Abstract
Polyhydroxyalkanoate (PHA)-producing Bacillus strains express class IV PHA synthase, which is composed of the subunits PhaR and PhaC. Recombinant Escherichia coli expressing PHA synthase from Bacillus cereus strain YB-4 (PhaRCYB-4) showed an unusual reduction of the molecular weight of PHA produced during the stationary phase of growth. Nuclear magnetic resonance analysis of the low-molecular-weight PHA revealed that its carboxy end structure was capped by ethanol, suggesting that the molecular weight reduction was the result of alcoholytic cleavage of PHA chains by PhaRCYB-4 induced by endogenous ethanol. This scission reaction was also induced by exogenous ethanol in both in vivo and in vitro assays. In addition, PhaRCYB-4 was observed to have alcoholysis activity for PHA chains synthesized by other synthases. The PHA synthase from Bacillus megaterium (PhaRCBm) from another subgroup of class IV synthases was also assayed and was shown to have weak alcoholysis activity for PHA chains. These results suggest that class IV synthases may commonly share alcoholysis activity as an inherent feature.
INTRODUCTION
Polyhydroxyalkanoates (PHAs) are a category of aliphatic polyesters synthesized by a wide variety of bacteria as an intracellular carbon and energy storage material in response to various environmental conditions. Recently, PHAs have attracted industrial attention because of their potential use as biodegradable and biocompatible thermoplastics (1). The biosynthesis of poly[(R)-3-hydroxybutyrate] [P(3HB)], the most commonly found natural PHA, has been well studied in Ralstonia eutropha and requires only three enzymes from acetyl coenzyme A (acetyl-CoA): 3-ketothiolase (PhaA), encoded by the phaA gene; acetoacetyl-CoA reductase (PhaB), encoded by the phaB gene; and PHA synthase (PhaC), encoded by the phaC gene (2). The coexpression of these three enzymes enables P(3HB)-negative bacteria like Escherichia coli to accumulate P(3HB) (3).
PHA synthases are grouped into four classes (classes I to IV) on the basis of subunit composition and substrate specificity (4). Class I synthases contain a single subunit, PhaC, and catalyze the polymerization of short-chain-length monomers (C3 to C5); the synthase from R. eutropha (PhaCRe) is an example of this class. Class II synthases, as represented by the synthase from Pseudomonas putida, also contain a single PhaC subunit but catalyze the polymerization of medium-chain-length monomers (C6 to C14). Class III synthases, as represented by the synthase from Allochromatium vinosum, contain two heterologous subunits, PhaE and PhaC, and catalyze the polymerization of short-chain-length monomers. Class IV synthases, a recently identified class of synthases, contain two heterologous subunits, PhaR and PhaC, and catalyze the polymerization of short-chain-length monomers (C3 to C5). On the basis of phylogenetic analysis (see Fig. S1 in the supplemental material), the PhaC subunits in class IV synthases can be further classified into two subgroups: the Bacillus megaterium subgroup and the Bacillus cereus subgroup (5–7). These PhaC subunits recognize PhaR subunits from different subgroups (7).
P(3HB) produced by E. coli expressing class I to IV synthases exhibits a variety of molecular weights, depending on the characteristics of the synthase expressed (8). It is noteworthy that the synthase from Delftia acidovorans (PhaCDa), a class I synthase, is capable of synthesizing high-molecular-weight P(3HB) in E. coli (9).
In previous studies (7, 10), it was shown that B. cereus strain YB-4 isolated from soil expresses a class IV PHA synthase consisting of two heterologous subunits, PhaRYB-4 (18.5 kDa) and PhaCYB-4 (41.7 kDa). Recombinant E. coli expressing PhaRCYB-4 showed an unusual decrease in the molecular weight of P(3HB) synthesized during cultivation, especially in the stationary phase of growth. In addition, kinetic analysis indicated that the decrease in molecular weight is the result of random scission of the polymer chain (7). The same phenomenon was observed with E. coli expressing the class IV synthase from Bacillus sp. strain INT005 (PhaRCBsp) (11). On the other hand, E. coli expressing PHA synthase from B. megaterium (PhaRCBm), another class IV synthase, did not produce P(3HB) with such a low molecular weight (7). A subunit recombination study of PhaR and PhaC from B. cereus YB-4 and B. megaterium revealed that the PhaCYB-4 subunit is responsible for the scission activity. However, unlike E. coli, the PHA-negative mutant R. eutropha PHB−4 produced high-molecular-weight P(3HB) even when PhaRCYB-4 was expressed (12). From these observations, we hypothesized that there might be regulatory mechanisms governing the P(3HB) scission activity of PhaCYB-4.
The aim of this study was to investigate the mechanism governing the P(3HB) scission activity of PhaRCYB-4. To this end, an in vivo scission assay was carried out, using E. coli JM109 as the host strain, by expressing PhaRCYB-4 from high- or low-copy-number plasmids. The results indicate that PhaRCYB-4 has alcoholytic cleavage activity for P(3HB) chains in the presence of both endogenous and exogenous ethanol. Moreover, the results from coexpression of PhaRCYB-4 with PhaCDa [PhaCDa produces a high-molecular-weight P(3HB) used as the scissile substrate in this experiment] indicate that PhaRCYB-4 is also able to cleave P(3HB) polymerized by another synthase. In addition, the alcoholytic cleavage activity of PhaRCYB-4 was assayed in vitro. This is the first study to document alcoholysis activity in PHA synthases.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli JM109 was used as a host strain for P(3HB) biosynthesis throughout the study. E. coli BL21(DE3) was used to produce His-tagged PhaRYB-4 and PhaCYB-4 from the expression plasmids pET15b-phaRYB-4 and pET15b-phaCYB-4, respectively. For preculturing, the recombinant bacteria were grown in lysogeny broth (LB) medium (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter NaCl). For maintenance of the plasmid within the cell, ampicillin (100 mg/liter) and/or kanamycin (50 mg/liter) was added to the medium as appropriate.
TABLE 1.
Bacterial strains and plasmids used in the study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli JM109 | recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) e14− (mcrA) supE44 relA1 Δ(lac-proAB)/F′ [traD36 proAB+ lacIq lacZΔM15] | TaKaRa Bio |
| E. coli BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm | Novagen |
| Plasmids | ||
| pBBR1MCS-2 | Broad-host-range vector, Kmr | 13 |
| pBBR1-phaRCYB-4AB | pBBR1MCS-2 derivative; phaRe promoter, phaRCYB-4 from B. cereus YB-4, and phaABRe from R. eutropha | 12 |
| pJRD215 | Broad-host-range vector, Kmr | 14 |
| pJRD-phaCDaAB | pJRD215 derivative; phaRe promoter, phaCDa from D. acidovorans DS-17, and phaABRe from R. eutropha H16 | This study |
| pGEM″ABex | pGEM-T derivative, phaRe promoter, phaABRe from R. eutropha H16, Apr | 15 |
| pGEM-phaCDaAB | pGEM″ABex derivative; phaCDa from D. acidovorans DS-17 (identical to pGEM″ABexCaC) | 16 |
| pGEM-phaCReAB | pGEM″ABex derivative; phaCRe from R. etutropha H16 | 17 |
| pGEM- phaRCYB-4AB | pGEM″ABex derivative; phaRCYB-4 from B. cereus YB-4 | 7 |
| pGEM-phaRCBmAB | pGEM″ABex derivative; phaRCBm from B. megaterium NBRC15308T | 7 |
| pGEM-phaRYB-4AB | pGEM″ABex derivative; phaRYB-4 from B. cereus YB-4 | 7 |
| pGEM-phaCYB-4AB | pGEM″ABex derivative; phaCYB-4 from B. cereus YB-4 | 7 |
| pGEM-phaRCYB-4 | pGEM-T derivative; phaRe promoter, phaRCYB-4 from B. cereus YB-4 | This study |
| pET15b | T7 promoter, His tag fusion protein | Novagen |
| pET15b-phaRYB-4 | pET15b derivative; phaRYB-4 from B. cereus YB-4 | This study |
| pET15b-phaCYB-4 | pET15b derivative; phaCYB-4 from B. cereus YB-4 | This study |
Plasmid construction.
To express PhaCDa, the plasmid pJRD-phaCDaAB was constructed as follows. Plasmid pGEM-phaCDaAB (16) was digested with BamHI, and a DNA fragment containing the phaRe promoter, the phaCDa gene, the phaABRe genes, and the rrnB T1T2 terminator was introduced into the same site in pJRD215 (14).
To remove the phaABRe genes from pGEM-phaRCYB-4AB, the plasmid was digested with SalI and NruI and self-ligated using a BKL kit (TaKaRa Bio Inc., Otsu, Japan) to yield pGEM-phaRCYB-4.
For construction of pET15b-phaRYB-4, the phaRYB-4 gene was amplified by PCR using genomic DNA as the template. The following PCR primers were used: forward primer 5′-AGG TCA TAT GAT TGA TCA AAA ATT CGA TCC-3′ and reverse primer 5′-CAA GCG GGA TCC TCA CTT TTT ATT TTC TGG-3′ (underlined sequences indicate the NdeI and BamHI sites, respectively). The PCR product was digested with NdeI and BamHI and inserted into the same site in pET15b. For construction of pET15b-phaCYB-4, the phaCYB-4 gene was amplified by PCR using genomic DNA as the template. The following PCR primers were used: forward primer 5′-TAG AAA GGA TCC TAC ATT CGC AAC AGA ATG-3′ and reverse primer 5′-TTG GAT CCT TTA TTT TTA ATT AGA ACG CTC-3′ (underlined sequences indicate the BamHI sites). The PCR product was digested with BamHI and inserted into the same site in pET15b under T7 promoter control.
P(3HB) synthesis and isolation.
Recombinant E. coli JM109 was cultivated in 500-ml shake flasks containing 100 ml LB medium with glucose (20 g/liter) on a reciprocal shaker (130 rpm) at 37°C for 12 to 72 h. After cultivation, cells were harvested, washed once with deionized water, and lyophilized. The polymers that had accumulated in the cells were extracted with chloroform for 72 h at room temperature and then purified with methanol. For ethanol supplementation, ethanol (Kanto Chemical, Tokyo, Japan) or [1-13C]ethanol (SI Science, Saitama, Japan) was used.
GC and GPC.
The P(3HB) content of the dried cells was determined by gas chromatography (GC). Samples for GC analysis were prepared by methanolysis using 15% (vol/vol) sulfuric acid (18). The number average molecular weight (Mn) and the weight average molecular weight (Mw) of P(3HB) synthesized by recombinant strains were determined by gel permeation chromatography (GPC). GPC measurements were performed at 40°C, using a Shimadzu 10A GPC system equipped with two Shodex K-806 M joint columns. Chloroform was used as the eluent at a flow rate of 0.8 ml/min. Samples for GPC analysis were prepared at a concentration of 1.0 mg/ml and passed through a 0.45-μm-pore-size filter. Molecular weights were determined using a standard curve calibrated with low-polydispersity polystyrenes.
NMR analysis.
The end-group structures of isolated P(3HB)s were analyzed by NMR spectroscopy. Each polymer (30 mg) was dissolved in CDCl3 (0.7 ml) and subjected to 1H and 13C nuclear magnetic resonance (NMR) analysis. NMR spectra were recorded using a JEOL LA500 spectrometer. For 1H NMR analysis, data were collected with a 7.2-ms pulse width (90° pulse angle), a 5-s pulse repetition, a 6,000-Hz spectrum width, and 16,000 data points and at 45°C. For 13C NMR analysis, data were collected with a 6.1-ms pulse width (90° pulse angle), a 5-s pulse repetition, a 27,000-Hz spectrum width, and 33,000 data points and at 45°C. Tetramethylsilane (Me4Si) was used as an internal chemical shift standard.
PHA synthase activity assay.
Cells were harvested, washed with deionized water, and stored at −80°C until the activity assay; it has been reported that cells can be stored under this condition without any detectable loss of activity (19). Cells were then resuspended in 50 mM potassium phosphate (KPi; pH 7.0), disrupted by sonication for 3 min, and centrifuged at 1,500 × g for 5 min at 4°C to obtain cell lysates containing PHA synthase bound to PHA granules. Protein concentrations of crude extracts were determined using a Quant-iT protein assay kit (Invitrogen, San Diego, CA).
Synthase activity in crude extracts was determined using the 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) method (20). An assay mixture containing 50 mM KPi (pH 7.0) and 0.6 mM (R)-3-hydroxybutyryl-CoA [(R)-3HB-CoA] was preincubated at 30°C for 2 min, and then the reaction was started by the addition of soluble extract. The mixture volume was 800 μl in total. After a 1-min incubation at 30°C, 100 μl of the reaction mixture was mixed with 100 μl of trichloroacetic acid (5%, wt/vol) to stop the reaction. This mixture was then centrifuged at 12,000 × g for 10 min at 4°C, and 150 μl of its supernatant was mixed with 750 μl of 1 mM DTNB in 500 mM KPi (pH 7.2). After incubation for 2 min at room temperature, the absorbance at 412 nm was measured. One unit was defined as the activity for production of 1 μmol of 2-nitrobenzoic acid (TNB) anion (corresponding to released CoA having a free thiol group) per min (ε412 = 14.5 × 103 M−1 · cm−1). The (R)-3HB-CoA used here was chemically synthesized as described previously (20).
Ethanol assay.
The ethanol concentration in the culture liquid was measured by the enzymatic method using an F-kit (Roche Diagnostics, Basel, Switzerland).
Western blot analysis.
For Western blot analysis of the PhaCYB-4 subunit, bacterial cells were diluted to an optical density at 600 nm of 1.0 and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by the standard procedure. Separated proteins were transferred to a polyvinylidene difluoride membrane using a Bio-Rad Trans-Blot SD semidry transfer cell. Detection of PhaCYB-4 was carried out with a rabbit antiserum specific for a C-terminal oligopeptide (N-ALLDHISSTDKQYVC-C) in PhaCYB-4. Protein bands were visualized using goat anti-rabbit IgG conjugated to horseradish peroxidase (Toyobo, Osaka, Japan) as a secondary antibody.
Enzyme purification.
His-tagged PhaRYB-4 and PhaCYB-4 were individually produced by E. coli BL21(DE3) harboring pET15b-phaRYB-4 and pET15b-phaCYB-4, respectively, as follows. Cells were cultivated at 30°C for 2.5 h in LB medium, and then isopropyl β-d-1-thiogalactopyranoside (IPTG; Wako Pure Chemical Industries, Osaka, Japan) was added to a final concentration of 0.1 mM. After 3 to 5 h of cultivation at 20°C, cells were harvested and stored at −80°C prior to use.
His-tagged proteins were purified using an AKTA system equipped with a HisTrap HP column (GE Healthcare, Little Chalfont, United Kingdom), as described previously (21). Aliquots of purified enzymes were frozen in liquid nitrogen and stored at −80°C. Enzyme concentrations were determined using a Quant-iT protein assay kit (Invitrogen, Carlsbad, CA). Protein purity was confirmed by SDS-PAGE by the standard procedure.
P(3HB) formation in vitro.
In vitro P(3HB) polymerization was performed in a total volume of 12 ml with 100 mM KPi (pH 7.0) containing 5 mM (R)-3HB-CoA and 0.27 U/ml of PhaRCYB-4 (36 nmol) for 3 min at 30°C. The reaction mixture was divided into three equal samples: A, B, and C. These were centrifuged at 15,000 × g for 5 min at 4°C to obtain a precipitate containing P(3HB) and synthase. The precipitate from sample A was dried and purified using chloroform and methanol without further incubation. The precipitates from samples B and C were resuspended in 4 ml of 100 mM KPi (pH 7.0) without and with ethanol (2.6 g/liter), respectively, and incubated at 30°C for 24 h. After that, P(3HB) was collected by centrifugation again, dried, and purified using chloroform and methanol. The molecular weight of the collected P(3HB) was determined by GPC as described above. The conversion ratio of (R)-3HB-CoA was calculated by determining the (R)-3HB-CoA concentration in the supernatant by high-pressure liquid chromatography (22).
RESULTS
End-group structure analysis of low-molecular-weight P(3HB).
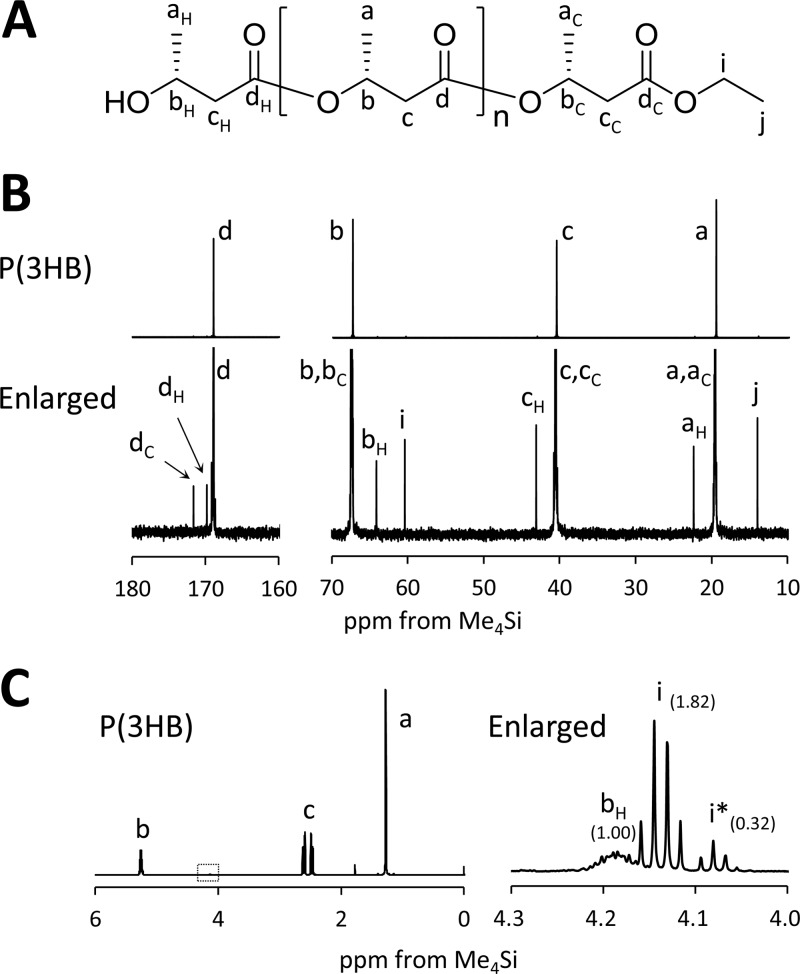
As reported previously (7), recombinant E. coli JM109 expressing PhaRCYB-4 showed an unusual reduction in the molecular weight of P(3HB) synthesized during the stationary phase of growth. A kinetic study suggested that the decrease in molecular weight was the result of random scission of the P(3HB) chain (7). In this study, to gain deeper insight into this mechanism, the end-group structures of low-molecular-weight P(3HB) were characterized by NMR spectroscopy. If the molecular weight of P(3HB) was reduced by hydrolysis, it would be expected that a free carboxy group would be detected as the end structure of the low-molecular-weight P(3HB). Low-molecular-weight P(3HB) (Mn = 20 × 103, polydispersity index = 2.1) extracted from E. coli JM109(pGEM-phaRCYB-4AB) cultured in 500-ml shake flasks containing 100 ml LB medium plus glucose (20 g/liter) at 37°C for 72 h was analyzed by 13C NMR. Signals derived from the terminal structure were detected as shown in Fig. 1B. Using authentic ethyl 3-hydroxybutyrate (ethyl 3HB) as a reference compound, the results suggest that the carboxy end was capped by ethanol. The 1H NMR analysis also suggested the existence of an ethanol-capped end (signal i) together with a minor end capped by a longer alcohol (signal i*), as shown in Fig. 1C. The molar ratio of the alcohol-capped end to all ends estimated from the peak intensities of signals i plus i* and signal bH, respectively, was almost 1:1. If the molecular weight had been reduced by hydrolysis, signals i and i* would be undetectable or relatively smaller than signal bH. Therefore, these results strongly suggest that the molecular weight was reduced not by hydrolytic cleavage but by alcoholytic cleavage. Because E. coli likely produces ethanol under anaerobic and microaerophilic culture conditions (23), host-produced ethanol might have a considerable influence on the degree of alcoholytic cleavage of the P(3HB) chain.
FIG 1.
(A) Chemical structure of ethanol-capped P(3HB). (B) Whole and enlarged 125-MHz 13C NMR spectra of low-molecular-weight P(3HB) synthesized by E. coli JM109 harboring pGEM-phaRCYB-4AB (Mn = 20 × 103; Mw/Mn = 2.1) cultured for 72 h at 37°C. (C) Whole and enlarged 500-MHz 1H NMR spectra of the low-molecular-weight P(3HB). i*, the methylene resonance for alcohols longer than ethanol. The values in parentheses show the intensity ratio of each peak.
Monitoring of ethanol concentration during cultivation.
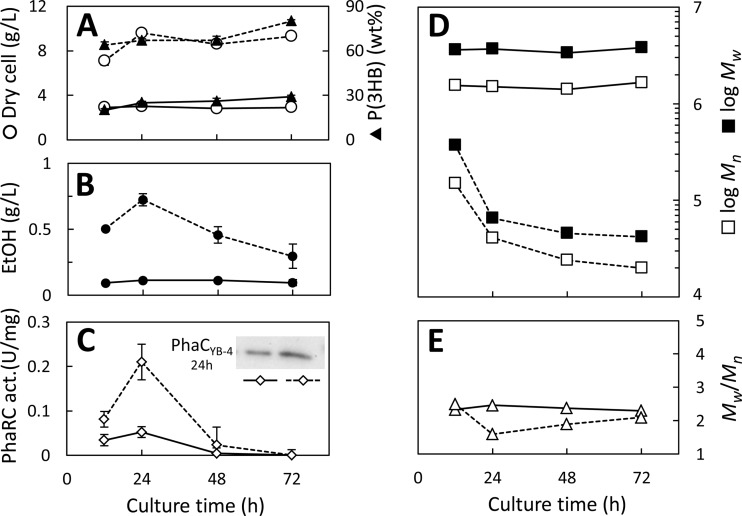
To understand the relationship between host-produced ethanol and P(3HB) molecular weight, the ethanol concentration in the culture liquid was monitored during the cultivation of strains harboring the high-copy-number plasmid pGEM-phaRCYB-4AB and the low-copy-number plasmid pBBR1-phaRCYB-4AB, which are produced at from 300 to 400 copies per cell (24) and approximately 30 to 40 copies per cell (25), respectively. Each recombinant strain was cultivated in LB medium containing glucose (20 g/liter) at 37°C. The culture results are shown in Fig. 2.
FIG 2.
P(3HB) synthesis by E. coli JM109 harboring the high-copy-number plasmid pGEM-phaRCYB-4AB (dashed line) and the low-copy-number plasmid pBBR1-phaRCYB-4AB (solid line). (A) Cell dry weight (open circles) and P(3HB) content (filled triangles); (B) ethanol (EtOH) concentration in the broth; (C) synthase activity (act.) and Western blot analysis of PhaCYB-4 after 24 h of cultivation; (D) Mn and Mw; (E) Mw/Mn. Cells were cultivated in 500-ml shake flasks containing 100 ml LB medium plus glucose (20 g/liter) at 37°C. Each experiment was carried out in triplicate.
In the case of the strain harboring pGEM-phaRCYB-4AB (high copy number), ethanol could be detected in the culture liquid and reached a maximum concentration of 0.72 g/liter after 24 h. This culture eventually grew to over 9 g/liter, with P(3HB) being present at 80% by weight (80 wt%) of its cell dry weight. The molecular weight of the P(3HB) synthesized by this strain was relatively high after 12 h of cultivation, and the Mn was equal to 150 × 103. However, the Mn of P(3HB) produced after 72 h of cultivation decreased to 24 × 103, which is consistent with the observation from our previous study (7).
In contrast, the strain harboring pBBR1-phaRCYB-4AB (low copy number) produced less ethanol (0.11 g/liter after 24 h) than the strain harboring pGEM-phaRCYB-4AB. The strain harboring pBBR1-phaRCYB-4AB grew to nearly 3 g/liter of cell dry weight by 12 h of cultivation but did not grow more after that. The P(3HB) accumulation in this strain at 72 h was 29 wt%, which is significantly lower than that from the strain harboring pGEM-phaRCYB-4AB (80 wt%). Notably, the Mns at 12 h and 72 h were equally high, at 1,600 × 103 and 1,700 × 103, respectively. The different behavior of this strain with respect to the molecular weight change might be attributed to its level of ethanol production.
The PHA synthase activity in these strains was assayed using cell extracts containing synthases bound to P(3HB) granules. The results are shown in Fig. 2C. Similar patterns of change in synthase activity could be observed for these two strains, but their maximum values were quite different. For each strain, the highest level of synthase activity, 0.21 ± 0.04 U/mg protein and 0.052 ± 0.015 U/mg protein for pGEM-phaRCYB-4AB and pBBR1-phaRCYB-4AB, respectively, was attained after 24 h of cultivation. Western blot analysis revealed that synthase (PhaCYB-4 subunit) concentrations in cells cultured for 24 h in the two strains were also different, corresponding to their respective levels of activity (Fig. 2C, inset). The results suggest that the plasmid copy number affected synthase expression, thereby affecting P(3HB) and ethanol production and the molecular weight of P(3HB).
Ethanol supplementation to induce alcoholytic cleavage.
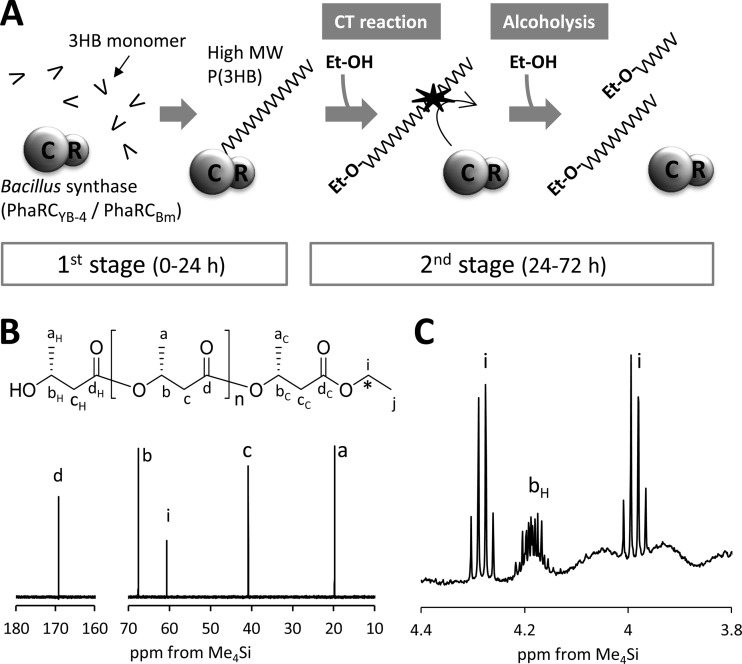
The strain expressing the low-copy-number plasmid pBBR1-phaRCYB-4AB did not produce either low-molecular-weight P(3HB) or significant amounts of ethanol (Fig. 2B and D). Using this transformant, we investigated whether alcoholytic cleavage of P(3HB) could be induced by adding ethanol to the culture medium. To avoid reducing the P(3HB) molecular weight during its biosynthesis via the chain transfer (CT) reaction, which terminates PHA chain elongation by transferring PHA chains from PhaC to a hydroxy compound (26), two-stage cultivation (Fig. 3A) was carried out as follows. For the first stage, cells were cultured in LB medium containing glucose (20 g/liter) for 24 h, to allow the accumulation of high-molecular-weight P(3HB). For the second stage, the cells were transferred into glucose-free M9 medium with or without ethanol supplementation (4 g/liter) and incubated for a further 48 h. The results of the two-stage cultivation procedure are shown in Table 2. The Mn of the culture without ethanol supplementation was 1,140 × 103, which is almost unchanged from that during the second stage, although the Mn with ethanol supplementation was as low as 42 × 103. The number of P(3HB) chains increased 36.1-fold in the second stage by the presence of ethanol. From these results, it is obvious that ethanol works as a trigger for a dramatic reduction of the P(3HB) Mn.
FIG 3.
(A) Schematic of the two-stage culture of E. coli JM109 harboring pBBR1-phaRCYB-4AB or pGEM-phaRCBmAB. In the first stage, cells were cultured in LB medium containing glucose (20 g/liter) at 37°C for 24 h for the production of high-molecular-weight (High MW) P(3HB). In the second stage, the cells were transferred into glucose-free M9 medium with or without ethanol (Et-OH) supplementation (4 g/liter) and incubated at 37°C for a further 48 h. Each experiment was carried out in triplicate. (B, C) NMR spectra of P(3HB) synthesized by E. coli JM109 harboring pBBR1-phaRCYB-4AB with [1-13C]ethanol supplementation (4 g/liter) during the second stage of culture. Asterisk in the chemical structure, 13C-labeled carbon. (B) The 125-MHz 13C NMR spectrum. (C) Enlarged 3.8- to 4.4-ppm region of the 500-MHz 1H NMR spectrum.
TABLE 2.
Effect of ethanol on P(3HB) molecular weight produced by E. coli JM109 expressing PhaRCYB-4 (pBBR1-phaRCYB-4AB) and PhaRCBm (pGEM-phaRCBmAB) in two-stage culturea
| Expressed synthaseb | Culture conditionsc |
Time of harvest (h) | Cell dry wt (g/liter) | P(3HB) content (wt%) | Mol wt |
Relative chain no. | ||
|---|---|---|---|---|---|---|---|---|
| First stage (0–24 h) | Second stage (24–72 h) | Mn (103) | Mw/Mn | |||||
| PhaRCYB-4 | LB + glucose | 24 | 3.3 ± 0.0 | 27 ± 0 | 1,250 ± 106 | 1.6 | 1.0 | |
| LB + glucose | M9 | 72 | 2.7 ± 0.0 | 31 ± 2 | 1,140 ± 61 | 2.8 | 1.0 | |
| LB + glucose | M9 + ethanol | 72 | 3.0 ± 0.0 | 36 ± 4 | 42 ± 2 | 11.1 | 36.1 | |
| PhaRCBm | LB + glucose | 24 | 4.2 ± 0.0 | 23 ± 1 | 1,050 ± 68 | 1.9 | 1.0 | |
| LB + glucose | M9 | 72 | 3.0 ± 0.0 | 33 ± 1 | 1,060 ± 72 | 2.0 | 1.0 | |
| LB + glucose | M9 + ethanol | 72 | 3.0 ± 0.0 | 35 ± 3 | 270 ± 28 | 4.8 | 4.2 | |
Results are means ± standard deviations from three separate experiments.
pBBR1-phaRCYB-4AB and pGEM-phaRCBmAB are low- and high-copy-number plasmids, respectively.
Cells were cultured in LB medium containing glucose (20 g/liter) for 24 h at 37°C and then incubated in glucose-free M9 medium with or without supplementation of ethanol (4 g/liter) for a further 48 h at 37°C.
The same experiment was conducted using 4 g/liter [1-13C]ethanol, resulting in the production of low-molecular-weight P(3HB). The 13C NMR spectrum of the P(3HB) synthesized in this experiment is shown in Fig. 3B. In this figure, the signal i at 60.6 ppm arising from the end-capped ethanol was enhanced (relative to signals a, b, c, and d arising from main-chain 3HB unit) compared to that seen in Fig. 1B, due to 13C enrichment. Additionally, in the 1H NMR spectrum, split methylene resonance arising from the coupling of 1H atoms to an adjoining 13C atom at the ethyl ester end was observed at 4.0 ppm and 4.3 ppm (Fig. 3C). These results strongly suggest that exogenous ethanol was incorporated into the carboxy end of P(3HB). On the basis of these results, it can be concluded that the reduction in P(3HB) molecular weight is caused by alcoholytic cleavage of the P(3HB) chain.
In addition, the same ethanol supplementation experiment was conducted for cultures of E. coli expressing PhaRCBm. The result from this experiment is also shown in Table 2. Unexpectedly, we found that expression of PhaRCBm leads to a reduction in P(3HB) molecular weight and an increase in P(3HB) chain number, although this result had not been seen in our previous study (7). This result suggests that PhaRCBm expresses weak cleavage activity with P(3HB) chains if ethanol is present.
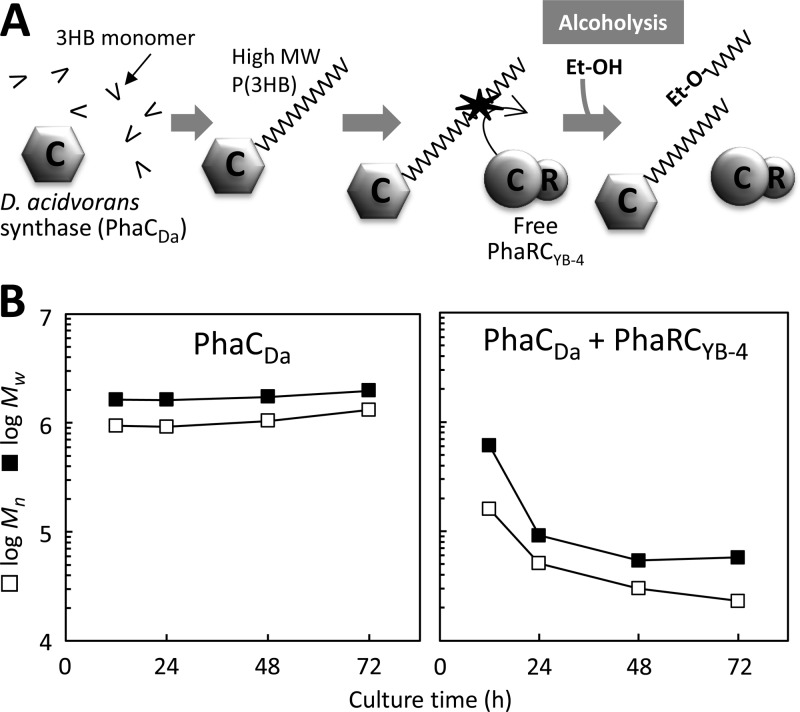
In vivo scission assay using PhaRCYB-4 and PhaCDa.
To investigate whether PhaRCYB-4 or its subunit could affect P(3HB) chains that had been polymerized by another PHA synthase in vivo, we coexpressed the synthase from B. cereus YB-4 (PhaRYB-4 and/or PhaCYB-4) and the synthase from Delftia acidovorans (PhaCDa) in E. coli JM109 (Fig. 4A). PhaCDa can synthesize high-molecular-weight P(3HB) (Mn > 1,000 × 103) (8, 9); thus, changes in the high-molecular-weight fraction of P(3HB) were monitored by GPC as an indicator for the scission reaction.
FIG 4.
(A) Schematic of the in vivo scission assay using E. coli JM109 as the host strain expressing PhaRCYB-4 (pGEM-phaRCYB-4) together with PhaCDa from pJRD-phaCDaAB, which provides the high-molecular-weight P(3HB) that serves as the scissile substrate in the cell. (B) Molecular weight changes of P(3HB) during cultivation in the in vivo assay.
Table 3 shows the results from the coexpression cultures. These cultures showed similar cell growth and P(3HB) accumulation, at 8.6 to 9.4 g/liter and 67 to 73%, respectively. In addition, these cultures produced approximately 0.2 g/liter ethanol by 72 h of cultivation. However, their behavior with respect to the P(3HB) molecular weight was quite different. By 24 h, the culture expressing PhaCDa alone produced P(3HB) with an Mn as high as 1,440 × 103, even though 0.3 g/liter ethanol was produced. Moreover, cultures coexpressing PhaCDa and PhaRYB-4 or PhaCYB-4 produced high-molecular-weight P(3HB), with Mns of 1,180 × 103 and 1,200 × 103, respectively. On the other hand, the culture coexpressing PhaCDa and PhaRCYB-4 produced low-molecular-weight P(3HB), with an Mn of 14 × 103. This value is much lower than that produced by the culture expressing PhaCDa alone. As shown in Fig. 4B, the molecular weight of P(3HB) produced by the strain coexpressing PhaCDa and PhaRCYB-4 continued to decrease even after 24 h of cultivation, by which point P(3HB) synthesis would have been finished.
TABLE 3.
P(3HB) production by various recombinant E. coli JM109 strains in shake flasksa
| Expressed synthase(s) | Cell dry wt (g/liter) | P(3HB) content (wt%) | Ethanol concn (g/liter) at: |
Mol wt |
||
|---|---|---|---|---|---|---|
| 24 h | 72 h | Mn (103) | Mw/Mn | |||
| PhaCDa | 9.4 ± 0.1 | 71 ± 2 | 0.30 ± 0.06 | 0.09 ± 0.01 | 1,440 ± 15 | 1.9 |
| PhaCDa, PhaRYB-4 | 8.7 ± 0.1 | 67 ± 5 | 0.47 ± 0.03 | 0.28 ± 0.00 | 1,180 ± 70 | 2.0 |
| PhaCDa, PhaCYB-4 | 9.3 ± 0.1 | 67 ± 2 | 0.41 ± 0.04 | 0.19 ± 0.01 | 1,200 ± 72 | 1.8 |
| PhaCDa, PhaRCYB-4 | 8.6 ± 0.1 | 73 ± 5 | 0.50 ± 0.03 | 0.30 ± 0.01 | 14 ± 1 | 2.1 |
| PhaRCYB-4 | 9.3 ± 0.1 | 80 ± 1 | 0.72 ± 0.05 | 0.24 ± 0.01 | 24 ± 1 | 1.6 |
Cells were cultivated in LB medium containing glucose (20 g/liter) at 37°C for 72 h. To express PhaRYB-4 and/or PhaCYB-4, derivatives of pGEM (a high-copy-number plasmid) were used. For expression of PhaCDa, pJRD-phaCDaAB (carrying phaCDa, phaARe, and phaBRe) was employed. Results are the averages ± standard deviations from three separate experiments.
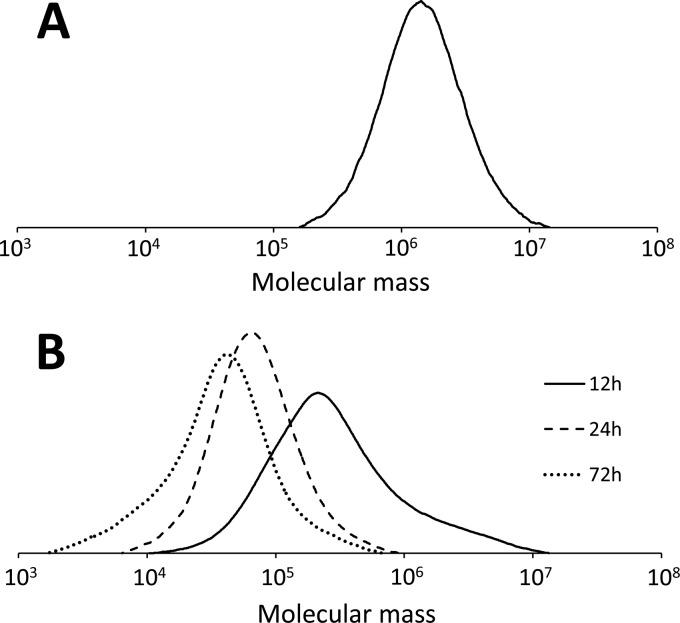
Figure 5A shows the molecular weight distribution of P(3HB) produced after 12 h of cultivation by the strain expressing PhaCDa. The Mn of this sample was 939 × 103, and the distribution of the molecular weight was unimodal. Figure 5B shows the molecular weight distribution of P(3HB) produced by a strain coexpressing PhaCDa and PhaRCYB-4 and demonstrates that there were remarkable changes in the molecular weight over the course of the cultivation period. The peak top of the curve shifted to the low-molecular-weight side, and the Mn decreased from 150 × 103 at 12 h to 14 × 103 at 72 h. In addition, there was a small shoulder in the high-molecular-weight side of the curve for the polymer at 12 h, which corresponds to the molecular weight of PhaCDa-synthesized P(3HB). This shoulder was diminished with increasing culture time, and the whole peak was shifted to the low-molecular-weight side. These observations suggest that the high-molecular-weight P(3HB) synthesized by PhaCDa can be broken down by PhaRCYB-4.
FIG 5.
Molecular weight distributions of P(3HB) extracted from E. coli JM109 expressing PhaCDa cultured for 12 h (A) and E. coli JM109 expressing PhaCDa and PhaRCYB-4 cultured for 12, 24, and 72 h (B). For expression of PhaCDa and PhaRCYB-4, pJRD-phaCDaAB (carrying phaCDa, phaARe, and phaBRe) and pGEM-phaRCYB-4 (carrying phaRCYB-4), respectively, were used. Cells were cultivated in LB medium containing glucose (20 g/liter) at 37°C.
Induction of alcoholysis activity of PhaRCYB-4 in vitro.
To demonstrate the alcoholysis activity of PhaRCYB-4 directly, an in vitro assay using purified PhaRCYB-4 and chemically synthesized (R)-3HB-CoA was performed. In vitro P(3HB) polymerization was allowed to progress for 3 min at 30°C. Then, the reaction mixture was divided into three samples, A, B, and C, by volume. These samples were then centrifuged to collect the precipitate containing P(3HB) and PhaRCYB-4. The precipitate from sample A was dried without further incubation. The precipitates from samples B and C were suspended in phosphate buffer (pH 7.0) without and with ethanol (2.6 g/liter), respectively, and incubated at 30°C for 24 h. These P(3HB) samples were then collected for molecular weight analysis.
Table 4 shows the results of the in vitro assay. The Mn of P(3HB) from the initial state of incubation (sample A) was 180 × 103. This value is close to the theoretical Mn (226 × 103) estimated from the (R)-3HB-CoA conversion ratio (55%) and the concentration of PhaRCYB-4 (36 nmol), using the following equation: absolute Mn = 0.7 × Mn(GPC), where Mn(GPC) is the Mn determined by GPC. In this calculation, it is assumed that the PhaRC heterotetramer forms an active site similar to that seen in a class III synthase (27). After 24 h of incubation, the Mn of P(3HB) incubated without ethanol (sample B) was almost unchanged from that of sample A. On the other hand, the Mn of P(3HB) incubated with ethanol (sample C, Mn = 17 × 103) was significantly lower than the Mns of the other two, indicating that the P(3HB) chain was degraded via alcoholysis catalyzed by PhaRCYB-4.
TABLE 4.
In vitro incubation of the P(3HB)–PhaRCYB-4 complex in phosphate buffer with or without ethanola
| Sample | Incubation with additive | Incubation time (h) | Mol wt of P(3HB) |
||
|---|---|---|---|---|---|
| Mn (103) | Mw (103) | Mw/Mn | |||
| A | No incubation | 0 | 180 ± 9 | 365 ± 30 | 2.0 |
| B | No additive | 24 | 193 ± 10 | 355 ± 16 | 1.8 |
| C | Ethanol | 24 | 17 ± 3 | 23 ± 7 | 1.4 |
P(3HB) polymerization using purified PhaRCYB-4 was performed for 3 min at 30°C, and the reaction mixture was divided into three equal samples, A, B, and C, by volume. Sample A represented the initial condition prior to incubation. Samples B and C were centrifuged, and the precipitates were incubated for 24 h at 30°C in fresh phosphate buffer (pH 7.0) without and with ethanol (2.6 g/liter), respectively. Results are the averages ± standard deviations from three separate experiments.
DISCUSSION
In earlier studies, it had been demonstrated that the molecular weight of PHA decreases with an increase in the synthase concentration in cells during PHA biosynthesis (28, 29). This experimental rule can be attributed to the monomer/synthase ratio, which is a well-known factor determining polymer molecular weight from organocatalytic living polymerization and is sometimes applicable to PHA biosynthesis. However, the results in this study cannot be explained by simple synthase amount-molecular weight relationships, because the molecular weight was observed to decrease even after polymer synthesis had finished. As an alternative mechanism to explain this decrease, we propose that random scission of P(3HB) chains takes place. This idea is supported by the results from previously reported kinetic analyses of the molecular weight change (7, 11). As there are no genes for P(3HB) polymerization and depolymerization in the E. coli genome, it has been hypothesized that the observed decrease in molecular weight was caused by exogenous PhaRC subunits. This study aimed to investigate the mechanisms underlying the unusual molecular weight decrease seen in P(3HB) produced by E. coli expressing PhaRCYB-4.
In this study, NMR analysis of the low-molecular-weight P(3HB) synthesized by PhaRCYB-4 showed that a high proportion of the carboxy end structures are capped by ethanol (Fig. 1). This finding led to the hypothesis that the reduction in P(3HB) molecular weight results from alcoholytic cleavage of P(3HB) chains. To test this hypothesis by showing that alcoholytic cleavage can be induced by the presence of ethanol, we first looked for culture conditions that would enable E. coli to produce high-molecular-weight P(3HB) even when PhaRCYB-4 was being expressed. Several culture conditions were tested, and it was found that transformation with the low-copy-number plasmid pBBR1-phaRCYB-4AB enabled E. coli to produce P(3HB) without a significant reduction in molecular weight. In addition, the strain harboring the low-copy-number plasmid had only a weak ability to produce ethanol (Fig. 2B). Our previous report (30) had shown that P(3HB)-producing E. coli produced ethanol as the main fermentation product, while plasmid-free E. coli strains produced acetate, providing clues to understanding the difference in ethanol production between strains harboring low- and high-copy-number plasmids. Indeed, in this study, E. coli harboring low-copy-number plasmid pBBR1-phaRCYB-4AB was observed to produce smaller amounts of P(3HB) and ethanol but a larger amount of acetate than E. coli harboring high-copy-number plasmid pGEM-phaRCYB-4AB) (see Table S1 in the supplemental material).
By adding ethanol to the culture of the strain harboring the low-copy-number plasmid, we examined whether alcoholytic cleavage of P(3HB) could be induced. The result clearly demonstrates that ethanol works as a trigger for the dramatic reduction of the P(3HB) molecular weight (Table 2). However, the concentration of exogenous ethanol required to induce the efficient reduction of the molecular weight of P(3HB) (4 g/liter) is much higher than that of endogenous, host-produced ethanol (0.2 to 0.8 g/liter). This may be because of differences between exogenous and endogenous ethanol; however, the concentration of intracellular ethanol might be similar under both conditions.
To obtain further evidence for the alcoholytic cleavage of the P(3HB) chain by PhaRCYB-4, an in vivo scission assay system was developed by coexpressing two synthases (PhaRCYB-4 and PhaCDa) in E. coli. PhaCDa synthesizes high-molecular-weight P(3HB) in E. coli. Thus, by monitoring the change in the high-molecular-weight fraction of P(3HB), the effect of PhaRCYB-4 on P(3HB) chains synthesized by PhaCDa was investigated. The result showed that the molecular weight of P(3HB) produced by a strain coexpressing the two synthases after 72 h of cultivation was as low as that produced by the strain expressing PhaRCYB-4 alone (Table 3). This observation strongly suggests that PhaRCYB-4 is able to cleave the chains of the high-molecular-weight P(3HB) synthesized by PhaCDa. The in vivo assay also revealed that the PhaRCYB-4 complex could induce scission of the P(3HB) chain, although the PhaCYB-4 or PhaRYB-4 subunit alone could not. Thus, scission activity, as well as polymerization activity, was shown to require both the PhaCYB-4 and PhaRYB-4 subunits.
Unlike E. coli, the PHA-negative mutant R. eutropha PHB−4 produces high-molecular-weight P(3HB) when expressing PhaRCYB-4 (12). Because the ethanol produced by this R. eutropha strain remains at undetectable levels (data not shown), the alcoholytic activity of PhaRCYB-4 could not be induced.
The alcoholysis activity of PhaRCYB-4 was directly demonstrated with an in vitro assay. We conducted in vitro synthesis of P(3HB) using purified PhaRCYB-4 and chemically synthesized (R)-3HB-CoA for 3 min, followed by 24 h of incubation of P(3HB)-PhaRCYB-4 complexes in phosphate buffer with or without ethanol. A significant decrease in the molecular weight of P(3HB) was observed only when ethanol was present. In addition, there was no detectable change in the molecular weight of P(3HB) during the incubation with ethanol under synthase-free conditions (data not shown). This observation provides convincing evidence for the alcoholysis activity of PhaRCYB-4.
Surprisingly, weak scission activity was also observed for PhaRCBm in vivo. This finding was unexpected, because a previous study demonstrated that PhaRCBm did not have P(3HB) chain scission activity in E. coli (7). Later, it was found that the strain expressing PhaRCBm produced less ethanol than is required for alcoholysis. Not only the PhaRC subunits from B. cereus YB-4 (7) and Bacillus sp. INT005 (11) but also the PhaRC subunit from B. megaterium were found to have scission activity for P(3HB) chains; therefore, class IV synthases might commonly have scission activity. As far as we have tested, the scission activities of class I synthases such as PhaCDa and PhaCRe are undetectable.
The alcoholysis activity of the PhaRC synthase was demonstrated in this study; however, a similar alcoholytic activity has been reported for lipases (31) and ester synthase/acyl-CoA:diacylglycerol acyltransferases (32). An alcoholysis activity of lipase toward triacylglycerols can be observed in the presence of excess alcohol. Recently, this reaction has attracted attention as a possible enzymatic method for biodiesel production. On the other hand, the alcoholysis activity of PhaRC could be used for the modification and functionalization of the carboxylic end of PHA. This report indicates that there are possible applications of PhaRC other than for PHA production. PhaC and lipases are members of the α/β-hydrolase fold family (2, 4); thus, the molecular mechanism underlying the alcoholic activity of these enzymes might be commonly shared.
It would be interesting if alcoholysis by PhaRCYB-4 were involved in the regulation of PHA molecular weight in B. cereus YB-4. In a previous study, we observed a decrease in the PHA molecular weight when B. cereus YB-4 was cultured for a long time (10). Thus, PhaRCYB-4 might be partly involved in this decrease. However, unlike E. coli, natural PHA producers are capable of mobilizing PHA by inherent PHA depolymerases. It is natural to think that PHA depolymerases would be mainly responsible for this decrease. The molecular weight decrease in B. cereus YB-4 might be associated with providing a rapid energy supply for spore and septum development (33) because low-molecular-weight PHA would be favored as a storage material to be mobilized quickly.
In summary, this study focused on the mechanisms underlying the unusual decrease in P(3HB) molecular weight observed in E. coli JM109 expressing PhaRCYB-4. The low-molecular-weight P(3HB) isolated from the cells had an ethanol-capped carboxy end. Scission of P(3HB) chains was observed in the presence of ethanol in both in vivo and in vitro assays. Thus, the molecular weight was reduced not by hydrolytic cleavage but by alcoholytic cleavage via PhaRCYB-4. This alcoholysis activity was also observed for P(3HB) chains synthesized by other synthases. Both the PhaRYB-4 and PhaCYB-4 subunits are essential for expression of the scission activity. Because the synthase from B. megaterium also exhibits scission activity, this capability might be an inherent feature of class IV synthases. The present study provides new insights into the catalytic properties of PHA synthase other than PHA polymerization.
Supplementary Material
ACKNOWLEDGMENTS
We thank Y. Nakamura (Tokyo Institute of Technology) for NMR analysis.
This work was supported by a Grant-in-Aid for Scientific Research (KAKENHI 23310060) to T. Tsuge. M. Hyakutake is the recipient of a JSPS young scientist fellowship (12J07940).
Footnotes
Published ahead of print 13 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03576-13.
REFERENCES
- 1.Sudesh K, Abe H, Doi Y. 2000. Synthesis, structure and properties of polyhydroxyalkanoates: biological polyesters. Prog. Polym. Sci. 25:1503–1555. 10.1016/S0079-6700(00)00035-6 [DOI] [Google Scholar]
- 2.Stubbe J, Tian J. 2003. Polyhydroxyalkanoate (PHA) homeostasis: the role of the PHA synthase. Nat. Prod. Rep. 20:445–457. 10.1039/b209687k [DOI] [PubMed] [Google Scholar]
- 3.Slater SC, Voige WH, Dennis DE. 1988. Cloning and expression in Escherichia coli of the Alcaligenes eutrophus H16 poly-β-hydroxybutyrate biosynthetic pathway. J. Bacteriol. 170:4431–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rehm BH. 2003. Polyester synthases: natural catalysts for plastics. Biochem. J. 376:15–33. 10.1042/BJ20031254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCool GJ, Cannon MC. 2001. PhaC and PhaR are required for polyhydroxyalkanoic acid synthase activity in Bacillus megaterium. J. Bacteriol. 183:4235–4243. 10.1128/JB.183.14.4235-4243.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satoh Y, Minamoto N, Tajima K, Munekata M. 2002. Polyhydroxyalkanoate synthase from Bacillus sp. INT005 is composed of PhaC and PhaR. J. Biosci. Bioeng. 94:343–350. 10.1016/S1389-1723(02)80175-X [DOI] [PubMed] [Google Scholar]
- 7.Tomizawa S, Hyakutake M, Saito Y, Agus J, Mizuno K, Abe H, Tsuge T. 2011. Molecular weight change of polyhydroxyalkanoate (PHA) caused by the PhaC subunit of PHA synthase from Bacillus cereus YB-4 in recombinant Escherichia coli. Biomacromolecules 12:2660–2666. 10.1021/bm2004687 [DOI] [PubMed] [Google Scholar]
- 8.Agus J, Kahar P, Abe H, Doi Y, Tsuge T. 2006. Molecular weight characterization of poly[(R)-3-hydroxybutyrate] synthesized by genetically engineered strains of Escherichia coli. Polym. Degrad. Stab. 91:1138–1146. 10.1016/j.polymdegradstab.2005.07.006 [DOI] [Google Scholar]
- 9.Hiroe A, Ushimaru K, Tsuge T. 2013. Characterization of polyhydroxyalkanoate (PHA) synthase derived from Delftia acidovorans DS-17 and the influence of PHA production in Escherichia coli. J. Biosci. Bioeng. 115:633–638. 10.1016/j.jbiosc.2012.12.015 [DOI] [PubMed] [Google Scholar]
- 10.Mizuno K, Ohta A, Hyakutake M, Ichinomiya Y, Tsuge T. 2010. Isolation of polyhydroxyalkanoate-producing bacteria from a polluted soil and characterization of the isolated strain Bacillus cereus YB-4. Polym. Degrad. Stab. 95:1335–1339. 10.1016/j.polymdegradstab.2010.01.033 [DOI] [Google Scholar]
- 11.Agus J, Kahar P, Hyakutake M, Tomizawa S, Abe H, Tsuge T, Satoh Y, Tajima K. 2010. Unusual change in molecular weight of polyhydroxyalkanoate (PHA) during cultivation of PHA-accumulating Escherichia coli. Polym. Degrad. Stab. 95:2250–2254. 10.1016/j.polymdegradstab.2010.09.009 [DOI] [Google Scholar]
- 12.Hyakutake M, Saito Y, Tomizawa S, Mizuno K, Tsuge T. 2011. Polyhydroxyalkanoate (PHA) synthesis by class IV PHA synthases employing Ralstonia eutropha PHB−4 as host strain. Biosci. Biotechnol. Biochem. 75:1615–1617. 10.1271/bbb.110229 [DOI] [PubMed] [Google Scholar]
- 13.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. 10.1016/0378-1119(95)00584-1 [DOI] [PubMed] [Google Scholar]
- 14.Davison J, Heusterspreute M, Chevalier N, Ha-Thi V, Brunei F. 1987. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene 51:275–280. 10.1016/0378-1119(87)90316-7 [DOI] [PubMed] [Google Scholar]
- 15.Takase K, Taguchi S, Doi Y. 2003. Enhanced synthesis of poly(3-hydroxybutyrate) in recombinant Escherichia coli by means of error-prone PCR mutagenesis, saturation mutagenesis, and in vitro recombination of the type II polyhydroxyalkanoate synthase gene. J. Biochem. 133:139–145. 10.1093/jb/mvg015 [DOI] [PubMed] [Google Scholar]
- 16.Tsuge T, Imazu S, Takase K, Taguchi S. 2004. An extra large insertion in the polyhydroxyalkanoate synthase from Delftia acidovorans DS-17: its deletion effects and relation to cellular proteolysis. FEMS Microbiol. Lett. 231:77–83. 10.1016/S0378-1097(03)00930-3 [DOI] [PubMed] [Google Scholar]
- 17.Matsusaki H, Abe H, Taguchi K, Fukui T, Doi Y. 2000. Biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyalkanoates) by recombinant bacteria expressing the PHA synthase gene phaC1 from Pseudomonas sp. 61-3. Appl. Microbiol. Biotechnol. 53:401–409. 10.1007/s002530051633 [DOI] [PubMed] [Google Scholar]
- 18.Kato M, Bao HJ, Kang CK, Fukui T, Doi Y. 1996. Production of a novel copolyester of 3-hydroxybutyric acid and medium-chain-length 3-hydroxyalkanoic acids by Pseudomonas sp. 61-3 from sugars. Appl. Microbiol. Biotechnol. 45:363–370. 10.1007/s002530050697 [DOI] [Google Scholar]
- 19.Gerngross TU, Snell KD, Peoples OP, Sinskey AJ. 1994. Overexpression and purification of the soluble polyhydroxyalkanoate synthase from Alcaligenes eutrophus: evidence for a required posttranslational modification for catalytic activity. Biochemistry 33:9311–9320 [DOI] [PubMed] [Google Scholar]
- 20.Valentin HE, Steinbüchel A. 1994. Application of enzymatically synthesized short-chain-length hydroxy fatty acid coenzyme A thioesters for assay of polyhydroxyalkanoic acid synthases. Appl. Microbiol. Biotechnol. 40:699–709. 10.1007/BF00173332 [DOI] [Google Scholar]
- 21.Ushimaru K, Sangiambut S, Thomson N, Sivaniah E, Tsuge T. 2013. New insights into activation and substrate recognition of polyhydroxyalkanoate synthase from Ralstonia eutropha. Appl. Microbiol. Biotechnol. 97:1175–1182. 10.1007/s00253-012-4089-x [DOI] [PubMed] [Google Scholar]
- 22.Tomizawa S, Sato S, Lan JCW, Nakamura Y, Abe H, Tsuge T. 2013. In vitro evidence of chain transfer to tetraethylene glycols in enzymatic polymerization of polyhydroxyalkanoate. Appl. Microbiol. Biotechnol. 97:4821–4829. 10.1007/s00253-013-4798-9 [DOI] [PubMed] [Google Scholar]
- 23.Dawes EA, Foster SM. 1956. The formation of ethanol in Escherichia coli. Biochim. Biophys. Acta 22:253–265. 10.1016/0006-3002(56)90148-2 [DOI] [PubMed] [Google Scholar]
- 24.Qiagen 2005. Qiagen® plasmid purification handbook. Qiagen, Valencia, CA [Google Scholar]
- 25.Antoine R, Locht C. 1992. Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from Gram-positive organisms. Mol. Microbiol. 6:1785–1799. 10.1111/j.1365-2958.1992.tb01351.x [DOI] [PubMed] [Google Scholar]
- 26.Kawaguchi Y, Doi Y. 1992. Kinetics and mechanism of synthesis and degradation of poly(3-hydroxybutyrate) in Alcaligenes eutrophus. Macromolecules 25:2324–2329. 10.1021/ma00035a007 [DOI] [Google Scholar]
- 27.Jia Y, Kappock J, Frick T, Sinskey AJ, Stubbe J. 2000. Lipases provide a new mechanistic model for polyhydroxybutyrate (PHB) synthases: characterization of the functional residues in Chromatium vinosum PHB synthase. Biochemistry 39:3927–3936. 10.1021/bi9928086 [DOI] [PubMed] [Google Scholar]
- 28.Sim SJ, Snell KD, Hogan SA, Stubbe J, Rha C, Sinskey AJ. 1997. PHA synthase activity controls the molecular weight and polydispersity of polyhydroxybutyrate in vivo. Nat. Biotechnol. 15:63–67. 10.1038/nbt0197-63 [DOI] [PubMed] [Google Scholar]
- 29.Hiroe A, Tsuge K, Nomura CT, Itaya M, Tsuge T. 2012. Rearrangement of gene order in the phaCAB operon leads to effective production of ultrahigh-molecular-weight poly[(R)-3-hydroxybutyrate] in genetically engineered Escherichia coli. Appl. Environ. Microbiol. 78:3177–3184. 10.1128/AEM.07715-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiroe A, Hyakutake M, Thomson NM, Sivaniah E, Tsuge T. 2013. Endogeneous ethanol affects biopolyester molecular weight in recombinant Escherichia coli. ACS Chem. Biol. 8:2568–2576. 10.1021/cb400465p [DOI] [PubMed] [Google Scholar]
- 31.Szczêsna Antczak M, Kubiak A, Antczak T, Bielecki S. 2009. Enzymatic biodiesel synthesis—key factors affecting efficiency of the process. Renew. Energy 34:1185–1194. 10.1016/j.renene.2008.11.013 [DOI] [Google Scholar]
- 32.Kalscheuer R, Stölting T, Steinbüchel A. 2006. Microdiesel: Escherichia coli engineered for fuel production. Microbiology 152(Pt 9):2529–2536. 10.1099/mic.0.29028-0 [DOI] [PubMed] [Google Scholar]
- 33.Slepecky RA, Law JH. 1961. Synthesis and degradation of poly-β-hydroxybutyric acid in connection with sporulation of Bacillus megaterium. J. Bacteriol. 82:37–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.