabolic fluxes during strong carbon catabolite repression by malate in Bacillus subtilis. J. Biol. Chem. 285:1587–1596. 10.1074/jbc.M109.061747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graumann P, Schroder K, Schmid R, Marahiel MA. 1996. Cold shock stress-induced proteins in Bacillus subtilis. J. Bacteriol. 178:4611–4619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volker U, Maul B, Hecker M. 1999. Expression of the σB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perego M, Hoch JA. 1996. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 93:1549–1553. 10.1073/pnas.93.4.1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parashar V, Konkol MA, Kearns DB, Neiditch MB. 2013. A plasmid-encoded phosphatase regulates Bacillus subtilis biofilm architecture, sporulation, and genetic competence. J. Bacteriol. 195:2437–2448. 10.1128/JB.02030-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sao-Jose C, Baptista C, Santos MA. 2004. Bacillus subtilis operon encoding a membrane receptor for bacteriophage SPP1. J. Bacteriol. 186:8337–8346. 10.1128/JB.186.24.8337-8346.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coros A, Callahan B, Battaglioli E, Derbyshire KM. 2008. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol. Microbiol. 69:794–808. 10.1111/j.1365-2958.2008.06299.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaimer C, Gonzalez-Pastor JE, Graumann PL. 2009. SpoIIIE and a novel type of DNA translocase, SftA, couple chromosome segregation with cell division in Bacillus subtilis. Mol. Microbiol. 74:810–825. 10.1111/j.1365-2958.2009.06894.x [DOI] [PubMed] [Google Scholar]
- 40.Ghigo JM. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442–445. 10.1038/35086581 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.