Abstract
Listeriosis is caused by the food-borne pathogen Listeria monocytogenes, which can be found in seafood and processing plants. To evaluate the risk to human health associated with seafood production in New Zealand, multi-virulence-locus sequence typing (MVLST) was used to define the sequence types (STs) of 31 L. monocytogenes isolates collected from seafood-processing plants, 15 from processed foods, and 6 from human listeriosis cases. The STs of these isolates were then compared with those from a collection of seafood isolates and epidemic strains from overseas. A total of 17 STs from New Zealand clustered into two lineages: seafood-related isolates in lineages I and II and all human isolates in lineage II. None of the New Zealand STs matched previously described STs from other countries. Isolates (belonging to ST01-N and ST03-N) from mussels and their processing environments, however, were identical to those of sporadic listeriosis cases in New Zealand. ST03-N isolates (16 from mussel-processing environments, 2 from humans, and 1 from a mussel) contained an inlA premature stop codon (PMSC) mutation. Therefore, the levels of invasiveness of 22 isolates from ST03-N and the three other common STs were compared using human intestinal epithelial Caco-2 cell lines. STs carrying inlA PMSCs, including ST03-N isolates associated with clinical cases, had a low invasion phenotype. The close relatedness of some clinical and environmental strains, as revealed by identical MVLST profiles, suggests that local and persistent environmental strains in seafood-processing environments pose a potential health risk. Furthermore, a PMSC in inlA does not appear to give L. monocytogenes a noninvasive profile.
INTRODUCTION
Listeria monocytogenes is an important food-borne pathogen that causes listeriosis (1). Listeriosis usually manifests itself as a gastrointestinal illness, but in some cases, more serious symptoms are observed, such as meningitis or septicemia (1). High-risk populations include immunocompromised individuals, infants, and the elderly. Infection in these populations results in a high mortality rate (1). In 2011, the reported incidence of listeriosis in New Zealand was 0.6/100,000 people (the highest rate in an English-speaking country), with 21 food-related cases leading to one death (2). The New Zealand Food Safety Authority (now within the Ministry for Primary Industries), has reviewed its risk management strategy across all food sectors with the aim of ensuring that the rate of listeriosis cases in the New Zealand population does not increase (3, 4).
Molecular subtyping has shown that isolates cluster into at least four evolutionary lineages (I, II, III, and IV) with different but overlapping ecological niches (5–7). The division into four lineages is consistent with serotype classification, which has separated L. monocytogenes into 13 serotypes (8). L. monocytogenes isolates belonging to lineage I serotypes 1/2b and 4b are the predominant causes of listeriosis in humans. Molecular epidemiology studies have identified three highly clonal lineage I serotype 4b strains, termed epidemic clones I, Ia, and II, which have been associated recurrently with outbreaks of the disease globally (7). Lineage II isolates, which are predominantly serotype 1/2a, have also been implicated in a number of recent outbreaks of listeriosis (9–12). Members of this highly diverse group, however, are also commonly isolated from the environment, including from food-processing plants and food samples. Lineage III and IV isolates, mostly belonging to serotypes 4a and 4c, are rare and are predominantly isolated from animal sources (8).
Various subtyping strategies discriminate closely related L. monocytogenes strains and are used to identify the sources of listeriosis outbreaks. Among them, pulsed-field gel electrophoresis (PFGE) is the most common and is used routinely in diagnostic laboratories throughout the world (13). Analysis of PFGE profiles, however, can be subjective, so DNA-sequencing-based techniques, such as multilocus sequence typing (MLST), have been developed to overcome the ambiguities of the fragment-based techniques and to enable easy exchange and comparison of data via publically available databases (14, 15). More recently, multi-virulence-locus sequence typing (MVLST) has been used as an alternative to MLST. Unlike MLST, which utilizes slowly diversifying housekeeping genes and can have low discriminatory power, MVLST targets more rapidly evolving virulence genes (16). The higher level of sequence polymorphisms associated with MVLST has enabled local epidemiological studies, often differentiating outbreak and epidemic strains from other isolates (17, 18). Cantinelli et al. (19), however, recently compared 125 L. monocytogenes strains and found that MVLST and MLST provided similar sequence types (STs), phylogenetic clustering, and discriminatory power.
Mutations in virulence genes are associated with a reduced invasion phenotype in L. monocytogenes. Perhaps the best-studied polymorphisms leading to virulence attenuation are those in the gene encoding the surface protein internalin A (InlA). InlA plays a critical role in the invasion of L. monocytogenes into human intestinal epithelial cells (20). Screening of L. monocytogenes isolated from human clinical cases, ready-to-eat (RTE) foods, and food-processing environments has revealed mutations in inlA leading to premature stop codons (PMSCs) (21–23), particularly among lineage II isolates of serotypes 1/2a and 1/2c (6). To date, at least 18 naturally occurring single nucleotide polymorphisms leading to a PMSC in inlA have been identified (23). Strains containing these PMSCs produce a truncated InlA protein and are generally less invasive than those without a PMSC (24).
Little is understood about the L. monocytogenes strains that exist in food-processing systems or about the sources of listeriosis cases in New Zealand. As a consequence, the risks that these strains pose to human health are unknown. Recently, a study of the prevalence and persistence of L. monocytogenes in seafood factories showed that in-house isolates are frequently found in the processing environment, but rarely in final products (25).
The aims of this study were (i) to investigate, using MVLST, the prevalence of L. monocytogenes STs in the seafood-processing environment and associated with food and human infection in New Zealand; (ii) to compare New Zealand L. monocytogenes STs with strains from different sources and geographic locations internationally, including key epidemic strains; (iii) to screen STs for the presence of previously described inlA PMSC mutations (types 1 to 18); and (iv) to establish the potential virulence of prevalent environmental STs in New Zealand based on their ability to invade human epithelial Caco-2 cells.
MATERIALS AND METHODS
Bacterial isolates.
A total of 52 L. monocytogenes isolates obtained from New Zealand seafood-processing environments (non-food contact), food, or human listeriosis cases were used in this study. Strains with the same PFGE profile (25) were included when they were obtained on different dates or from different locations within a seafood-processing plant. Clinical and nonseafood food isolates were supplied by Environmental Science & Research (ESR), Christchurch, New Zealand. All bacterial isolates were stored at −85°C on beads in cryopreservative medium containing 1% NaCl (Merck, Damstadt, Germany), 0.8% nutrient broth (Difco, BD, Sparks, MD), and 1.5% glycerol (Merck). Isolates were recovered by inoculation in Trypticase soya broth (Difco) enriched with 0.6% (wt/vol) yeast extract (Difco) (TSB-YE). The New Zealand isolates used in this study are described in Table 1.
TABLE 1.
New Zealand L. monocytogenes strains analyzed in this study
| Strain | Lineage | Serotypea | Pulsotypea | STb | Truncated InlA (amino acid)c | Source (yr of isolation) |
|---|---|---|---|---|---|---|
| 15A01 | II | 1/2a or 3a | 3814 | 01-N | N | Seafood-processing environment (2007)d |
| 15A04 | II | 1/2a or 3a | 3814 | 01-N | N | Seafood-processing environment (2007)d |
| 15A10 | II | 1/2a or 3a | 3814 | 01-N | N | Seafood-processing environment (2007)d |
| 15B03 | II | 1/2a or 3a | 3814 | 01-N | N | Seafood-processing environment (2007)d |
| 15G10 | II | 1/2a or 3a | 3832 | 01-N | N | Seafood-processing environment (2008)d |
| 15I03 | II | 1/2a or 3a | 3814 | 01-N | N | Seafood-processing environment (2008)d |
| 15J02 | II | 1/2a or 3a | 6514 | 01-N | N | Seafood-processing environment (2008)d |
| 15J08 | II | 1/2a or 3a | 3814 | 01-N | N | Seafood-processing environment (2008)d |
| 15K01 | II | 1/2a or 3a | 4814 | 01-N | N | Seafood-processing environment (2008)d |
| 16A01 | II | 1/2a or 3a | 3860 | 01-N | N | Seafood, outbreak (1992)e |
| 18F03 | II | 1/2a | 3860 | 01-N | N | Human, outbreak (1992)e |
| 16A03 | I | 1/2b or 3b or 7 | 3527 | 02-N | N | Seafood (1991)d |
| 16C03 | I | 1/2b or 3b or 7 | 9006 | 02-N | N | Other food (2005)d |
| 16C10 | I | 1/2a or 3a | 6502 | 02-N | N | Seafood-processing environment (2008)d |
| 15B09 | II | 1/2a or 3a | 8779 | 03-N | Y (700) | Seafood-processing environment (2007)d |
| 15C02 | II | 1/2a or 3a | 5132 | 03-N | Y (700) | Seafood-processing environment (2007)d |
| 15C05 | II | 1/2a or 3a | 5132 | 03-N | Y (700) | Seafood-processing environment (2007)d |
| 15C08 | II | 1/2a or 3a | 5132 | 03-N | Y (700) | Seafood-processing environment (2007)d |
| 15D01 | II | 1/2a or 3a | 5132 | 03-N | Y (700) | Seafood-processing environment (2007)d |
| 15D04 | II | 1/2a or 3a | 5132 | 03-N | Y (700) | Seafood-processing environment (2007)d |
| 15D07 | II | 1/2a or 3a | 5176 | 03-N | Y (700) | Seafood-processing environment (2007)d |
| 15D10 | II | 1/2a or 3a | 5132 | 03-N | Y (700) | Seafood-processing environment (2007)d |
| 15E03 | II | 1/2a or 3a | 5104 | 03-N | Y (700) | Seafood (2007)d |
| 15E06 | II | 1/2a or 3a | 5132 | 03-N | Y (700) | Seafood-processing environment (2008)d |
| 15E09 | II | 1/2a or 3a | 5132 | 03-N | Y (700) | Seafood-processing environment (2008)d |
| 15F02 | II | 1/2a or 3a | 5132 | 03-N | Y (700) | Seafood-processing environment (2008)d |
| 15F05 | II | 1/2a or 3a | 5182 | 03-N | Y (700) | Seafood-processing environment (2008)d |
| 15F08 | II | 1/2a or 3a | 5182 | 03-N | Y (700) | Seafood-processing environment (2008)d |
| 15G01 | II | 1/2a or 3a | 5132 | 03-N | Y (700) | Seafood-processing environment (2008)d |
| 15G04 | II | 1/2a or 3a | 5132 | 03-N | Y (700) | Seafood-processing environment (2008)d |
| 15G07 | II | 1/2a or 3a | 5132 | 03-N | Y (700) | Seafood-processing environment (2008)d |
| 18D09 | II | 1/2a | 5132 | 03-N | Y (700) | Human, sporadic (1999)e |
| 18E05 | II | 1/2a | 0101 | 03-N | Y (700) | Human, sporadic (2004)e |
| 15I06 | II | 1/2a or 3a | 3814 | 04-N | N | Seafood-processing environment (2008)d |
| 15B06 | II | 1/2a or 3a | 6502 | 05-N | N | Seafood (2008)d |
| 15I09 | II | 1/2a or 3a | 3814 | 05-N | N | Seafood-processing environment (2008)d |
| 15J05 | II | 1/2a or 3a | 3802 | 05-N | N | Seafood-processing environment (2008)d |
| 06B03 | II | 1/2a or 3a | 8841 | 06-N | N | Seafood (1999)d |
| 08A08 | II | 1/2a or 3a | 0202 | 07-N | N | Other food (1999)d |
| 16B05 | II | 1/2c or 3c | 4658 | 08-N | N | Other food (2005)d |
| 16B08 | I | 4b or 4d or 4e | 7806 | 09-N | N | Other food (2005)d |
| 16C06 | I | 4b or 4d or 4e | 2312 | 10-N | N | Other food (2005)d |
| 15A07 | II | 1/2a or 3a | 6502 | 11-N | N | Seafood-processing environment (2007)d |
| 06B04 | I | 1/2b or 3b | 6437 | 12-N | N | Other food (1997)d |
| 08A06 | II | 1/2a or 3a | 0202 | 13-N | N | Other food (1997)d |
| 08A10 | II | 1/2a or 3a | 0202 | 13-N | N | Seafood-processing environment (1999)d |
| 08A07 | I | 4b or 4d or 4e | 2361 | 14-N | N | Other food (1997)d |
| 18E01 | II | 1/2a | 0203 | 15-N | N | Human, sporadic (2000)e |
| 18E09 | II | 1/2a | 0202 | 15-N | N | Human, sporadic (1999) |
| 18E03 | II | 1/2a | 0101 | 16-N | Y (700) | Other food (2006) |
| 18E07 | II | 1/2a | 0101 | 16-N | Y (700) | Other food (2005) |
| 18F01 | II | 1/2a | 3846 | 17-N | N | Human, sporadic (2004) |
Data obtained in a previous study (6).
STs of MVLST groups were assigned based on concatenated DNA sequences derived from inlB, inlC, dal, lisR, clpP, and prfA. Different numbers represent different STs.
N, absence of inlA mutation; Y, presence of inlA mutation type 3; amino acid, amino acid position of inlA mutation type 3.
Cultures were isolated in our previous study (25).
Cultures were supplied by ESR.
MVLST.
Bacterial genomic DNA was extracted using a DNeasy tissue kit (Qiagen). For each bacterial strain, the intragenic regions of three virulence genes (inlB, inlC, and prfA) and three virulence-associated genes (lisR, clpP, and dal) were amplified using primers designed in previous studies (16, 17). PCRs were performed in a total volume of 25 μl containing 1.0 μl template DNA (1 to 10 ng/μl). The final concentrations of individual PCR components were as follows: 1× buffer, 0.16 mM deoxyribonucleotide triphosphates, 1.5 mM MgCl2, 0.8 μM appropriate primers, and 0.05 unit Taq DNA polymerase (Invitrogen). Reaction conditions were as follows: initial denaturation at 94°C for 4 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min and a final extension step of 72°C for 5 min. PCR fragments were purified with a QIAquick PCR purification kit (Qiagen) and were subsequently sequenced in both directions using an Automatic Sequencer 3730x (Macrogen Inc.). The primers used for amplification of each PCR product were also used in sequencing reactions. DNA sequences were then assembled and trimmed with Sequencher 4.5 (Gene Codes) and deposited in GenBank.
DNA sequence analysis.
Multiple alignments of DNA sequences for inlB, inlC, prfA, lisR, clpP, and dal were performed using ClustalX v 1.81 (26). For each gene, DNA sequences with at least 1 nucleotide difference were assigned as a unique allele. For each strain, the combination of six alleles defined its allelic profile, and a unique profile was designated an ST using a numeric system. A neighbor-joining (NJ) tree was then constructed for the STs of L. monocytogenes using the heuristic search algorithm in PAUP v 4.0 (27). Confidence measures for NJ branch points were generated by performing 1,000 bootstrap replicates. DNA sequences from the International Life Sciences Institute North America (ILSI NA) collection (28) were retrieved from GenBank for comparison (Table 2). The sequences comprised those from 44 L. monocytogenes strains representing 25 different PFGE profiles, including 21 human and food isolates from nine major human listeriosis outbreaks that occurred worldwide between 1981 and 2002. DNA sequences from 59 L. monocytogenes isolates collected from seafood products in Japan between 2004 and 2005 (29) were also used in comparisons (Table 2). DNA sequences for lineage III isolates were used to root the NJ tree, as they are more distantly related to lineages I and II (30).
TABLE 2.
International L. monocytogenes strains used for DNA sequence comparisons
| Strain | Lineagea | Serotypea | STb | Truncated InlA (amino acid)c | Source (isolation)a |
|---|---|---|---|---|---|
| FSLJ1-031 | III | 4a | 16-I | N | Human (sporadic; Canada; 1991) |
| FSLJ1-049 | I | 1/2b | 14-I | N | Human (sporadic; USA) |
| FSLJ1-094 | II | 1/2c | 10-I | N | Human (sporadic) |
| FSLJ1-101 | II | 1/2a | 01-I | − | Human (sporadic; USA; 1989) |
| FSLJ1-119 | I | 4b | 18-I | − | Human (epidemic; USA; 1985) |
| FSLJ1-158 | III | 4b | 26-I | N | Animal (USA; 1997) |
| FSLJ1-168 | III | 4a | 15-I | N | Human (sporadic) |
| FSLJ1-169 | I | 3b | 13-I | N | Human (sporadic) |
| FSLJ1-177 | I | 1/2b | 07-I | N | Human (sporadic; USA; 1997) |
| FSLJ1-225 | I | 4b | 22-I | − | Human (epidemic; USA; 1983) |
| FSLJ2-031 | II | 1/2a | 02-I | N | Animal (USA; 1996) |
| FSLJ2-064 | I | 1/2b | 09-I | N | Animal (cow) |
| FSLN1-225 | I | 4b | 19-I | − | Human (epidemic; USA; 1998) |
| FSLN3-013 | I | 4b | 22-I | N | Food (epidemic; UK; 1988) |
| FSLR2-500 | I | 4b | 30-I | N | Food (epidemic; USA; 2000) |
| FSLR2-502 | I | 3c | 14-I | − | Food (epidemic; USA; 1994) |
| FSLW1-110 | III | 4c | 27-I | N | Unknown |
| FSLW1-111 | III | 4c | 28-I | N | Unknown |
| 2_9 | II | 1/2a | 18-J | N | Seafood (Japan; 2002) |
| 5_2 | II | 1/2a | 14-J | N | Seafood (Japan; 2002) |
| 5_4 | II | 1/2a | 26-J | N | Seafood (Japan; 2002) |
| 9_17 | I | 3b | 03-J | N | Seafood (Japan; 2003) |
| 13_19 | I | 1/2b | 02-J | N | Seafood (Japan; 2003) |
| 20_5_1 | I | 4b | 06-J | N | Seafood (Japan; 2004) |
| 20_7_1 | II | 1/2a | 15-J | N | Seafood (Japan; 2004) |
| 22_13_3 | II | 1/2a | 13-J | N | Seafood (Japan; 2004) |
| 22_18_5 | II | 1/2a | 20-J | N | Seafood (Japan; 2004) |
| 23_4_1 | I | 1/2b | 01-J | N | Seafood (Japan; 2004) |
| 25_8_1 | II | 1/2a | 22-J | N | Seafood (Japan; 2004) |
| 25_15_1 | II | 1/2a | 25-J | N | Seafood (Japan; 2004) |
| 26_26_2 | II | 1/2a | 08-J | N | Seafood (Japan; 2005) |
| 29_10_1 | I | 1/2b | 04-J | N | Seafood (Japan; 2005) |
| 29_13_1 | I | 1/2b | 05-J | N | Seafood (Japan; 2005) |
| 30_8_1 | II | 1/2a | 10-J | N | Seafood (Japan; 2005) |
| 30_11_1 | II | 1/2a | 12-J | N | Seafood (Japan; 2005) |
| 30_25_1 | II | 3a | 23-J | N | Seafood (Japan; 2005) |
| 32_27_1 | II | 1/2a | 24-J | N | Seafood (Japan; 2005) |
| 34_9_1 | II | 3a | 17-J | N | Seafood (Japan; 2005) |
| 34_18_2 | I | 4b | 07-J | N | Seafood (Japan; 2005) |
| 36_6_1 | II | 1/2a | 11-J | N | Seafood (Japan; 2005) |
| 36_17_1 | II | 1/2a | 16-J | N | Seafood (Japan; 2005) |
| 36_25_1 | II | 1/2a | 21-J | Y (526) | Seafood (Japan; 2005) |
| 38_16_3 | II | 1/2a | 19-J | N | Seafood (Japan; 2005) |
| 40_4_1 | II | 1/2a | 09-J | N | Seafood (Japan; 2005) |
Data obtained from the ILSI NA L. monocytogenes collection (28) and RTE seafood products from Japan (29).
Different codes represent different STs. The STs for the international (28) and Japanese (29) collections are marked with an I or J, respectively.
N, absence of inlA mutation; Y, presence of inlA mutation type 3; −, not typed; amino acid, amino acid position of inlA mutation type 3.
Screening for inlA PMSCs.
Partial inlA DNA sequences spanning regions associated with previously defined PMSCs (mutation types 1 to 18) (23, 24) were obtained for the New Zealand clinical and environmental isolates. PCRs were performed as previously described using primers inlA-F and inlA-R (24). Each PCR amplicon was sequenced in both directions, as previously described, and the presence of PMSCs was established by the identification of in-frame stop codons within the resulting inlA gene sequence.
Cell culture invasion assay.
Invasion assays were performed on Caco-2 cells using a selection of strains from the most prevalent New Zealand STs, as previously described by Gaillard et al. (31). Assays were conducted using Caco-2 cells, as efficient invasion of these human intestinal epithelial cells requires InlA and is indicative of virulence (32). Caco-2 cells were grown using minimal essential medium (MEM) (Life Technologies, Carlsbad, CA) supplemented with 1% sodium pyruvate (Life Technologies), 1% nonessential amino acids (Life Technologies), and 10% fetal bovine serum (Life Technologies). For each experiment, 1.2 × 105 Caco-2 cells were seeded into each well of a 24-well plate (Corning, Lowell, MA) in MEM and incubated for 24 h at 37°C under 5% CO2 to form a monolayer. At the same time, each L. monocytogenes isolate was grown in brain heart infusion (BHI) broth (Difco) overnight at 37°C with shaking at 200 rpm. The following day, a 10-fold dilution of each overnight culture was prepared, and the bacteria were grown under the same incubation conditions until logarithmic phase (optical density at 600 nm [OD600] = 1.0). An aliquot (100 μl) of each bacterial suspension (ca. 1 × 107) was then added to a single well containing a Caco-2 monolayer to obtain a final multiplicity of infection (MOI) of 100:1 (bacteria/cell). The resulting 24-well plate was incubated for 30 min at 37°C, and each well was washed twice with phosphate-buffered saline (PBS) (Life Technologies). Fresh MEM containing 100 μg/ml of gentamicin (Life Technologies) was then added to each well, and the plate was incubated for 1 h at 37°C prior to washing twice with PBS. The cells were lysed by the addition of 1% Triton X-100 (Sigma-Aldrich, St. Louis, MO) and vigorous pipetting. For each isolate, the numbers of viable bacteria in the initial inoculum and of those released from lysed Caco-2 cells after incubation were compared by plating a dilution series of each on TSB-YE agar using the drop plate technique (5 drops of 10 μl each). CFU were counted after 24 h at 37°C. The invasion efficiency was calculated as the percentage of the initial inoculum recovered from a Caco-2 monolayer. The relative invasion efficiency was subsequently calculated by comparison of the invasion efficiency for each strain to that of the control strain, Scott A (defined as 100%). L. monocytogenes Scott A has been widely used as a reference strain for testing the efficacy of preservation techniques (33) and in virulence studies (34), and its genome was recently published (35). Three independent assays were performed, and each strain was tested in duplicate in each assay.
Statistical analysis.
Analysis of variance was used to compare the capacities of L. monocytogenes strains to invade Caco-2 cells. The invasiveness data were square-root transformed to stabilize variability before analysis. Tukey's test was used to compare the means to identify which were significantly different (P < 0.05).
Nucleotide sequence accession numbers.
The DNA sequences assembled in this study were deposited in GenBank under accession number KC290711 and PMSCs within the inlA gene sequence under accession numbers KC594235 to KC594285.
RESULTS
MVLST confirms the similarity of environmental and human isolates in New Zealand and distinguishes them from international strains.
A total of 301 single nucleotide polymorphisms spanning three virulence genes and three virulence-associated genes were identified among environmental-, food-, and human-related L. monocytogenes isolates. The numbers of unique alleles from the selected genes ranged from 10 for lisR to 22 for dal, and the percentages of polymorphic sites varied between 8.32% for prfA and 18.46% for dal (Table 3).
TABLE 3.
Allelic polymorphisms in the six virulence-associated gene fragments analyzed in the study
| Gene | No. of alleles | No. of polymorphic sites | % polymorphic sites |
|---|---|---|---|
| clpP | 12 | 38 | 9.07 |
| dal | 22 | 84 | 18.46 |
| lisR | 10 | 52 | 11.61 |
| inlB | 18 | 48 | 11.09 |
| inlC | 16 | 40 | 9.57 |
| prfA | 18 | 39 | 8.32 |
| Total | 96 | 301 | 11.39 |
The 301 nucleotide polymorphisms differentiated the 52 New Zealand L. monocytogenes isolates into 17 STs (Table 1). Just seven of the STs represented all the isolates collected from the seafood-processing environment (n = 31) in New Zealand. Of these STs, ST01-N and ST03-N comprised the majority (80.6%) of the isolates, indicating that they are widespread and recurrent in seafood-processing plants in New Zealand. The human isolates were seemingly more diverse, with six strains represented by four STs. Three of the human isolates, however, had STs identical to those of prevalent environmental strains (one ST01-N and two ST03-N) (Table 1). Thirteen STs represented the 15 food-related isolates, some overlapping the STs for isolates collected from the seafood-processing environment and clinical samples. All STs for New Zealand isolates were distinct from those associated with epidemic strains or with food production overseas, including those from Japanese RTE seafood products. The STs for all overseas strains are described in Table 2.
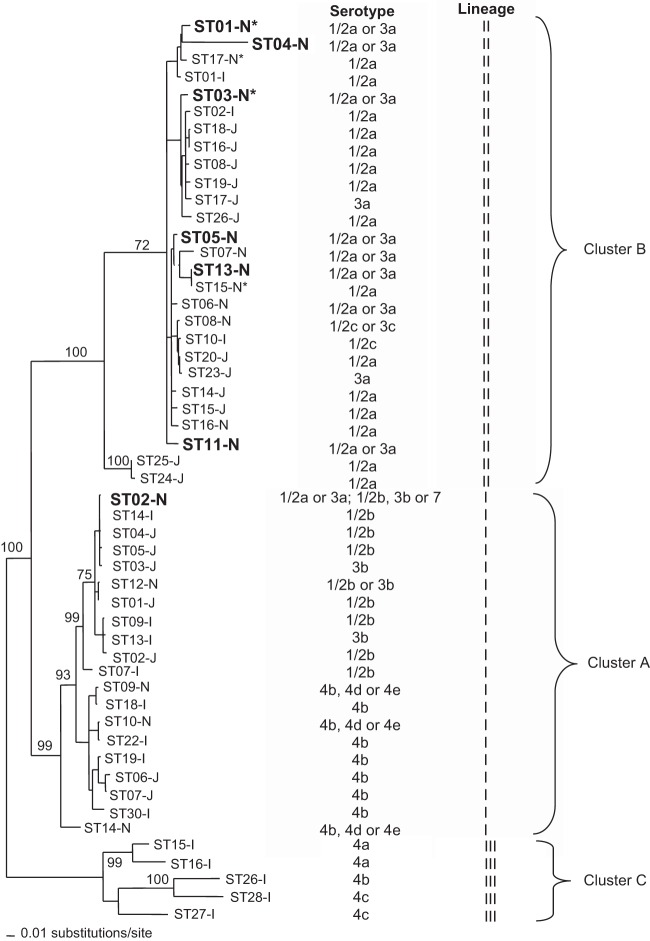
Cluster analysis differentiated the 17 STs of L. monocytogenes from New Zealand and overseas into three main groups: cluster A, cluster B, and cluster C (Fig. 1). Cluster A corresponds to lineage I, cluster B to lineage II, and cluster C to lineage III. STs collected from New Zealand seafood-processing plants clustered within lineages I and II and represented serotypes 1/2a or 3a. All STs from seafood products also clustered within lineages I and II and, with the exception of ST02-N, were of the same serotype. Furthermore, all but one ST from other foods in New Zealand clustered closely with the STs of isolates from seafood and seafood-processing plants. All four STs associated with human isolates from New Zealand belonged to lineage II and clustered with those from processing environments and food products.
FIG 1.
NJ tree of the STs identified from New Zealand and international L. monocytogenes strains based on the p distances of the polymorphic nucleotides for a concatenated sequence containing clpP, dal, lisR, inlB, inlC, and prfA (p distance is the proportion of nucleotide sites at which two compared sequences are different). Bootstrap support values are shown as node labels (if greater than 70). The STs of the New Zealand seafood environmental isolates are shown in boldface. The asterisks indicate STs that include a human isolate from New Zealand.
Although isolates from Japanese RTE seafood products had different STs, with the exception of ST25-J and ST24-J, all clustered closely (with strong bootstrap support) with the New Zealand STs within lineages I and II. They did, however, represent serotypes 1/2b, 3b, and 4b, as well as 1/2a and 3a (29). In contrast, none of the human epidemic STs from overseas, which belong to lineage I and serotype 4b, clustered strongly with STs from New Zealand seafood or seafood-processing environments. Furthermore, only one ST (ST01-I) associated with a sporadic human infection overseas clustered with STs from New Zealand, but interestingly, this ST was most similar to two of the four STs for clinical isolates in New Zealand.
Presence of PMSC mutation type 3 in prevalent STs and human isolates.
Partial DNA sequencing of the inlA gene from all isolates of L. monocytogenes from New Zealand revealed only one nonsense mutation in inlA, at amino acid position 700. This mutation corresponded to PMSC mutation type 3. In total, 21 of the 52 (40.4%) New Zealand isolates carried a PMSC type 3 mutation (Table 1). The majority (90%) belonged to ST01-N. Most of the isolates encoding a truncated form of InlA were from environmental samples (n = 16; 76.2% of the total truncated strains), although some were obtained from human samples (n = 2; 9.5%) or from food, including seafood (n = 3; 14.3%).
Invasiveness of L. monocytogenes STs prevalent in New Zealand.
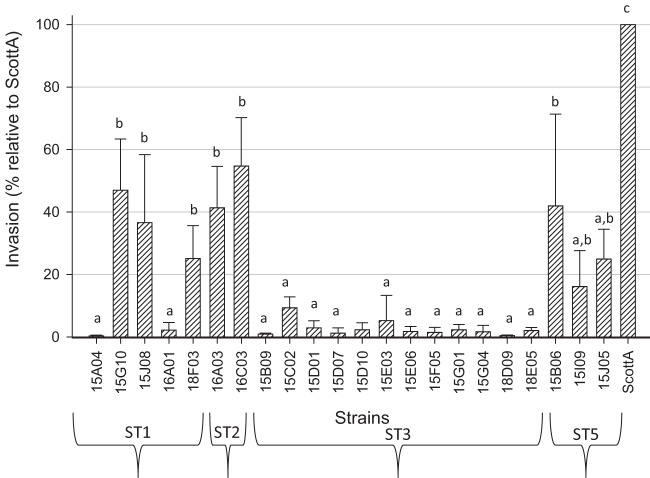
All isolates belonging to ST03-N showed an invasion capacity of less than 10% in Caco-2 human intestinal epithelial cells compared with the Scott A strain (P < 0.05), independent of their sources of isolation (Table 1 and Fig. 2). In contrast, ST01-N isolates, which did not contain known virulence-attenuating mutations in the partially sequenced inlA gene, varied in virulence: some strains presented less than 10% invasiveness (similar to PMSC inlA strains; P < 0.05), whereas others reached approximately 50% of the Scott A strain value (P < 0.05) (Fig. 2). There appeared to be no relationship between invasiveness and the source of the isolates (environment, food, and/or human) (P > 0.05). Isolates 16A01 and 18F03 were obtained from smoked mussels and a human clinical sample, respectively, and were linked to the only New Zealand seafood-related listeriosis outbreak that occurred in 1992 (36). Although both belong to ST01-N, these strains showed different invasion capacities (P < 0.05), with the human isolate expressing 10 times greater invasion than the seafood isolate.
FIG 2.
Invasiveness of selected L. monocytogenes strains belonging to the most common STs found in this study. The bars represent means and standard deviations (SD) of invasiveness (percent) in relation to reference strain Scott A (designated 100%). Different letters above the bars represent significant differences (P < 0.05) between strains determined using Tukey's test.
DISCUSSION
In this study, MVLST identified common STs for seafood-related and clinical L. monocytogenes isolates, which were unique to New Zealand. MVLST also showed that seafood-related isolates were homogeneous and recurrent within the factories, while human isolates were relatively diverse. A large number of the seafood-related isolates and several clinical isolates carried a type 3 inlA PMSC with reduced invasiveness, supporting the hypothesis that expression of InlA may result in a fitness cost to the bacterium. Furthermore, L. monocytogenes may cause infection in an inlA-independent manner.
MVLST is a method that uses highly discriminatory loci that have been shown to differentiate epidemic strains in a manner comparable with PFGE (16, 17). Of the six virulence-related genes established for typing L. monocytogenes, dal, lisR, and inlB are the most discriminatory. Between 12 and 19% of the nucleotides in these genes are polymorphic (17). In particular, the inclusion of inlB contributes to the differentiation of strains of serotypes 1/2a and 4b, as well as epidemiologically unrelated strains within serotypes. Consistent with these observations, the most polymorphic gene in our study was dal (18% of the nucleotides in the gene were polymorphic), followed by lisR (12%) and inlB (11%).
MVLST identified 17 STs from a selection of L. monocytogenes isolates collected from seafood-processing environments, seafood, other foods, and New Zealand clinical cases. The differentiation of the 52 isolates into 17 STs was consistent with their discrimination using PFGE. However, PFGE was more discriminatory than MVLST, as PFGE identified 21 pulsotypes. Regardless, the more subjective nature of PFGE and the ease in comparing DNA sequence data from multiple studies indicates that MVLST may be more useful for defining L. monocytogenes strains, their prevalences, and their associations with clinical cases of listeriosis. For example, MVLST distinguished one isolate linked to listeriosis (ST15-N) from food and environmental isolates (ST13-N). This was not possible using PFGE.
Several STs of L. monocytogenes are widespread in seafood-processing plants in New Zealand, yet none of them match those of the international isolates, which include isolates from Japanese seafood products (29) and a collection of epidemic strains (28). Chenal-Francisque et al. (37) also observed that clones of L. monocytogenes that were widely distributed in one country are often distinct, with few strains from different geographical regions in the world having the same genotype. The unique genotypes of isolates in different countries indicate that spread of L. monocytogenes globally is limited and that the highest risk associated with the microorganism is most likely from local strains.
Even though genotypes of L. monocytogenes are largely restricted to distinct geographical localities, a previous study conducted with 300 L. monocytogenes strains from 42 different countries on five continents using MLST showed that a subset of clones of clinical isolates appear to be distributed worldwide (37). A clear genotypic partition can be observed between these clinical isolates and those from food or the environment. In our MVLST study, environmental, food, and human isolates had the same or similar STs, consistent with their closely related or identical pulsotypes (25). Identical genotypes of isolates from these sources confirm that local, environmentally persistent strains are epidemiologically important in listeriosis in New Zealand. In particular, the identification of isolates from the seafood-processing environments with the same STs as those isolated from sporadic cases of listeriosis demonstrates the potential health risk associated with seafood. Furthermore, the close relationship of ST04-N (from the seafood-processing environment) and clinical STs from New Zealand (ST01-N and ST17-N) with a sporadic human isolate from overseas (ST01-I) suggests some particular subtypes of L. monocytogenes lineage II might be more virulent to humans than others.
InlA is a key virulence factor that supports infection of epithelial cells by L. monocytogenes (38). Nonsense mutations in inlA are reported to be at least partially responsible for an attenuated invasion phenotype (7, 24, 39–41). These mutations may have accumulated as a result of selection for environmentally adapted ecotypes with reduced virulence, which do not require a full-length InlA protein for ecological success (22). Nightingale et al. (24) and Gray et al. (42) showed that inlA PMSCs were associated with about 40% of L. monocytogenes strains isolated from RTE foods in the United States. In France, 35% of food isolates, including seafood isolates, had a truncated InlA (43). Felicio et al. (41) also reported similar rates from L. monocytogenes strains isolated from Alheira in Portugal. Controversially, Handa-Miya et al. (29) found inlA PMSCs in only 1.7% of L. monocytogenes strains isolated from RTE seafood in Japan. In accordance with the majority of these studies, 20% of L. monocytogenes strains from food and 67% of environment-related isolates in New Zealand have a PMSC in inlA. These strains might represent a low-risk group of L. monocytogenes strains.
L. monocytogenes isolates carrying inlA PMSCs have rarely been recovered from human listeriosis samples. A truncated InlA was not evident in any of the 61 L. monocytogenes isolates from abortion cases in France (43). Furthermore, Van Stelten et al. (23) found that only 5.1% of human isolates have a PMSC in inlA, while two other studies in the United States provided further evidence that an inlA PMSC is uncommon in strains that cause listeriosis (22, 42). Nightingale et al. (24), however, revealed that 36.5% of the 52 human clinical isolates selected from ribotypes commonly associated with PMSCs carried an inlA PMSC, placing in doubt previous assumptions that a functional InlA protein is required for effective invasion. In our study, type 3 PMSC mutations were ubiquitous in ST03-N, which includes both environmental and clinical isolates, yet neither ST04-N from seafood-processing environments or clinical strains from New Zealand (ST01-N and ST17-N) had a PMSC. Furthermore, the invasiveness of ST03-N isolates was low compared with L. monocytogenes Scott A in a Caco-2 cell line assay, whereas ST01-N isolates had variable invasiveness. The reasons for the apparent underrepresentation of PMSCs in inlA in strains isolated from clinical samples and the mechanisms by which InlA-deficient strains cause disease remain to be determined. If InlA is important, but not essential, for invasion by L. monocytogenes, the low incidence of PMSCs detected may be related to the reduced virulence of clinical isolates carrying a truncated inlA gene.
Nightingale et al. (40) suggested that mutations in inlA are related to the genotype of L. monocytogenes (i.e., ribotype, ST, or pulsotype) rather than to the source of isolation (i.e., they are more likely to be due to founder effects and not causally linked to invasiveness). Orsi et al. (6) also suggest that infrequent reporting of the PMSCs in listeriosis-related strains may have resulted in the overrepresentation of 4b isolates in clinical samples in comparison to 1/2a strains (harboring inlA-PMSC), while the PMSC may have resulted in positive selection for strains with a truncated InlA protein during environmental adaptation. In general, 1/2a strains appear to be more likely to acquire genetic information by horizontal transfer or to undergo recombination events that may afford rapid adaptation to niche-specific stresses (44). The increased mutability of these strains may have led to truncation of InlA as part of their niche adaptation.
Similar to our finding that isolates from human cases can harbor a PMSC, isolates with PMSCs have previously been associated with food-related outbreaks of listeriosis (e.g., the 1985 cheese outbreak in the United States and the 1988 pâté outbreak in the United Kingdom [40, 45]). These outbreak strains showed attenuated invasion abilities when tested in Caco-2 cell lines, leading to the suggestion that their capacity to cause listeriosis may have been due to inlA-independent properties (e.g., better survival and growth in food and food-associated environments). Holch et al. (46) also showed that environmental L. monocytogenes strains harboring inlA-PMSC could cross the placental barrier in a similar way to a strain expressing a full-length inlA in pregnant mice and guinea pigs. These data suggest an inlA-independent pathway for invasion of certain tissues (e.g., the placenta and fetus). Given that ST03-N is apparently widespread in New Zealand, its ability to survive in food-associated environments may lead to sporadic cases, even though it is less virulent than other STs (e.g., ST04-N).
Eighteen PMSCs have previously been reported, but only one (PMSC type 3) was detected in L. monocytogenes from New Zealand. This PMSC was present in ST03-N, the most prevalent of our STs (Table 1). Previous studies have shown that 11% of PMSCs in L. monocytogenes in RTE foods in the United States are PMSC type 3, while mutation type 1 accounts for 30% (24, 42). Van Stelten et al. (23) also showed that PMSC mutation types 1, 3, and 4 represented >90% of the inlA mutations detected in 1,009 isolates, with PMSC type 3 the most common. These data indicate that L. monocytogenes isolates carrying PMSCs in inlA are distributed worldwide and that PMSC type 3 is relatively common among isolates (23, 40, 47). PMSC type 3 is specifically associated with lineage II serotypes 1/2a and 1/2c (6, 8).
Our results indicate that a PMSC mutation type 3 in inlA may produce an invasion capacity of strains in human cell lines lower than those of isolates that do not have such a PMSC. This might be expected to confer a significant attenuation of virulence on isolates carrying the PMSC. We also discovered, however, that two of the six L. monocytogenes isolates from clinical cases contained identical inlA PMSCs and were the likely causes of listeriosis. The isolation of identical STs in clinical and environmental isolates and the revelation that, in contrast to some other studies, a third of the isolates from cases of listeriosis carried a type 3 PMSC in inlA confirm that other factors are probably involved in the development of listeriosis. Careful assessment of possible pathogenicity markers other than inlA is required to ensure that the risk associated with environmentally persistent strains is fully understood.
In summary, our findings suggest New Zealanders are commonly exposed to virulence-attenuated L. monocytogenes strains. Although the risk associated with these strains is likely to be far lower than that of more invasive strains, some risk still exists that they may result in listeriosis. To gain a greater understanding of the risks associated with exposure to virulence-attenuated isolates, the genotypes of more isolates will need to be assessed. Their capacities to invade various cell lines will also need to be investigated more thoroughly.
ACKNOWLEDGMENTS
We thank Jessicah Win, Sandra Visnovsky, and Tom Nelson for technical assistance and Brent Gilpin for providing the human and nonseafood food isolates.
This work was supported by the Foundation for Research, Science and Technology, New Zealand (contract number CAWX0703).
Footnotes
Published ahead of print 20 December 2013
REFERENCES
- 1.Painter J, Stutsker L. 2007. Listeriosis in humans, p 19 In Ryser TE, Marth HE. (ed), Listeria, listeriosis and food safety, vol 1 CRC Press, Boca Raton, FL [Google Scholar]
- 2.Lim E, Lopez L, Borman A, Cressey P, Pirie R. 2012. Annual report concerning foodborne disease in New Zealand 2011. http://foodsafety.govt.nz/elibrary/industry/foodborne-disease-nz-doc.pdf [Google Scholar]
- 3.Crerar SK, Castle M, Hassel S, Schumacher D. 2011. Recent experiences with Listeria monocytogenes in New Zealand and development of a food control risk-based strategy. Food Control 22:1510–1512. 10.1016/j.foodcont.2010.07.016 [DOI] [Google Scholar]
- 4.New Zealand Food Safety Authority 2008. Listeria monocytogenes risk management strategy 2008–2013. Ministry for Primary Industries, Wellington, New Zealand: http://www.foodsafety.govt.nz/elibrary/industry/Listeria_Risk-Aims_Help.pdf [Google Scholar]
- 5.Nightingale K. 2010. Listeria monocytogenes: knowledge gained through DNA sequence-based subtyping, implications, and future considerations. J. AOAC Int. 93:1275–1286 [PubMed] [Google Scholar]
- 6.Orsi RH, den Bakker HC, Wiedmann M. 2011. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301:79–96. 10.1016/j.ijmm.2010.05.002 [DOI] [PubMed] [Google Scholar]
- 7.Van Stelten A, Simpson JM, Chen Y, Scott VN, Whiting RC, Ross WH, Nightingale KK. 2011. Significant shift in median guinea pig infectious dose shown by an outbreak-associated Listeria monocytogenes epidemic clone strain and a strain carrying a premature stop codon mutation in inlA. Appl. Environ. Microbiol. 77:2479–2487. 10.1128/AEM.02626-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nadon CA, Woodward DL, Young C, Rodgers FG, Wiedmann M. 2001. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 39:2704–2707. 10.1128/JCM.39.7.2704-2707.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CDC 2011. Investigation update: multistate outbreak of listeriosis linked to whole cantaloupes from Jensen Farms, Colorado. CDC, Atlanta, GA [Google Scholar]
- 10.CDC 2012. Multistate outbreak of listeriosis linked to imported Frescolina Marte brand ricotta salata cheese. CDC, Atlanta, GA [Google Scholar]
- 11.Gaul LK, Farag NH, Shim T, Kingsley MA, Silk BJ, Hyytia-Trees E. 2013. Hospital-acquired listeriosis outbreak caused by contaminated diced celery—Texas, 2010. Clin. Infect. Dis. 56:20–26. 10.1093/cid/cis817 [DOI] [PubMed] [Google Scholar]
- 12.Knabel SJ, Reimer A, Verghese B, Lok M, Ziegler J, Farber J, Pagotto F, Graham M, Nadon CA, Canadian Public Health Laboratory. Gilmour MW. 2012. Sequence typing confirms that a predominant Listeria monocytogenes clone caused human listeriosis cases and outbreaks in Canada from 1988 to 2010. J. Clin. Microbiol. 50:1748–1751. 10.1128/JCM.06185-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graves LM, Swaminathan B. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65:55–62. 10.1016/S0168-1605(00)00501-8 [DOI] [PubMed] [Google Scholar]
- 14.Chan MS, Maiden MC, Spratt BG. 2001. Database-driven multi locus sequence typing (MLST) of bacterial pathogens. Bioinformatics 17:1077–1083. 10.1093/bioinformatics/17.11.1077 [DOI] [PubMed] [Google Scholar]
- 15.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140–3145. 10.1073/pnas.95.6.3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Jayarao BM, Knabel SJ. 2004. Multi-virulence-locus sequence typing of Listeria monocytogenes. Appl. Environ. Microbiol. 70:913–920. 10.1128/AEM.70.2.913-920.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Zhang W, Knabel SJ. 2005. Multi-virulence-locus sequence typing clarifies epidemiology of recent listeriosis outbreaks in the United States. J. Clin. Microbiol. 43:5291–5294. 10.1128/JCM.43.10.5291-5294.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lomonaco S, Chen Y, Knabel J. 2008. Analysis of additional virulence genes and virulence gene regions in Listeria monocytogenes confirms the epidemiologic relevance of multi-virulence-locus sequence typing. J. Food Prot. 71:2559–2566 [DOI] [PubMed] [Google Scholar]
- 19.Cantinelli T, Chenal-Francisque V, Diancourt L, Frezal L, Leclercq A, Wirth T, Lecuit M, Brisse S. 2013. “Epidemic clones” of Listeria monocytogenes are widespread and ancient clonal groups. J. Clin. Microbiol. 51:3770–3779. 10.1128/JCM.01874-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lecuit M, Ohayon H, Braun L, Mengaud J, Cossart P. 1997. Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect. Immun. 65:5309–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miya S, Takahashi H, Ishikawa T, Fujii T, Kimura B. 2010. Risk of Listeria monocytogenes contamination of raw ready-to-eat seafood products available at retail outlets in Japan. Appl. Environ. Microbiol. 76:3383–3386. 10.1128/AEM.01456-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nightingale KK, Windham K, Wiedmann M. 2005. Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J. Bacteriol. 187:5537–5551. 10.1128/JB.187.16.5537-5551.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Stelten A, Simpson JM, Ward TJ, Nightingale KK. 2010. Revelation by single-nucleotide polymorphism genotyping that mutations leading to a premature stop codon in inlA are common among Listeria monocytogenes isolates from ready-to-eat foods but not human listeriosis cases. Appl. Environ. Microbiol. 76:2783–2790. 10.1128/AEM.02651-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nightingale KK, Windham K, Martin KE, Yeung M, Wiedmann M. 2005. Select Listeria monocytogenes subtypes commonly in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 71:8764–8772. 10.1128/AEM.71.12.8764-8772.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruz CD, Fletcher GC. 2011. Prevalence and biofilm-forming ability of Listeria monocytogenes in New Zealand mussel (Perna canaliculus) processing plants. Food Microbiol. 28:1387–1393. 10.1016/j.fm.2011.06.014 [DOI] [PubMed] [Google Scholar]
- 26.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers JS, Swofford DL. 1998. A fast method for approximating maximum likelihoods of phylogenetic trees from nucleotide sequences. Syst. Biol. 47:77–89. 10.1080/106351598261049 [DOI] [PubMed] [Google Scholar]
- 28.Fugett E, Fortes E, Nnoka C, Wiedmann M. 2006. International Life Sciences Institute North America Listeria monocytogenes strain collection: development of standard Listeria monocytogenes strain sets for research and validation studies. J. Food Prot. 69:2929–2938 [DOI] [PubMed] [Google Scholar]
- 29.Handa-Miya S, Kimura B, Takahashi H, Sato M, Ishikawa T, Igarashi K, Fujii T. 2007. Nonsense-mutated inlA and prfA not widely distributed in Listeria monocytogenes isolates from ready-to-eat seafood products in Japan. Int. J. Food Microbiol. 117:312–318. 10.1016/j.ijfoodmicro.2007.05.003 [DOI] [PubMed] [Google Scholar]
- 30.Wiedmann M, Bruce JL, Keating C, Johnson AE, McDonough PL, Batt CA. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaillard JL, Berche P, Mounier J, Richard S, Sansonetti P. 1987. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect. Immun. 55:2822–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H, Marquis H, Boor KJ. 2005. SigmaB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology 151:3215–3222. 10.1099/mic.0.28070-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guenther S, Huwyler D, Richard S, Loessner MJ. 2009. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl. Environ. Microbiol. 75:93–100. 10.1128/AEM.01711-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lammerding AM, Glass KA, Gendron-Fitzpatrick A, Doyle MP. 1992. Determination of virulence of different strains of Listeria monocytogenes and Listeria innocua by oral inoculation of pregnant mice. Appl. Environ. Microbiol. 58:3991–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Briers Y, Klumpp J, Schuppler M, Loessner MJ. 2011. Genome sequence of Listeria monocytogenes Scott A, a clinical isolate from a food-borne listeriosis outbreak. J. Bacteriol. 193:4284–4285. 10.1128/JB.05328-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brett MS, Short P, McLauchlin J. 1998. A small outbreak of listeriosis associated with smoked mussels. Int. J. Food Microbiol. 43:223–229. 10.1016/S0168-1605(98)00116-0 [DOI] [PubMed] [Google Scholar]
- 37.Chenal-Francisque V, Lopez J, Cantinelli T, Caro V, Tran C, Leclercq A, Lecuit M, Brisse S. 2011. Worldwide distribution of major clones of Listeria monocytogenes. Emerg. Infect. Dis. 17:1110–1112. 10.3201/eid/1706.101778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lecuit M, Vandormael-Pournin S, Lefort J, Huerre M, Gounon P, Dupuy C, Babinet C, Cossart P. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292:1722–1725. 10.1126/science.1059852 [DOI] [PubMed] [Google Scholar]
- 39.Roberts AJ, Williams SK, Wiedmann M, Nightingale KK. 2009. Some Listeria monocytogenes outbreak strains demonstrate significantly reduced invasion, inlA transcript levels, and swarming motility in vitro. Appl. Environ. Microbiol. 75:5647–5658. 10.1128/AEM.00367-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nightingale KK, Ivy RA, Ho AJ, Fortes ED, Njaa BL, Peters RM, Wiedmann M. 2008. inlA premature stop codons are common among Listeria monocytogenes isolates from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Appl. Environ. Microbiol. 74:6570–6583. 10.1128/AEM.00997-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Felicio MT, Hogg T, Gibbs P, Teixeira P, Wiedmann M. 2007. Recurrent and sporadic Listeria monocytogenes contamination in alheiras represents considerable diversity, including virulence-attenuated isolates. Appl. Environ. Microbiol. 73:3887–3895. 10.1128/AEM.02912-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gray MJ, Zadoks RN, Fortes ED, Dogan B, Cai S, Chen Y, Scott VN, Gombas DE, Boor KJ, Wiedmann M. 2004. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833–5841. 10.1128/AEM.70.10.5833-5841.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacquet C, Doumith M, Gordon JI, Martin PM, Cossart P, Lecuit M. 2004. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189:2094–2100. 10.1086/420853 [DOI] [PubMed] [Google Scholar]
- 44.Kovacevic J, Arguedas-Villa C, Wozniak A, Tasara T, Allen KJ. 2013. Examination of food chain-derived Listeria monocytogenes strains of different serotypes reveals considerable diversity in inlA genotypes, mutability, and adaptation to cold temperatures. Appl. Environ. Microbiol. 79:1915–1922. 10.1128/AEM.03341-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nightingale KK, Milillo SR, Ivy RA, Ho AJ, Oliver HF, Wiedmann M. 2007. Listeria monocytogenes F2365 carries several authentic mutations potentially leading to truncated gene products, including inlB, and demonstrates atypical phenotypic characteristics. J. Food Prot. 70:482–488 [DOI] [PubMed] [Google Scholar]
- 46.Holch A, Ingmer H, Licht TR, Gram L. 2013. Listeria monocytogenes strains encoding premature stop codons (PMSC) in inlA invade mice and guinea pig fetuses in orally dosed dams. J. Med. Microbiol. 62:1799–1806. 10.1099/jmm.0.057505-0 [DOI] [PubMed] [Google Scholar]
- 47.Olier M, Rousseaux S, Piveteau P, Lemaitre JP, Rousset A, Guzzo J. 2004. Screening of glutamate decarboxylase activity and bile salt resistance of human asymptomatic carriage, clinical, food, and environmental isolates of Listeria monocytogenes. Int. J. Food Microbiol. 93:87–99. 10.1016/j.ijfoodmicro.2003.10.010 [DOI] [PubMed] [Google Scholar]